Transcriptome Analysis Reveals the Central Role of the Transcription Factor MYB in Regulating Anthocyanin Accumulation in Economic Grape Species (Vitis vinifera)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Experimental Treatment
2.2. Physiological Measurements
2.3. Determination of Total Anthocyanins
2.4. RNA Extraction and Transcriptome Sequencing
2.5. Analysis of Differential Expression, Transcriptome Alignment, and Functional Enrichment
2.6. Analysis of Transcription Factor Binding Site in Promoter Region and Co-Expression Pattern Cluster
2.7. RNA Isolation and Real-Time Quantitative PCR (RT-qPCR) Validation
2.8. Subcellular Localization of VvMYB2
2.9. Instantaneous Transformation of Callus
2.10. Statistical Analysis
3. Results
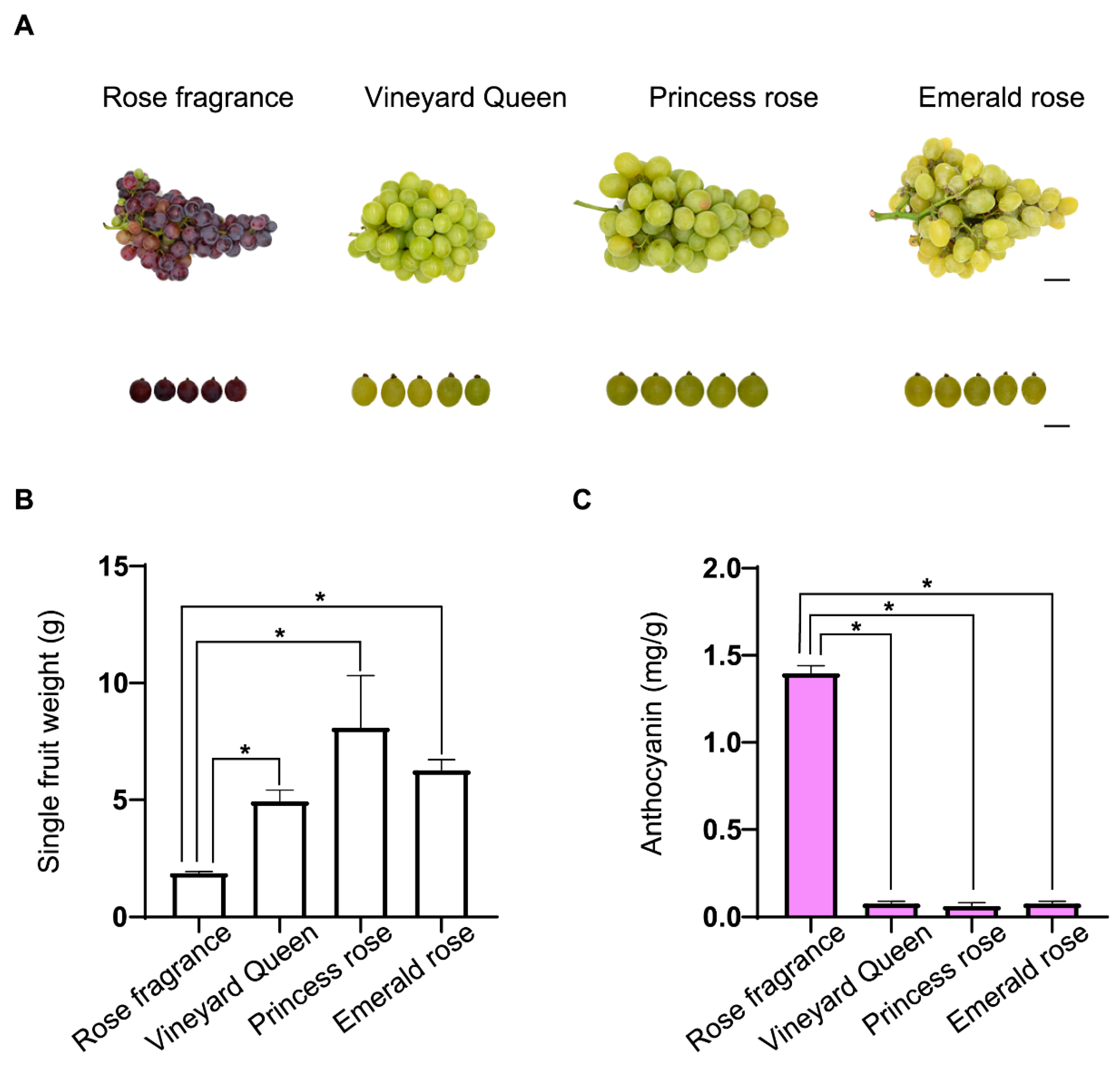
3.1. Physiological Indicator Analysis
3.2. Global Transcriptome Analysis and Functional Classification
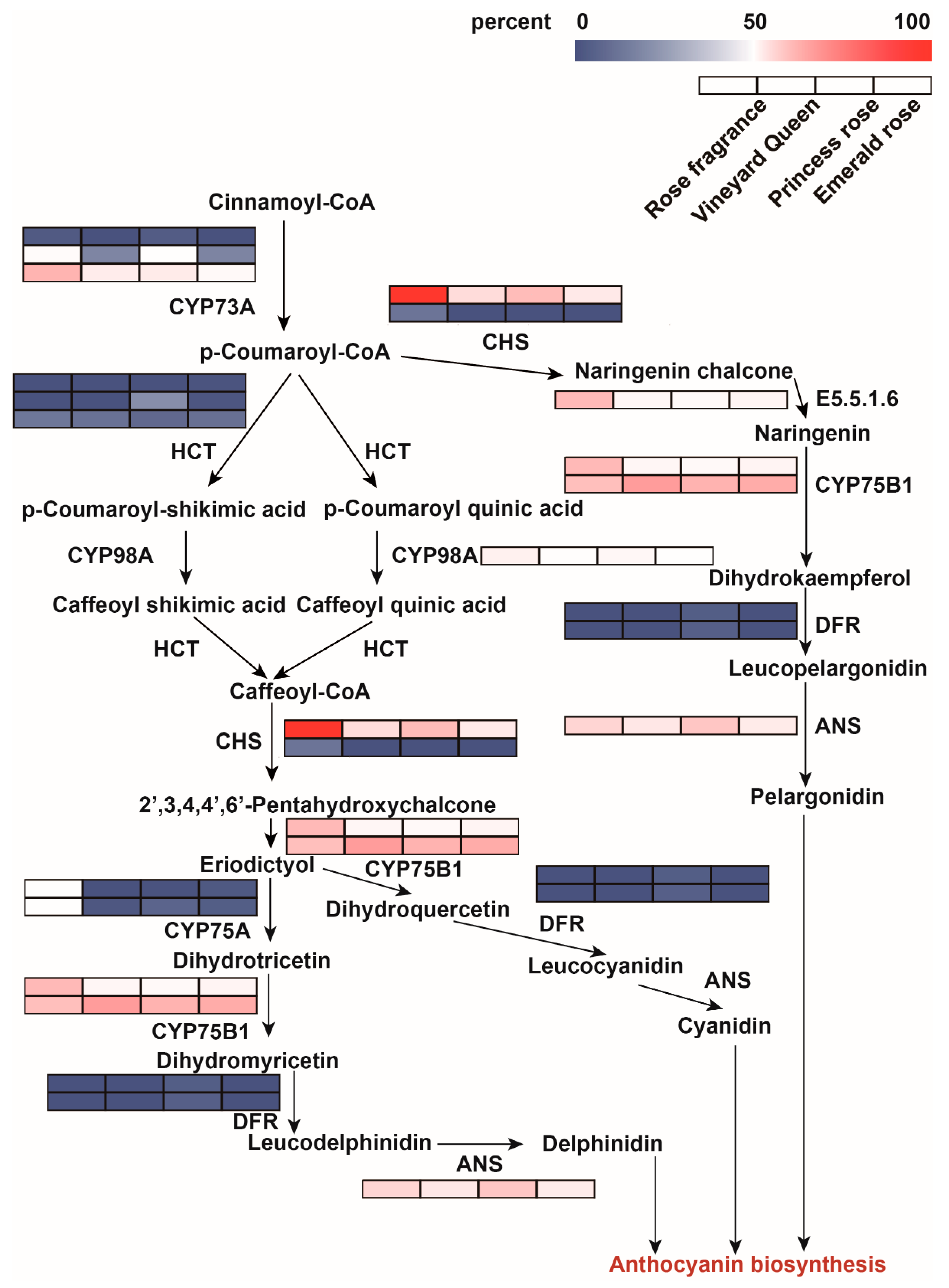
3.3. Screening of Enzyme Genes in Flavonoid Biosynthesis Pathway
3.4. Screening of Transcription Factors in Flavonoid Biosynthesis Pathway
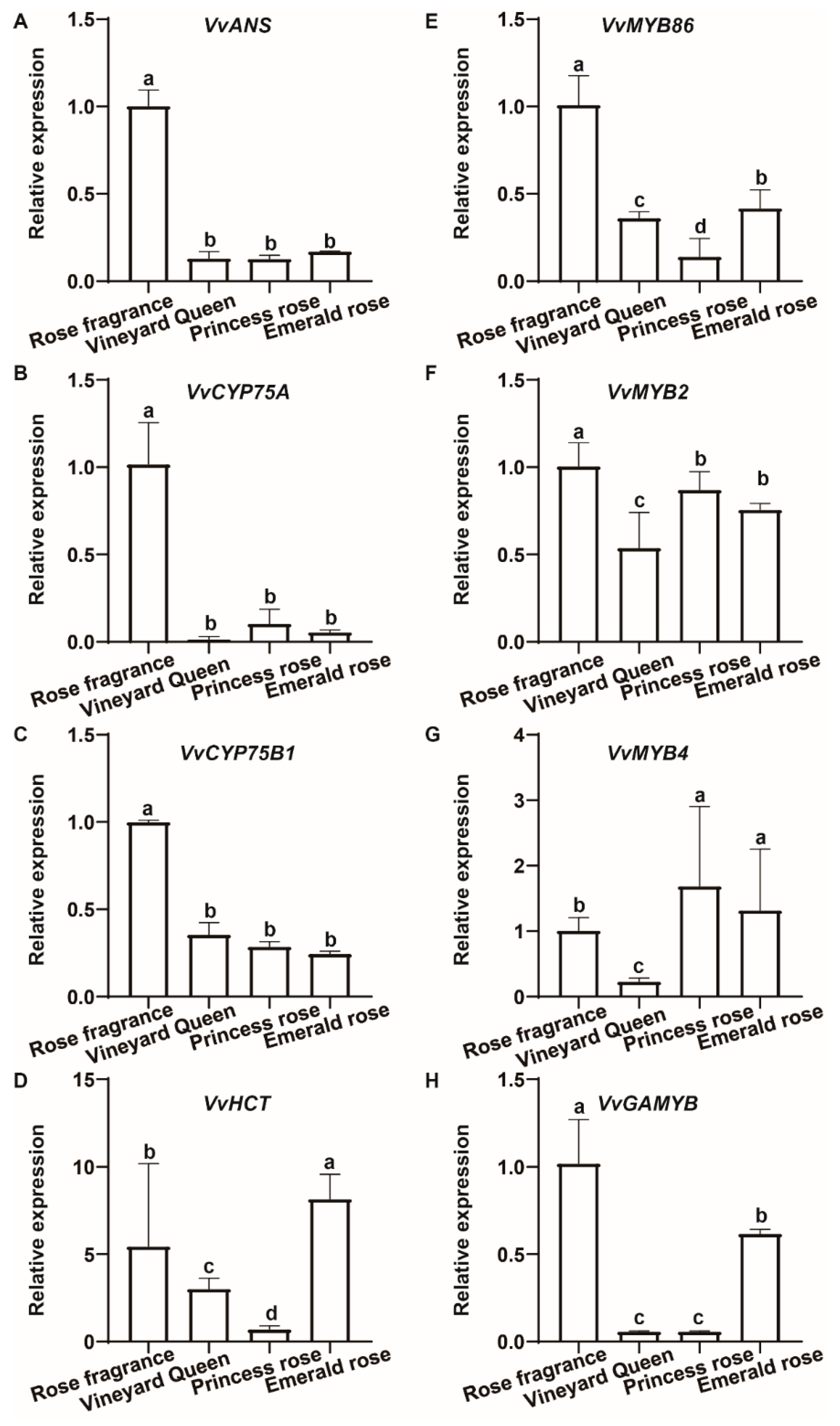
3.5. Verification of DEGs by RT-qPCR
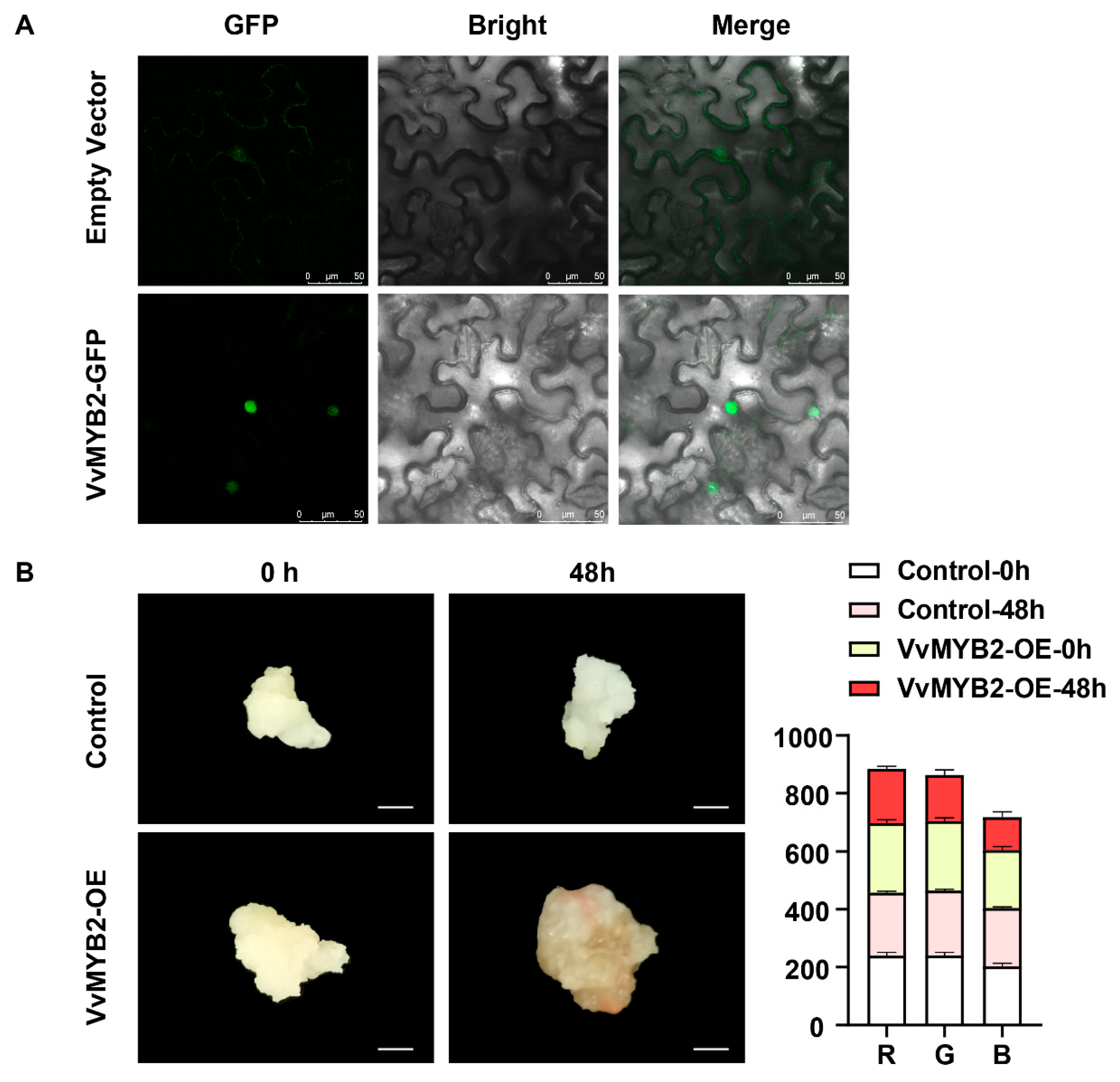
3.6. Subcellular Localization and Callus Transient Transformation Analysis of VvMYB2 Function
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Girard, B.; Yuksel, D.; Cliff, M.; Delaquis, P.; Reynolds, A. Vinification effects on the sensory, colour and GC profiles of Pinot noir wines from British Columbia. Food Res. Int. 2001, 34, 483–499. [Google Scholar] [CrossRef]
- Shaw, P.E.; Wilson III, C.W. Importance of nootkatone to the aroma of grapefruit oil and the flavor of grapefruit juice. J. Agric. Food Chem. 1981, 29, 677–679. [Google Scholar] [CrossRef]
- Slegers, A.; Angers, P.; Ouellet, É.; Truchon, T.; Pedneault, K. Volatile compounds from grape skin, juice and wine from five interspecific hybrid grape cultivars grown in Québec (Canada) for wine production. Molecules 2015, 20, 10980–11016. [Google Scholar] [CrossRef] [PubMed]
- Fredes, C.; Montenegro, G.; Zoffoli, J.P.; Gómez, M.; Robert, P. Polyphenol content and antioxidant activity of maqui (Aristotelia chilensis Molina Stuntz) during fruit development and maturation in Central Chile. Chil. J. Agric. Res. 2012, 72, 582–589. [Google Scholar] [CrossRef]
- Outchkourov, N.S.; Carollo, C.A.; Gomez-Roldan, V.; de Vos, R.C.; Bosch, D.; Hall, R.D.; Beekwilder, J. Control of anthocyanin and non-flavonoid compounds by anthocyanin-regulating MYB and bHLH transcription factors in Nicotiana benthamiana leaves. Front. Plant Sci. 2014, 5, 519. [Google Scholar] [CrossRef] [PubMed]
- Naing, A.H.; Kim, C.K. Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plant. 2021, 172, 1711–1723. [Google Scholar] [CrossRef]
- Giampieri, F.; Tulipani, S.; Alvarez-Suarez, J.M.; Quiles, J.L.; Mezzetti, B.; Battino, M. The strawberry: Composition, nutritional quality, and impact on human health. Nutrition 2012, 28, 9–19. [Google Scholar] [CrossRef]
- Simona, O. A review of the current knowledge of thermal stability of anthocyanins and approaches to their stabilization to heat. Antioxidants 2021, 10, 1337. [Google Scholar]
- Pantelidis, G.E.; Vasilakakis, M.; Manganaris, G.A.; Diamantidis, G. Antioxidant capacity, phenol, anthocyanin and ascorbic acid contents in raspberries, blackberries, red currants, gooseberries and Cornelian cherries. Food Chem. 2007, 102, 777–783. [Google Scholar] [CrossRef]
- Quattrocchio, F.; Wing, J.F.; van der Woude, K.; Mol, J.; Koes, R. Analysis of bHLH and MYB domain proteins: Species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. Cell Mol. Biol. 1998, 13, 475–488. [Google Scholar] [CrossRef]
- Wang, Z.; Meng, D.; Wang, A.; Li, T.; Jiang, S.; Cong, P.; Li, T. The methylation of the PcMYB10 promoter is associated with green-skinned sport in Max Red Bartlett pear. Plant Physiol. 2013, 162, 885–896. [Google Scholar] [CrossRef]
- Matsui, K.; Umemura, Y.; Ohme-Takagi, M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008, 55, 954–967. [Google Scholar] [CrossRef]
- Abrahams, S.; Lee, E.; Walker, A.R.; Tanner, G.J.; Larkin, P.J.; Ashton, A.R. The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J. 2003, 35, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Song, Z.; Dong, B.; Niu, L.; Cao, H.; Li, H.; Du, T.; Liu, T.; Yang, W.; Meng, D. Hyperoside regulates its own biosynthesis via MYB30 in promoting reproductive development and seed set in okra. Plant Physiol. 2021, 185, 951–968. [Google Scholar] [CrossRef] [PubMed]
- XIE, X.B.; Li, S.; ZHANG, R.F.; Zhao, J.; CHEN, Y.C.; Zhao, Q.; YAO, Y.X.; YOU, C.X.; ZHANG, X.S.; HAO, Y.J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.; Hrazdina, G. Interaction of nitrogen availability during bloom and light intensity during veraison. II. Effects on anthocyanin and phenolic development during grape ripening. Am. J. Enol. Vitic. 1998, 49, 341–349. [Google Scholar] [CrossRef]
- Shepard, J.F. The regeneration of potato plants from leaf-cell protoplasts. Sci. Am. 1982, 246, 154–169. [Google Scholar] [CrossRef]
- Polashock, J.; Zelzion, E.; Fajardo, D.; Zalapa, J.; Georgi, L.; Bhattacharya, D.; Vorsa, N. The American cranberry: First insights into the whole genome of a species adapted to bog habitat. BMC Plant Biol. 2014, 14, 165. [Google Scholar] [CrossRef]
- Cacho, J.; Fernández, P.; Ferreira, V.; Castells, J.E. Evolution of five anthocyanidin-3-glucosides in the skin of the Tempranillo, Moristel, and Garnacha grape varieties and influence of climatological variables. Am. J. Enol. Vitic. 1992, 43, 244–248. [Google Scholar] [CrossRef]
- Kim, B.G.; Kim, J.H.; Min, S.Y.; Shin, K.-H.; Kim, J.H.; Kim, H.Y.; Ryu, S.N. Anthocyanin content in rice is related to expression levels of anthocyanin biosynthetic genes. J. Plant Biol. 2007, 50, 156–160. [Google Scholar] [CrossRef]
- Ben-Simhon, Z.; Judeinstein, S.; Trainin, T.; Harel-Beja, R.; Bar-Ya’akov, I.; Borochov-Neori, H.; Holland, D. A “White” anthocyanin-less pomegranate (Punica granatum L.) caused by an insertion in the coding region of the leucoanthocyanidin dioxygenase (LDOX.; ANS) gene. PLoS ONE 2015, 10, e0142777. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.U.; Park, J.-I.; Jung, H.-J.; Hur, Y.; Nou, I.-S. Anthocyanin biosynthesis for cold and freezing stress tolerance and desirable color in Brassica rapa. Funct. Integr. Genom. 2015, 15, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Castellarin, S.D.; Di Gaspero, G.; Marconi, R.; Nonis, A.; Peterlunger, E.; Paillard, S.; Adam-Blondon, A.-F.; Testolin, R. Colour variation in red grapevines (Vitis vinifera L.): Genomic organisation, expression of flavonoid 3’-hydroxylase, flavonoid 3’, 5’-hydroxylase genes and related metabolite profiling of red cyanidin-/blue delphinidin-based anthocyanins in berry skin. BMC Genom. 2006, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Yang, H.; Yu, B.; Dare, A.; Varkonyi-Gasic, E.; Wang, J.; Zhang, J. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016, 67, 2159–2176. [Google Scholar] [CrossRef] [PubMed]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Ma, L.; Sang, Z. Preliminary study on formation of flower color in Magnolia wufengensis. Acta Bot. Boreali-Occident. Sin. 2010, 30, 2252–2257. [Google Scholar]
- Fournand, D.; Vicens, A.; Sidhoum, L.; Souquet, J.-M.; Moutounet, M.; Cheynier, V. Accumulation and extractability of grape skin tannins and anthocyanins at different advanced physiological stages. J. Agric. Food Chem. 2006, 54, 7331–7338. [Google Scholar] [CrossRef]
- Kong, J.-M.; Chia, L.-S.; Goh, N.-K.; Chia, T.-F.; Brouillard, R. Analysis and biological activities of anthocyanins. Phytochemistry 2003, 64, 923–933. [Google Scholar] [CrossRef]
- Escribano-Bailón, M.T.; Santos-Buelga, C.; Rivas-Gonzalo, J.C. Anthocyanins in cereals. J. Chromatogr. A 2004, 1054, 129–141. [Google Scholar] [CrossRef]
- Vatai, T.; Škerget, M.; Knez, Ž. Extraction of phenolic compounds from elder berry and different grape marc varieties using organic solvents and/or supercritical carbon dioxide. J. Food Eng. 2009, 90, 246–254. [Google Scholar] [CrossRef]
- Ravaglia, D.; Espley, R.V.; Henry-Kirk, R.A.; Andreotti, C.; Ziosi, V.; Hellens, R.P.; Costa, G.; Allan, A.C. Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 2013, 13, 68. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, L.; Zhou, X.; Zhou, M.; Lu, Y.; Ma, L.; Ma, H.; Zhang, Z. Transgenic wheat expressing Thinopyrum intermedium MYB transcription factor TiMYB2R-1 shows enhanced resistance to the take-all disease. J. Exp. Bot. 2013, 64, 2243–2253. [Google Scholar] [CrossRef] [PubMed]
- Himi, E.; Maekawa, M.; Noda, K. Differential expression of three flavanone 3-hydroxylase genes in grains and coleoptiles of wheat. Int. J. Plant Genom. 2011, 2011, 369460. [Google Scholar] [CrossRef]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.A.; Grayer, R.J. Anthocyanins and other flavonoids. Nat. Prod. Rep. 2004, 21, 539–573. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.H.; Van Ha, C.; Nishiyama, R.; Watanabe, Y.; Leyva-González, M.A.; Fujita, Y.; Tran, U.T.; Li, W.; Tanaka, M.; Seki, M. Arabidopsis type B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. Proc. Natl. Acad. Sci. USA 2016, 113, 3090–3095. [Google Scholar] [CrossRef]
- Castellarin, S.D.; Matthews, M.A.; Di Gaspero, G.; Gambetta, G.A. Water deficits accelerate ripening and induce changes in gene expression regulating flavonoid biosynthesis in grape berries. Planta 2007, 227, 101–112. [Google Scholar] [CrossRef]
- Song, Z.; Dong, B.; Yang, Q.; Niu, L.; Li, H.; Cao, H.; Amin, R.; Meng, D.; Yujie, F. Screening of CBL genes in pigeon pea with focus on the functional analysis of CBL4 in abiotic stress tolerance and flavonoid biosynthesis. Environ. Exp. Bot. 2020, 177, 104102. [Google Scholar] [CrossRef]
- Yang, W.; Li, N.; Fan, Y.; Dong, B.; Song, Z.; Cao, H.; Du, T.; Liu, T.; Qi, M.; Niu, L. Transcriptome analysis reveals abscisic acid enhancing drought resistance by regulating genes related to flavonoid metabolism in pigeon pea. Environ. Exp. Bot. 2021, 191, 104627. [Google Scholar] [CrossRef]
- Yang, Q.; Dong, B.; Wang, L.; Song, Z.; Niu, L.; Li, H.; Cao, H.; Meng, D.; Fu, Y. AeCDPK6 phosphorylates and stabilizes AeMYB30 promoting hyperoside biosynthesis to prolong full-blooming period in okra. J. Exp. Bot. 2020, 71, 4042–4056. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, L.; Qi, M.; Meng, D.; Yang, Q.; Wang, Y.; Ren, F.; Yang, L.; Chen, Y.; Liu, L.; Tang, M.; et al. Transcriptome Analysis Reveals the Central Role of the Transcription Factor MYB in Regulating Anthocyanin Accumulation in Economic Grape Species (Vitis vinifera). Forests 2023, 14, 416. https://doi.org/10.3390/f14020416
Su L, Qi M, Meng D, Yang Q, Wang Y, Ren F, Yang L, Chen Y, Liu L, Tang M, et al. Transcriptome Analysis Reveals the Central Role of the Transcription Factor MYB in Regulating Anthocyanin Accumulation in Economic Grape Species (Vitis vinifera). Forests. 2023; 14(2):416. https://doi.org/10.3390/f14020416
Chicago/Turabian StyleSu, Ling, Meng Qi, Dong Meng, Qing Yang, Yongmei Wang, Fengshan Ren, Liying Yang, Yingchun Chen, Liyuan Liu, Meiling Tang, and et al. 2023. "Transcriptome Analysis Reveals the Central Role of the Transcription Factor MYB in Regulating Anthocyanin Accumulation in Economic Grape Species (Vitis vinifera)" Forests 14, no. 2: 416. https://doi.org/10.3390/f14020416
APA StyleSu, L., Qi, M., Meng, D., Yang, Q., Wang, Y., Ren, F., Yang, L., Chen, Y., Liu, L., Tang, M., Song, Y., & Gong, L. (2023). Transcriptome Analysis Reveals the Central Role of the Transcription Factor MYB in Regulating Anthocyanin Accumulation in Economic Grape Species (Vitis vinifera). Forests, 14(2), 416. https://doi.org/10.3390/f14020416





