Long-Term Simulated Nitrogen Deposition Has Moderate Impacts on Soil Microbial Communities across Three Bioclimatic Domains of the Eastern Canadian Forest
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Experimental Design, Sampling Procedure, and Chemical Analyses
2.3. DNA Isolation, Amplicon Sequencing, and Taxonomy
2.4. Statistical Analyses
3. Results
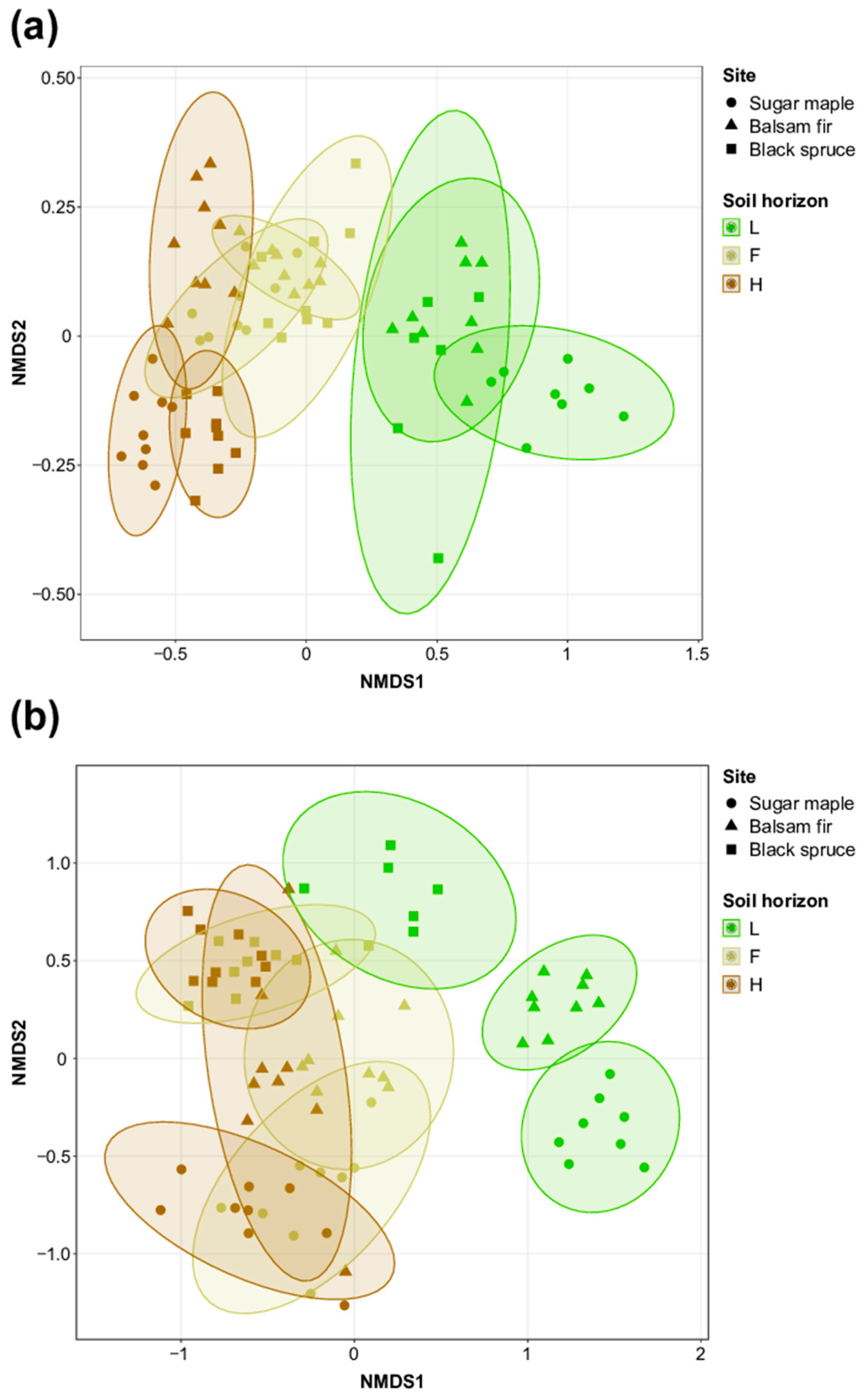
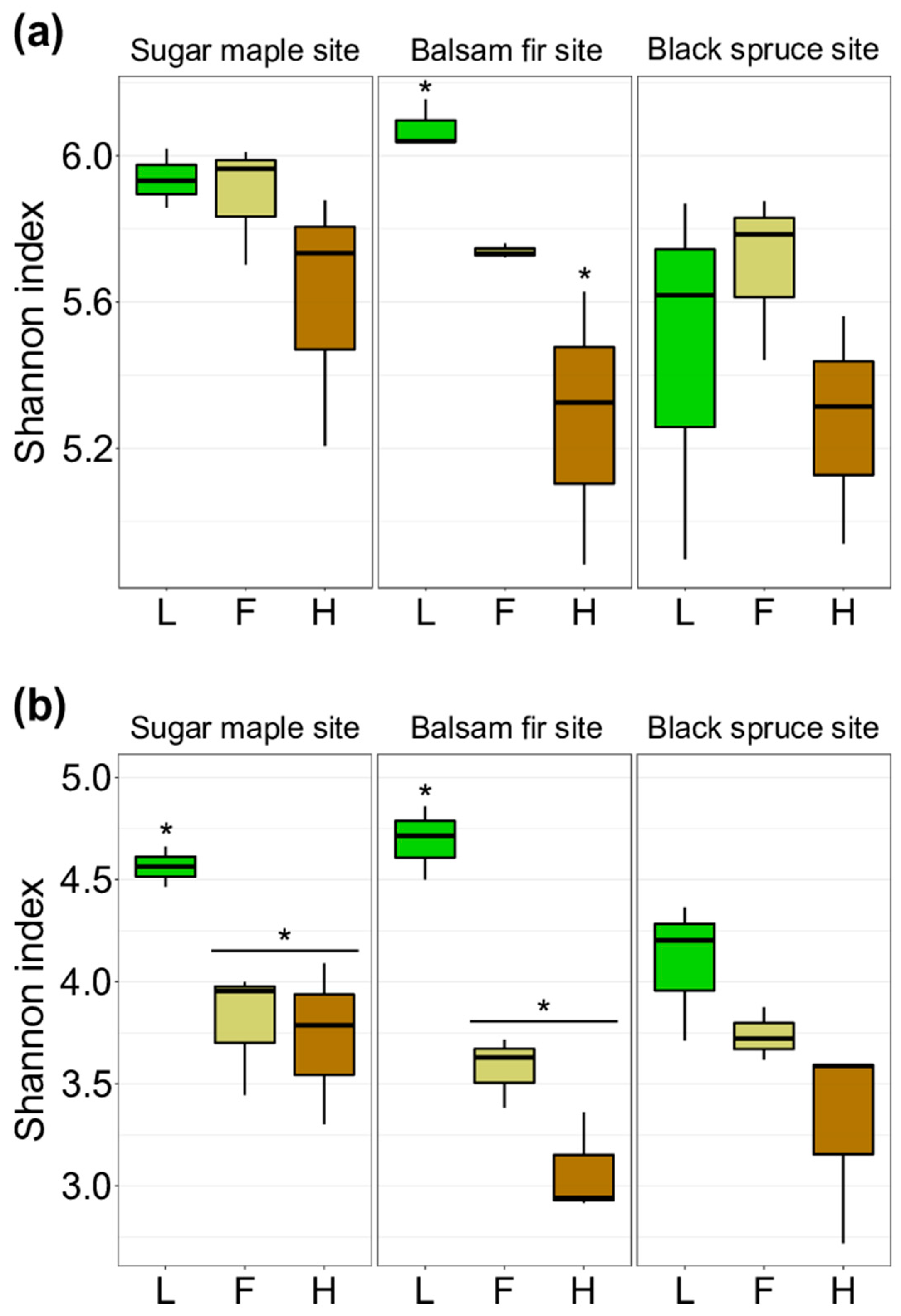
3.1. Characterization of Soil Bacterial and Fungal Communities in L, F, and H Horizons from Three Contrasted Forest Bioclimatic Domains
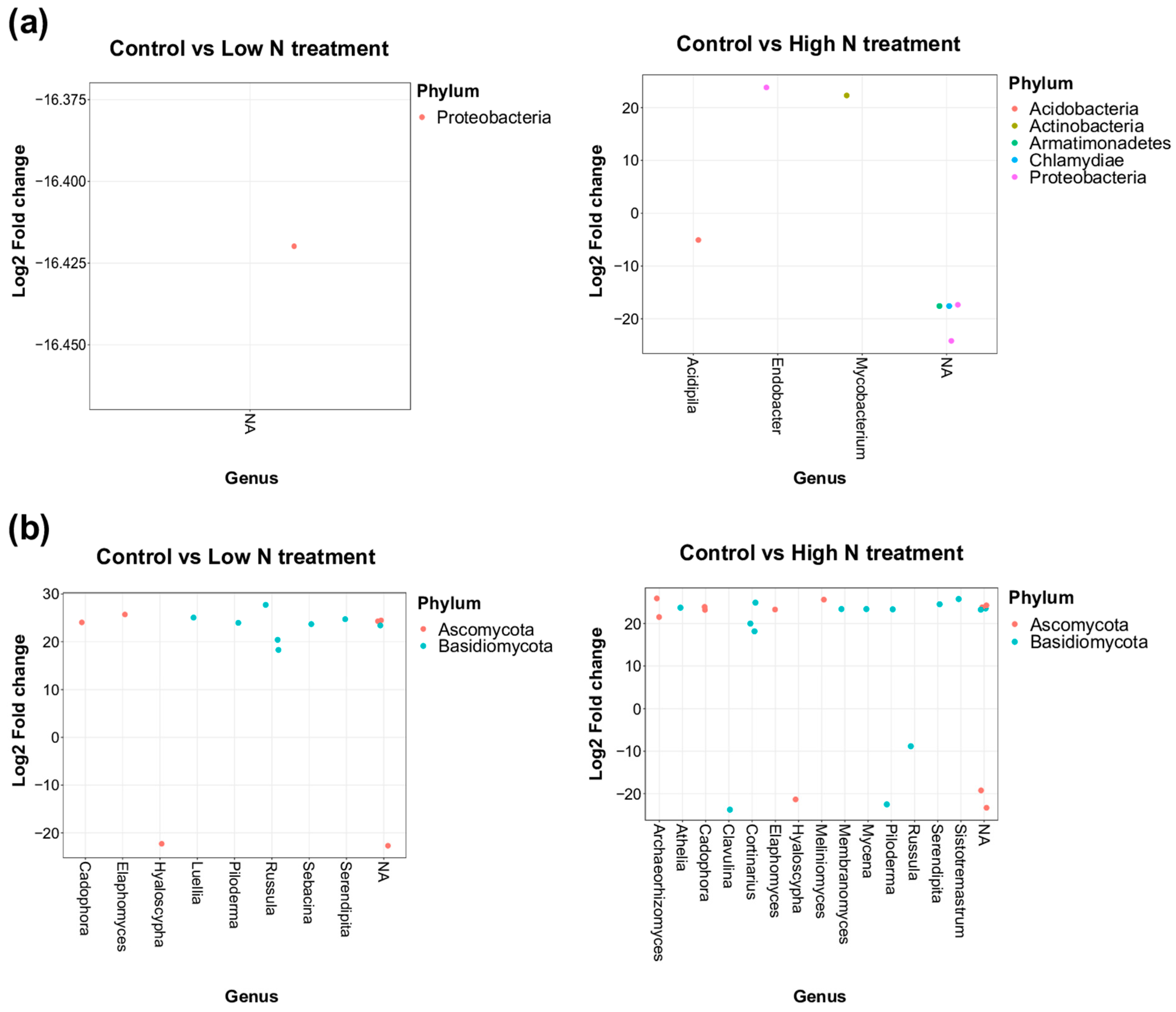
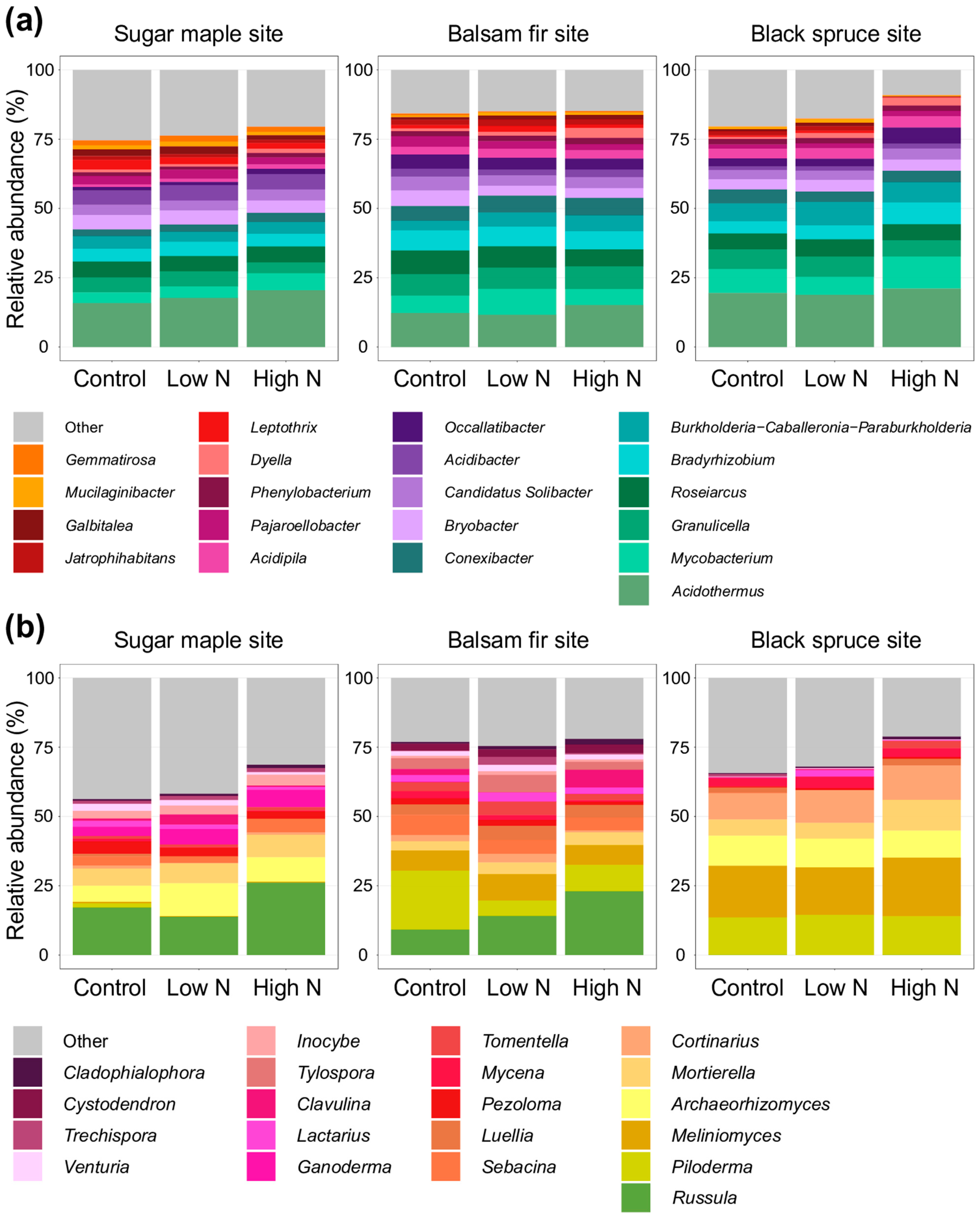
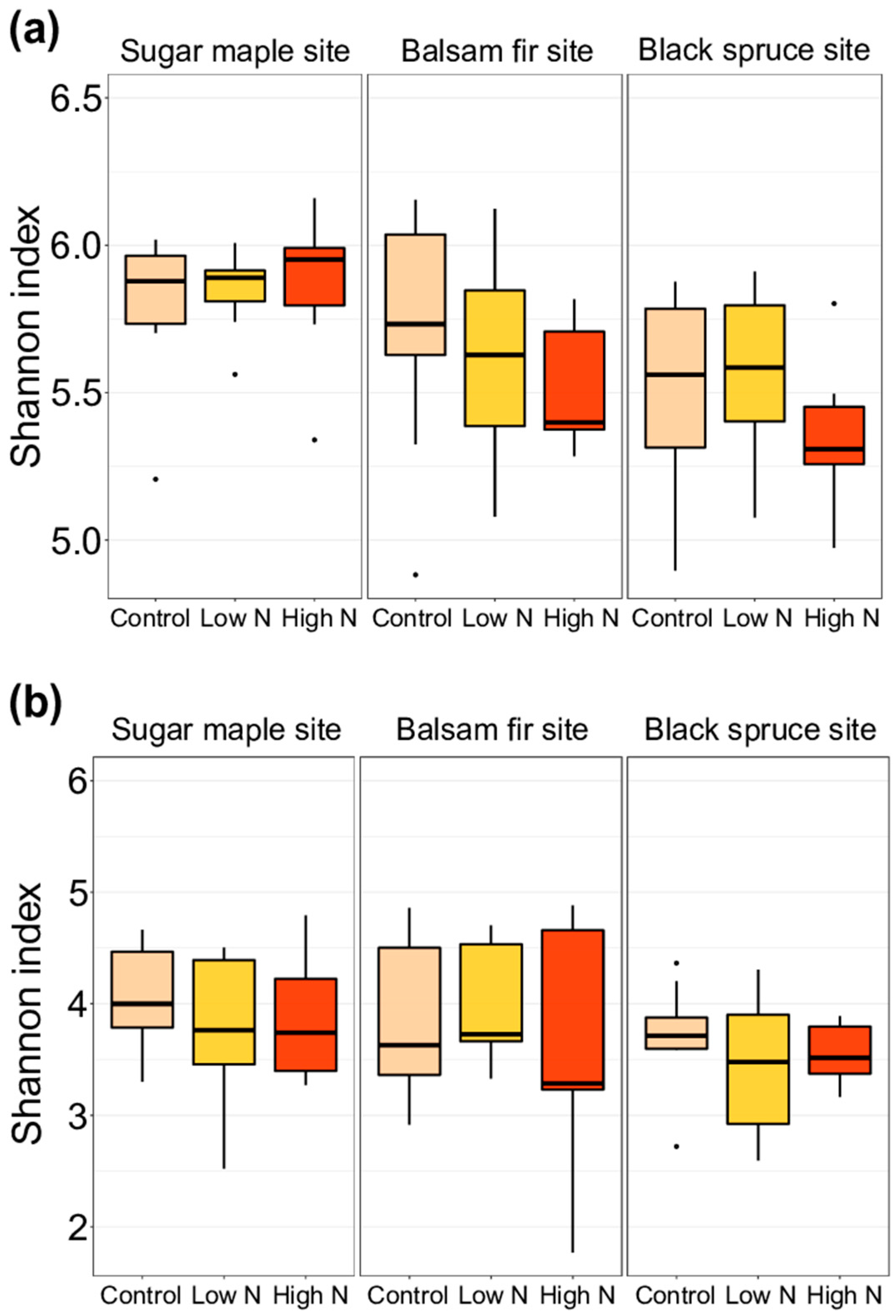
3.2. Impacts of Nitrogen Addition on Soil Microbiome and Chemistry at the Site and at the Horizon Level
4. Discussion
4.1. Spatial Location as the Main Driver of Forest Soil Microbial Community Composition
4.2. Moderate Effects of Long-Term Increased N Deposition on Forest Soil Microbial Communities
4.3. Potential Consequences for Northern Temperate and Boreal Forest Ecosystems
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Der Heijden, M.G.A.; Bardgett, R.D.; Van Straalen, N.M. The Unseen Majority: Soil Microbes as Drivers of Plant Diversity and Productivity in Terrestrial Ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Reich, P.B.; Trivedi, C.; Eldridge, D.J.; Abades, S.; Alfaro, F.D.; Bastida, F.; Berhe, A.A.; Cutler, N.A.; Gallardo, A.; et al. Multiple Elements of Soil Biodiversity Drive Ecosystem Functions across Biomes. Nat. Ecol. Evol. 2020, 4, 210–220. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the Unknown: Disentangling the Complexities of the Soil Microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Serrano-Silva, N.; Sarria-Guzmán, Y.; Dendooven, L.; Luna-Guido, M. Methanogenesis and Methanotrophy in Soil: A Review. Pedosphere 2014, 24, 291–307. [Google Scholar] [CrossRef]
- López-Mondéjar, R.; Zühlke, D.; Becher, D.; Riedel, K.; Baldrian, P. Cellulose and Hemicellulose Decomposition by Forest Soil Bacteria Proceeds by the Action of Structurally Variable Enzymatic Systems. Sci. Rep. 2016, 6, 25279. [Google Scholar] [CrossRef]
- Zheng, M.; Zhou, Z.; Luo, Y.; Zhao, P.; Mo, J. Global Pattern and Controls of Biological Nitrogen Fixation under Nutrient Enrichment: A Meta-Analysis. Glob. Chang. Biol. 2019, 25, 3018–3030. [Google Scholar] [CrossRef]
- Byers, A.K.; Condron, L.; Donavan, T.; O’Callaghan, M.; Patuawa, T.; Waipara, N.; Black, A. Soil Microbial Diversity in Adjacent Forest Systems-Contrasting Native, Old Growth Kauri (Agathis australis) Forest with Exotic Pine (Pinus radiata) Plantation Forest. FEMS Microbiol. Ecol. 2020, 96, fiaa047. [Google Scholar] [CrossRef]
- Mukhtar, H.; Lin, C.M.; Wunderlich, R.F.; Cheng, L.C.; Ko, M.C.; Lin, Y.P. Climate and Land Cover Shape the Fungal Community Structure in Topsoil. Sci. Total Environ. 2021, 751, 141721. [Google Scholar] [CrossRef]
- Lladó, S.; López-Mondéjar, R.; Baldrian, P. Drivers of Microbial Community Structure in Forest Soils. Appl. Microbiol. Biotechnol. 2018, 102, 4331–4338. [Google Scholar] [CrossRef]
- Jeanbille, M.; Buée, M.; Bach, C.; Cébron, A.; Frey-Klett, P.; Turpault, M.P.; Uroz, S. Soil Parameters Drive the Structure, Diversity and Metabolic Potentials of the Bacterial Communities Across Temperate Beech Forest Soil Sequences. Microb. Ecol. 2016, 71, 482–493. [Google Scholar] [CrossRef]
- Yan, G.; Xing, Y.; Xu, L.; Wang, J.; Dong, X.; Shan, W.; Guo, L.; Wang, Q. Effects of Different Nitrogen Additions on Soil Microbial Communities in Different Seasons in a Boreal Forest. Ecosphere 2017, 8, e01879. [Google Scholar] [CrossRef]
- Tian, D.; Niu, S. A Global Analysis of Soil Acidification Caused by Nitrogen Addition. Environ. Res. Lett. 2015, 10, 024019. [Google Scholar] [CrossRef]
- Geisseler, D.; Lazicki, P.A.; Scow, K.M. Mineral Nitrogen Input Decreases Microbial Biomass in Soils under Grasslands but Not Annual Crops. Appl. Soil Ecol. 2016, 106, 1–10. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, C.; Zheng, M.; Jiang, L.; Luo, Y. Patterns and Mechanisms of Responses by Soil Microbial Communities to Nitrogen Addition. Soil Biol. Biochem. 2017, 115, 433–441. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, C.; Yu, W.W.; Turak, A.; Chen, D.; Huang, Y.; Ao, J.; Jiang, Y.; Huang, Z. Effects of Nitrogen and Phosphorus Inputs on Soil Bacterial Abundance, Diversity, and Community Composition in Chinese Fir Plantations. Front. Microbiol. 2018, 9, 1543. [Google Scholar] [CrossRef]
- Zhang, T.; Chen, H.Y.H.; Ruan, H. Global Negative Effects of Nitrogen Deposition on Soil Microbes. ISME J. 2018, 12, 1817–1825. [Google Scholar] [CrossRef]
- Rossi, S.; Bordeleau, A.; Houle, D.; Morin, H. Effect of Chronic Ammonium Nitrate Addition on the Ectomycorrhizal Community in a Black Spruce Stand. Can. J. For. Res. 2012, 42, 1204–1212. [Google Scholar] [CrossRef]
- Wang, C.; Liu, D.; Bai, E. Decreasing Soil Microbial Diversity Is Associated with Decreasing Microbial Biomass under Nitrogen Addition. Soil Biol. Biochem. 2018, 120, 126–133. [Google Scholar] [CrossRef]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative Metagenomic, Phylogenetic and Physiological Analyses of Soil Microbial Communities across Nitrogen Gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef]
- Janssens, I.A.; Dieleman, W.; Luyssaert, S.; Subke, J.A.; Reichstein, M.; Ceulemans, R.; Ciais, P.; Dolman, A.J.; Grace, J.; Matteucci, G.; et al. Reduction of Forest Soil Respiration in Response to Nitrogen Deposition. Nat. Geosci. 2010, 3, 315–322. [Google Scholar] [CrossRef]
- Xia, N.; Du, E.; Wu, X.; Tang, Y.; Wang, Y.; de Vries, W. Effects of Nitrogen Addition on Soil Methane Uptake in Global Forest Biomes. Environ. Pollut. 2020, 264, 114751. [Google Scholar] [CrossRef] [PubMed]
- Compton, J.E.; Watrud, L.S.; Porteous, L.A.; DeGrood, S. Response of Soil Microbial Biomass and Community Composition to Chronic Nitrogen Additions at Harvard Forest. For. Ecol. Manag. 2004, 196, 143–158. [Google Scholar] [CrossRef]
- Lu, M.; Yang, Y.; Luo, Y.; Fang, C.; Zhou, X.; Chen, J.; Yang, X.; Li, B. Responses of Ecosystem Nitrogen Cycle to Nitrogen Addition: A Meta-Analysis. New Phytol. 2011, 189, 1040–1050. [Google Scholar] [CrossRef] [PubMed]
- Zak, D.R.; Argiroff, W.A.; Freedman, Z.B.; Upchurch, R.A.; Entwistle, E.M.; Romanowicz, K.J. Anthropogenic N Deposition, Fungal Gene Expression, and an Increasing Soil Carbon Sink in the Northern Hemisphere. Ecology 2019, 100, e02804. [Google Scholar] [CrossRef] [PubMed]
- Bonner, M.T.; Castro, D.; Schneider, A.N.; Sundström, G.; Hurry, V.; Street, N.R.; Näsholm, T. Why Does Nitrogen Addition to Forest Soils Inhibit Decomposition? Soil Biol. Biochem. 2019, 137, 107570. [Google Scholar] [CrossRef]
- Hasegawa, S.; Marshall, J.; Sparrman, T.; Näsholm, T. Decadal Nitrogen Addition Alters Chemical Composition of Soil Organic Matter in a Boreal Forest. Geoderma 2021, 386, 114906. [Google Scholar] [CrossRef]
- Galloway, J.N.; Dentener, F.J.; Capone, D.G.; Boyer, E.W.; Howarth, R.W.; Seitzinger, S.P.; Asner, G.P.; Cleveland, C.C.; Green, P.A.; Holland, E.A.; et al. Nitrogen cycles: Past, present, and future. Biogeochemistry 2004, 70, 153–226. [Google Scholar] [CrossRef]
- Houle, D.; Marty, C.; Duchesne, L. Response of Canopy Nitrogen Uptake to a Rapid Decrease in Bulk Nitrate Deposition in Two Eastern Canadian Boreal Forests. Oecologia 2015, 177, 29–37. [Google Scholar] [CrossRef]
- Ackerman, D.; Millet, D.B.; Chen, X. Global Estimates of Inorganic Nitrogen Deposition Across Four Decades. Glob. Biogeochem. Cycles 2019, 33, 100–107. [Google Scholar] [CrossRef]
- Yang, Y.; Cheng, H.; Gao, H.; An, S. Response and Driving Factors of Soil Microbial Diversity Related to Global Nitrogen Addition. Land Degrad. Dev. 2019, 31, 190–204. [Google Scholar] [CrossRef]
- Li, Y.; Wang, C.; Gao, S.; Wang, P.; Qiu, J.; Shang, S. Impacts of Simulated Nitrogen Deposition on Soil Enzyme Activity in a Northern Temperate Forest Ecosystem Depend on the Form and Level of Added Nitrogen. Eur. J. Soil Biol. 2021, 103, 103287. [Google Scholar] [CrossRef]
- Tian, D.; Du, E.; Jiang, L.; Ma, S.; Zeng, W.; Zou, A.; Feng, C.; Xu, L.; Xing, A.; Wang, W.; et al. Responses of Forest Ecosystems to Increasing N Deposition in China: A Critical Review. Environ. Pollut. 2018, 243, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Birdsey, R.A.; Fang, J.; Houghton, R.; Kauppi, P.E.; Kurz, W.A.; Philipps, O.L.; Shvidenko, A.; Lewis, S.L.; Canadell, J.G.; et al. A large and persistent carbon sink in the world’s forests. Science 2011, 333, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Lladó, S.; López-Mondéjar, R.; Baldrian, P. Forest Soil Bacteria: Diversity, Involvement in Ecosystem Processes, and Response to Global Change. Microbiol. Mol. Biol. Rev. 2017, 81, e00063-16. [Google Scholar] [CrossRef] [PubMed]
- Shao, P.; Han, H.; Sun, J.; Xie, H. Effects of Global Change and Human Disturbance on Soil Carbon Cycling in Boreal Forest: A Review. Pedosphere 2022, 33, 194–211. [Google Scholar] [CrossRef]
- Boonstra, R.; Andreassen, H.P.; Boutin, S.; Hušek, J.; Ims, R.A.; Krebs, C.J.; Skarpe, C.; Wabakken, P. Why Do the Boreal Forest Ecosystems of Northwestern Europe Differ from Those of Western North America? Bioscience 2016, 66, 722–734. [Google Scholar] [CrossRef]
- Ramirez, K.S.; Craine, J.M.; Fierer, N. Consistent Effects of Nitrogen Amendments on Soil Microbial Communities and Processes across Biomes. Glob. Chang. Biol. 2012, 18, 1918–1927. [Google Scholar] [CrossRef]
- Marty, C.; Houle, D.; Courchesne, F.; Gagnon, C. Soil C:N Ratio Is the Main Driver of Soil δ15N in Cold and N-Limited Eastern Canadian Forests. Catena 2019, 172, 285–294. [Google Scholar] [CrossRef]
- Lovett, G.M.; Goodale, C.L. A New Conceptual Model of Nitrogen Saturation Based on Experimental Nitrogen Addition to an Oak Forest. Ecosystems 2011, 14, 615–631. [Google Scholar] [CrossRef]
- Soil Classification Working Group. The Canadian System of Soil Classification, 3rd ed.; Agriculture and Agri-Food Canada Publication: Ottawa, ON, Canada, 1998; Volume 1646, 187p. [Google Scholar]
- Ste-Marie, C.; Houle, D. Forest Floor Gross and Net Nitrogen Mineralization in Three Forest Types in Quebec, Canada. Soil Biol. Biochem. 2006, 38, 2135–2143. [Google Scholar] [CrossRef]
- Magill, A.H.; Aber, J.D.; Currie, W.S.; Nadelhoffer, K.J.; Martin, M.E.; McDowell, W.H.; Melillo, J.M.; Steudler, P. Ecosystem Response to 15 Years of Chronic Nitrogen Additions at the Harvard Forest LTER, Massachusetts, USA. For. Ecol. Manag. 2004, 196, 7–28. [Google Scholar] [CrossRef]
- Moore, J.D.; Houle, D. Chemistry of Soil and Foliage in Declining Sugar Maple Stands over 13 Years of Nitrogen Addition. For. Ecol. Manag. 2023, 535, 120897. [Google Scholar] [CrossRef]
- Reysenbach, A.-L.; Pace, N.R. Reliable amplification of hyperthermophilic archaeal 16S rRNA genes by the polymerase chain reaction. In Archaea: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1995; pp. 101–107. [Google Scholar]
- Chelius, M.K.; Triplett, E.W. The Diversity of Archaea and Bacteria in Association with the Roots of Zea mays L. Microb. Ecol. 2001, 41, 252–263. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S rRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 20 April 2023).
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vázquez-Baeza, Y.; Birmingham, A.; et al. Normalization and Microbial Differential Abundance Strategies Depend upon Data Characteristics. Microbiome 2017, 5, 27. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Abarenkov, K.; Zirk, A.; Piirmann, T.; Pöhönen, R.; Ivanov, F.; Nilsson, R.H.; Kõljalg, U. UNITE General FASTA Release for Fungi 2; Version 04.02.2020; UNITE Community: Tartu, Estonia, 2020. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Põlme, S.; Abarenkov, K.; Henrik Nilsson, R.; Lindahl, B.D.; Clemmensen, K.E.; Kauserud, H.; Nguyen, N.; Kjøller, R.; Bates, S.T.; Baldrian, P.; et al. FungalTraits: A User-Friendly Traits Database of Fungi and Fungus-like Stramenopiles. Fungal Divers. 2020, 105, 1–16. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Volume 35, ISBN 978-0-387-98140-6. [Google Scholar]
- Wickham, H.; Girlich, M. tidyr: Tidy Messy Data. R Package Version 1.2.0. 2022. Available online: https://CRAN.R-project.org/package=tidyr (accessed on 20 April 2023).
- Oksanen, J.; Kindt, R.; O’Hara, B. Vegan: Community Ecology Package. R Package Version 2.5-7. 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 20 April 2023).
- Anders, S.; Huber, W. Differential Expression Analysis for Sequence Count Data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Deng, Y.; Shen, L.; Wen, C.; Yan, Q.; Ning, D.; Qin, Y.; Xue, K.; Wu, L.; He, Z.; et al. Temperature Mediates Continental-Scale Diversity of Microbes in Forest Soils. Nat. Commun. 2016, 7, 12083. [Google Scholar] [CrossRef] [PubMed]
- Urbanová, M.; Šnajdr, J.; Baldrian, P. Composition of Fungal and Bacterial Communities in Forest Litter and Soil Is Largely Determined by Dominant Trees. Soil Biol. Biochem. 2015, 84, 53–64. [Google Scholar] [CrossRef]
- Nacke, H.; Goldmann, K.; Schöning, I.; Pfeiffer, B.; Kaiser, K.; Villamizar, G.A.C.; Schrumpf, M.; Buscot, F.; Daniel, R.; Wubet, T. Fine Spatial Scale Variation of Soil Microbial Communities under European Beech and Norway Spruce. Front. Microbiol. 2016, 7, 2067. [Google Scholar] [CrossRef]
- Shen, C.; Xiong, J.; Zhang, H.; Feng, Y.; Lin, X.; Li, X.; Liang, W.; Chu, H. Soil PH Drives the Spatial Distribution of Bacterial Communities along Elevation on Changbai Mountain. Soil Biol. Biochem. 2013, 57, 204–211. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Cajthaml, T.; Põlme, S.; Hiiesalu, I.; Anslan, S.; Harend, H.; Buegger, F.; Pritsch, K.; Koricheva, J.; et al. Tree Diversity and Species Identity Effects on Soil Fungi, Protists and Animals Are Context Dependent. ISME J. 2016, 10, 346–362. [Google Scholar] [CrossRef]
- Talbot, J.M.; Bruns, T.D.; Taylor, J.W.; Smith, D.P.; Branco, S.; Glassman, S.I.; Erlandson, S.; Vilgalys, R.; Liao, H.L.; Smith, M.E.; et al. Endemism and Functional Convergence across the North American Soil Mycobiome. Proc. Natl. Acad. Sci. USA 2014, 111, 6341–6346. [Google Scholar] [CrossRef]
- Bastida, F.; Torres, I.F.; Andrés-Abellán, M.; Baldrian, P.; López-Mondéjar, R.; Větrovský, T.; Richnow, H.H.; Starke, R.; Ondoño, S.; García, C.; et al. Differential Sensitivity of Total and Active Soil Microbial Communities to Drought and Forest Management. Glob. Chang. Biol. 2017, 23, 4185–4203. [Google Scholar] [CrossRef]
- Lindahl, B.D.; Ihrmark, K.; Boberg, J.; Trumbore, S.E.; Högberg, P.; Stenlid, J.; Finlay, R.D. Spatial Separation of Litter Decomposition and Mycorrhizal Nitrogen Uptake in a Boreal Forest. New Phytol. 2007, 173, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Baldrian, P.; Kolaiřík, M.; Štursová, M.; Kopecký, J.; Valášková, V.; Větrovský, T.; Žifčáková, L.; Šnajdr, J.; Rídl, J.; Vlček, Č.; et al. Active and Total Microbial Communities in Forest Soil Are Largely Different and Highly Stratified during Decomposition. ISME J. 2012, 6, 248–258. [Google Scholar] [CrossRef] [PubMed]
- López-Mondéjar, R.; Voříšková, J.; Větrovský, T.; Baldrian, P. The Bacterial Community Inhabiting Temperate Deciduous Forests Is Vertically Stratified and Undergoes Seasonal Dynamics. Soil Biol. Biochem. 2015, 87, 43–50. [Google Scholar] [CrossRef]
- Nie, Y.; Wang, M.; Zhang, W.; Ni, Z.; Hashidoko, Y.; Shen, W. Ammonium Nitrogen Content Is a Dominant Predictor of Bacterial Community Composition in an Acidic Forest Soil with Exogenous Nitrogen Enrichment. Sci. Total Environ. 2018, 624, 407–415. [Google Scholar] [CrossRef]
- Houle, D.; Moore, J.D. Soil Solution, Foliar Concentrations and Tree Growth Response to 8 years of Ammonium-Nitrate Additions in Two Boreal Forests of Quebec, Canada. For. Ecol. Manag. 2019, 437, 263–271. [Google Scholar] [CrossRef]
- Sponseller, R.A.; Gundale, M.J.; Futter, M.; Ring, E.; Nordin, A.; Näsholm, T.; Laudon, H. Nitrogen Dynamics in Managed Boreal Forests: Recent Advances and Future Research Directions. Ambio 2016, 45, 175–187. [Google Scholar] [CrossRef]
- Li, H.; Xu, Z.; Yang, S.; Li, X.; Top, E.M.; Wang, R.; Zhang, Y.; Cai, J.; Yao, F.; Han, X.; et al. Responses of Soil Bacterial Communities to Nitrogen Deposition and Precipitation Increment Are Closely Linked with Aboveground Community Variation. Microb. Ecol. 2016, 71, 974–989. [Google Scholar] [CrossRef]
- Olsson, B.A.; Hansson, K.; Persson, T.; Beuker, E.; Helmisaari, H.S. Heterotrophic Respiration and Nitrogen Mineralisation in Soils of Norway Spruce, Scots Pine and Silver Birch Stands in Contrasting Climates. For. Ecol. Manag. 2012, 269, 197–205. [Google Scholar] [CrossRef]
- Herrmann, A.M.; Coucheney, E. Temperature Responses of Nitrogen Processes Cannot Be Inferred from Carbon Turnover in a Boreal Norway Spruce Forest. J. Plant Nutr. Soil Sci. 2019, 182, 934–944. [Google Scholar] [CrossRef]
- Allison, S.D.; Martiny, J.B.H. Resistance, Resilience, and Redundancy in Microbial Communities. Proc. Natl. Acad. Sci. USA 2008, 105, 11512–11519. [Google Scholar] [CrossRef]
- Moore, J.A.M.; Anthony, M.A.; Pec, G.J.; Trocha, L.K.; Trzebny, A.; Geyer, K.M.; van Diepen, L.T.A.; Frey, S.D. Fungal Community Structure and Function Shifts with Atmospheric Nitrogen Deposition. Glob. Chang. Biol. 2021, 27, 1349–1364. [Google Scholar] [CrossRef] [PubMed]
- Frey, S.D.; Ollinger, S.; Nadelhoffer, K.; Bowden, R.; Brzostek, E.; Burton, A.; Caldwell, B.A.; Crow, S.; Goodale, C.L.; Grandy, A.S.; et al. Chronic Nitrogen Additions Suppress Decomposition and Sequester Soil Carbon in Temperate Forests. Biogeochemistry 2014, 121, 305–316. [Google Scholar] [CrossRef]
- Maaroufi, N.I.; Nordin, A.; Hasselquist, N.J.; Bach, L.H.; Palmqvist, K.; Gundale, M.J. Anthropogenic Nitrogen Deposition Enhances Carbon Sequestration in Boreal Soils. Glob. Chang. Biol. 2015, 21, 3169–3180. [Google Scholar] [CrossRef] [PubMed]
- Zak, D.R.; Freedman, Z.B.; Upchurch, R.A.; Steffens, M.; Kögel-Knabner, I. Anthropogenic N Deposition Increases Soil Organic Matter Accumulation without Altering Its Biochemical Composition. Glob. Chang. Biol. 2017, 23, 933–944. [Google Scholar] [CrossRef]
- Li, W.; Jin, C.; Guan, D.; Wang, Q.; Wang, A.; Yuan, F.; Wu, J. The Effects of Simulated Nitrogen Deposition on Plant Root Traits: A Meta-Analysis. Soil Biol. Biochem. 2015, 82, 112–118. [Google Scholar] [CrossRef]
- Gundale, M.J.; Bach, L.H.; Nordin, A. The Impact of Simulated Chronic Nitrogen Deposition on the Biomass and N2-Fixation Activity of Two Boreal Feather Moss-Cyanobacteria Associations. Biol. Lett. 2013, 9, 20130797. [Google Scholar] [CrossRef]
- Sorensen, P.L.; Lett, S.; Michelsen, A. Moss-Specific Changes in Nitrogen Fixation Following Two Decades of Warming, Shading, and Fertilizer Addition. Plant Ecol. 2012, 213, 695–706. [Google Scholar] [CrossRef]
- Xing, A.; Xu, L.; Shen, H.; Du, E.; Liu, X.; Fang, J. Long Term Effect of Nitrogen Addition on Understory Community in a Chinese Boreal Forest. Sci. Total Environ. 2019, 646, 989–995. [Google Scholar] [CrossRef]
- Blok, D.; Heijmans, M.M.P.D.; Schaepman-Strub, G.; van Ruijven, J.; Parmentier, F.J.W.; Maximov, T.C.; Berendse, F. The Cooling Capacity of Mosses: Controls on Water and Energy Fluxes in a Siberian Tundra Site. Ecosystems 2011, 14, 1055–1065. [Google Scholar] [CrossRef]
- Koranda, M.; Michelsen, A. Mosses Reduce Soil Nitrogen Availability in a Subarctic Birch Forest via Effects on Soil Thermal Regime and Sequestration of Deposited Nitrogen. J. Ecol. 2021, 109, 1424–1438. [Google Scholar] [CrossRef]
- Liu, J.; Wu, N.; Wang, H.; Sun, J.; Peng, B.; Jiang, P.; Bai, E. Nitrogen Addition Affects Chemical Compositions of Plant Tissues, Litter and Soil Organic Matter. Ecology 2016, 97, 1796–1806. [Google Scholar] [CrossRef] [PubMed]

| (a) Bacterial Communities b | (b) Fungal Communities c | |||||||
|---|---|---|---|---|---|---|---|---|
| Df | F | R2 | p a | Df | F | R2 | p a | |
| Site | 2 | 18.36 | 0.177 | 0.001 * | 2 | 18.14 | 0.251 | 0.001 * |
| Horizon | 2 | 32.81 | 0.316 | 0.001 * | 2 | 8.40 | 0.116 | 0.001 * |
| Treatment | 2 | 2.21 | 0.021 | 0.006 * | 2 | 1.70 | 0.023 | 0.012 * |
| Site × Horizon | 4 | 7.42 | 0.143 | 0.001 * | 4 | 5.40 | 0.149 | 0.001 * |
| Site × Treatment | 4 | 1.55 | 0.030 | 0.032 * | 4 | 1.62 | 0.045 | 0.002 * |
| Horizon × Treatment | 4 | 1.45 | 0.028 | 0.049 * | 4 | 0.84 | 0.023 | 0.861 n.s. |
| Site × Horizon × Treatment | 7 | 1.11 | 0.038 | 0.215 n.s. | 7 | 0.83 | 0.040 | 0.954 n.s. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Renaudin, M.; Khlifa, R.; Legault, S.; Kembel, S.W.; Kneeshaw, D.; Moore, J.-D.; Houle, D. Long-Term Simulated Nitrogen Deposition Has Moderate Impacts on Soil Microbial Communities across Three Bioclimatic Domains of the Eastern Canadian Forest. Forests 2023, 14, 1124. https://doi.org/10.3390/f14061124
Renaudin M, Khlifa R, Legault S, Kembel SW, Kneeshaw D, Moore J-D, Houle D. Long-Term Simulated Nitrogen Deposition Has Moderate Impacts on Soil Microbial Communities across Three Bioclimatic Domains of the Eastern Canadian Forest. Forests. 2023; 14(6):1124. https://doi.org/10.3390/f14061124
Chicago/Turabian StyleRenaudin, Marie, Rim Khlifa, Simon Legault, Steven W. Kembel, Daniel Kneeshaw, Jean-David Moore, and Daniel Houle. 2023. "Long-Term Simulated Nitrogen Deposition Has Moderate Impacts on Soil Microbial Communities across Three Bioclimatic Domains of the Eastern Canadian Forest" Forests 14, no. 6: 1124. https://doi.org/10.3390/f14061124
APA StyleRenaudin, M., Khlifa, R., Legault, S., Kembel, S. W., Kneeshaw, D., Moore, J.-D., & Houle, D. (2023). Long-Term Simulated Nitrogen Deposition Has Moderate Impacts on Soil Microbial Communities across Three Bioclimatic Domains of the Eastern Canadian Forest. Forests, 14(6), 1124. https://doi.org/10.3390/f14061124






