A Method for Extracting the Tree Feature Parameters of Populus tomentosa in the Leafy Stage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Methodology Overview
2.2. Study Area and Data Collection
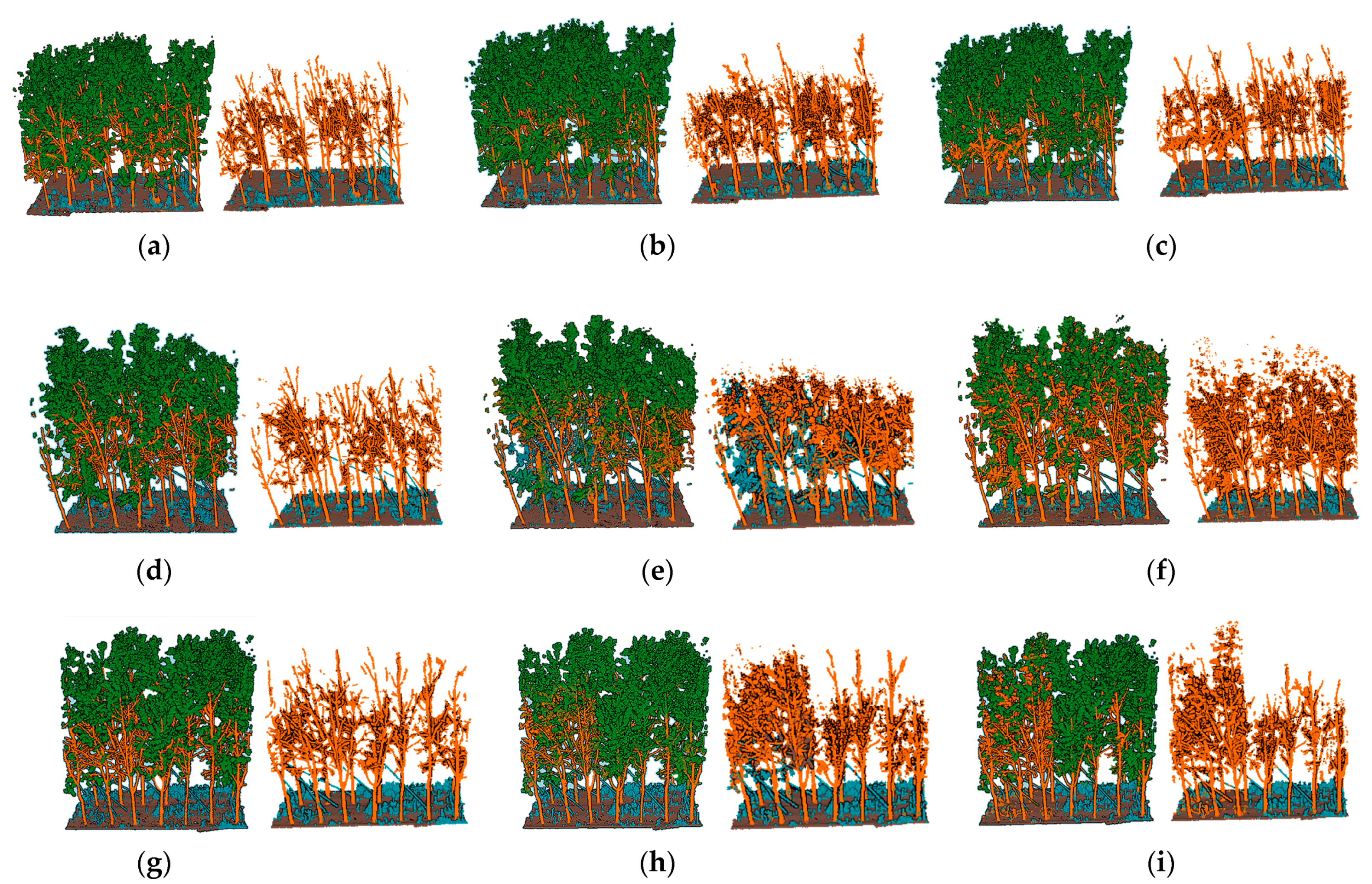
2.3. Class Selection Approach
2.4. Point Cloud Pre-Processing
2.4.1. Training Data and Validation Data
2.4.2. Testing Data
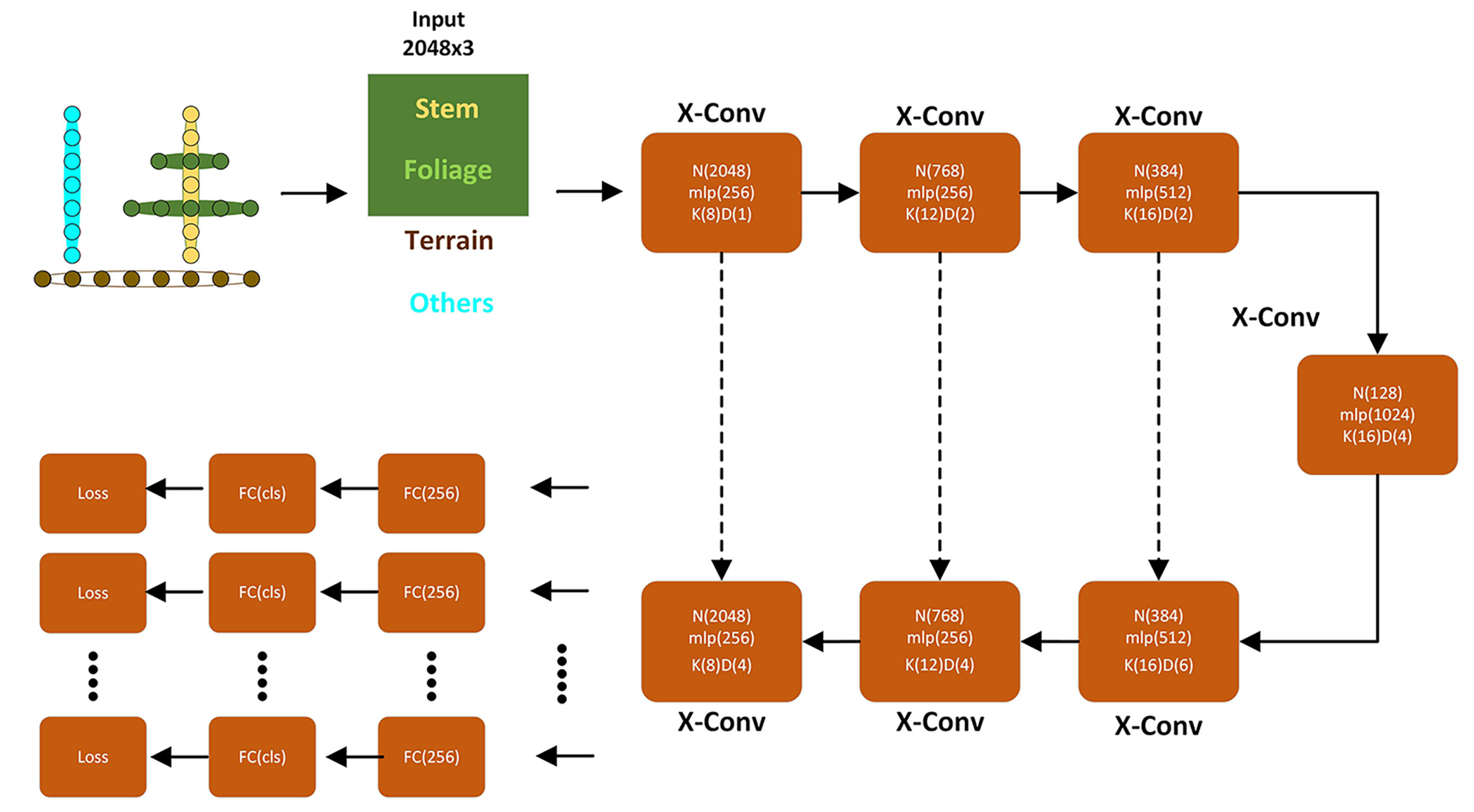
2.5. The PointCNN Deep Learning Network
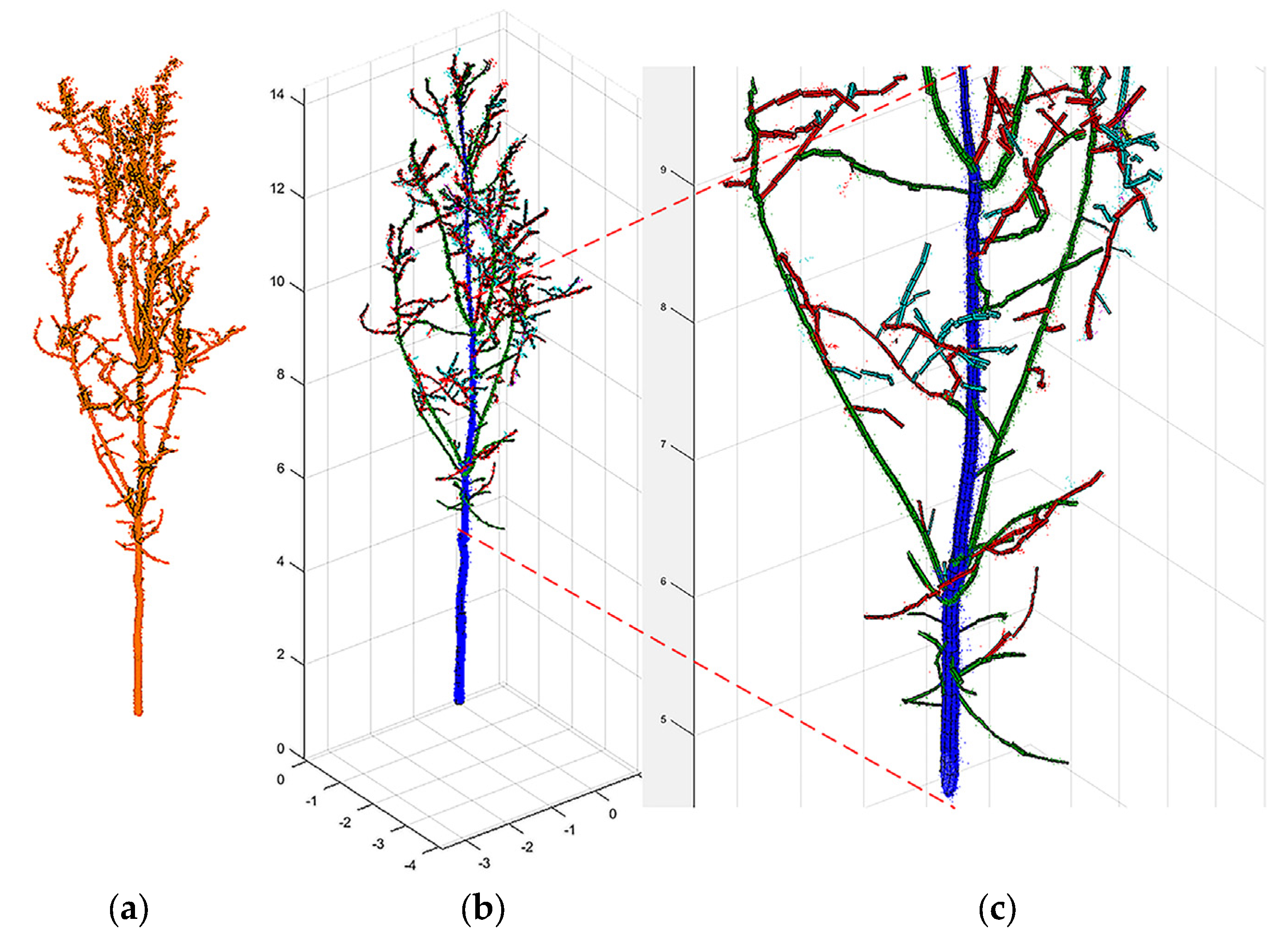
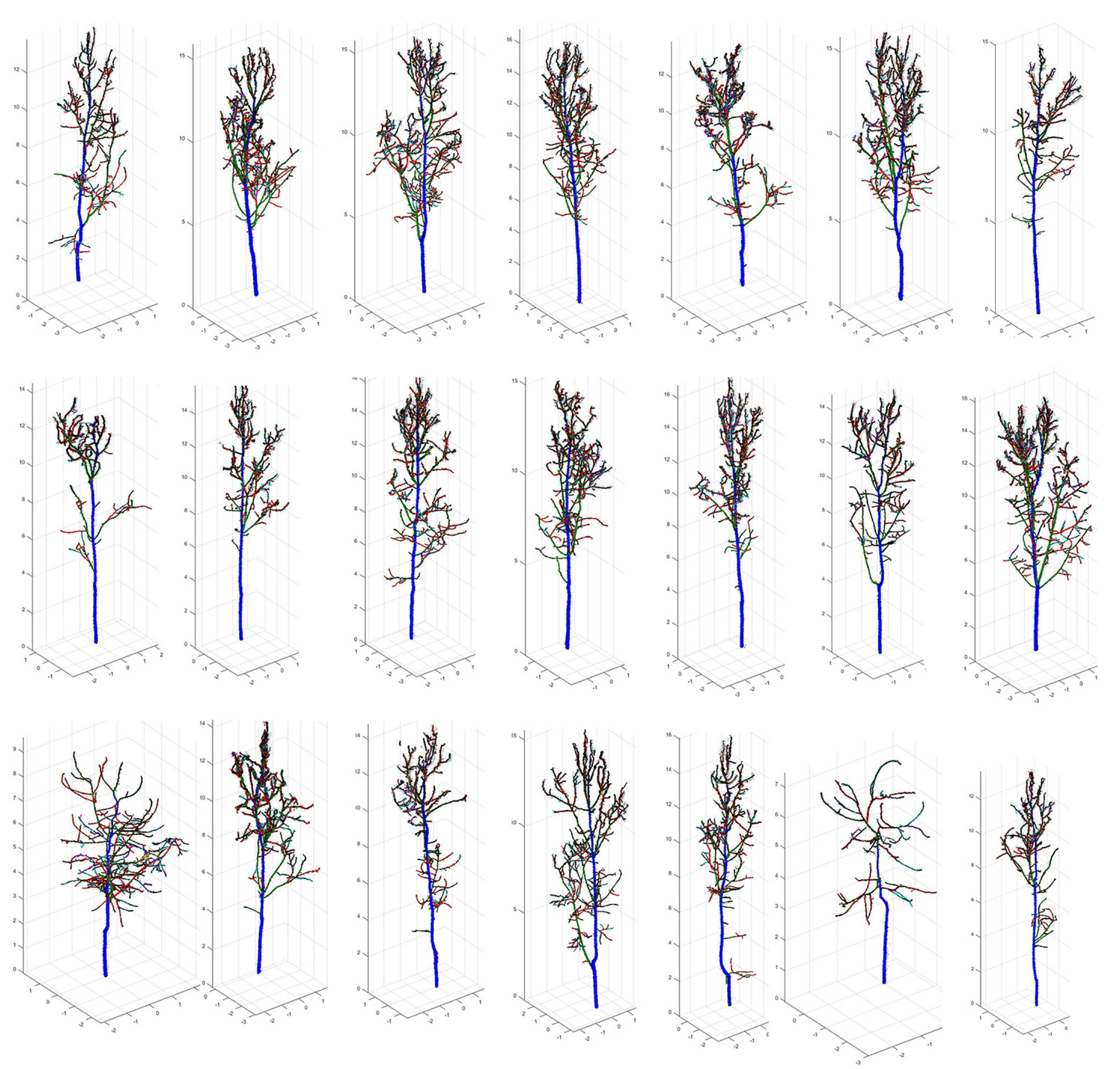
2.6. QSM Formation and Tree Feature Parameter Extraction
2.7. Training and Performance Measures
3. Results
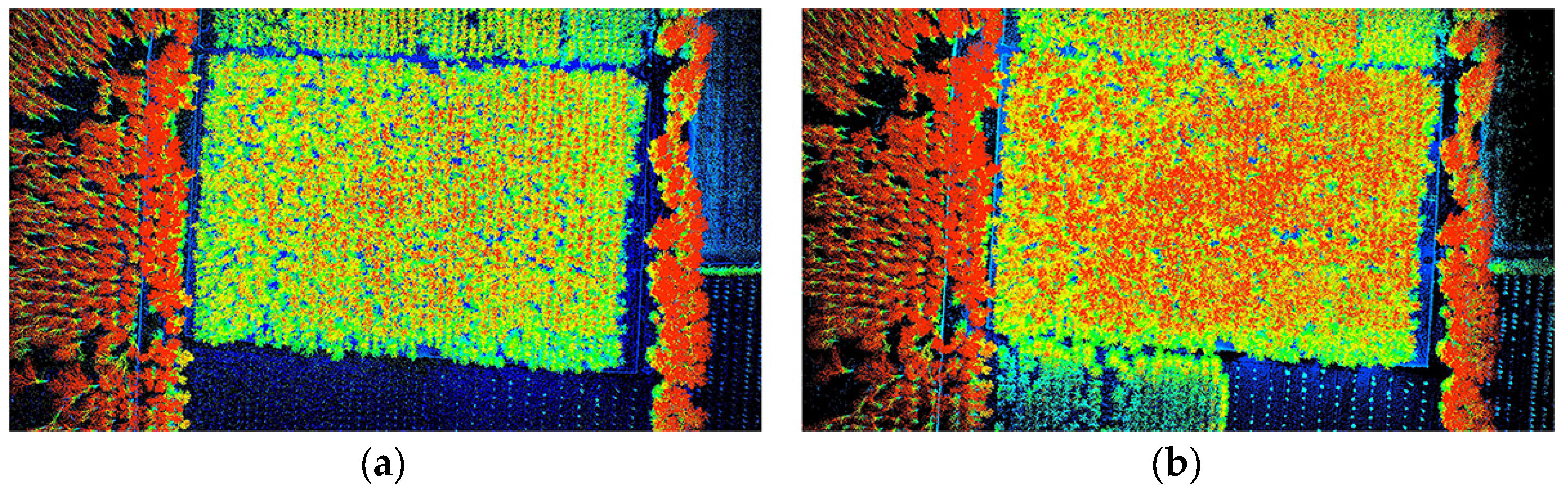
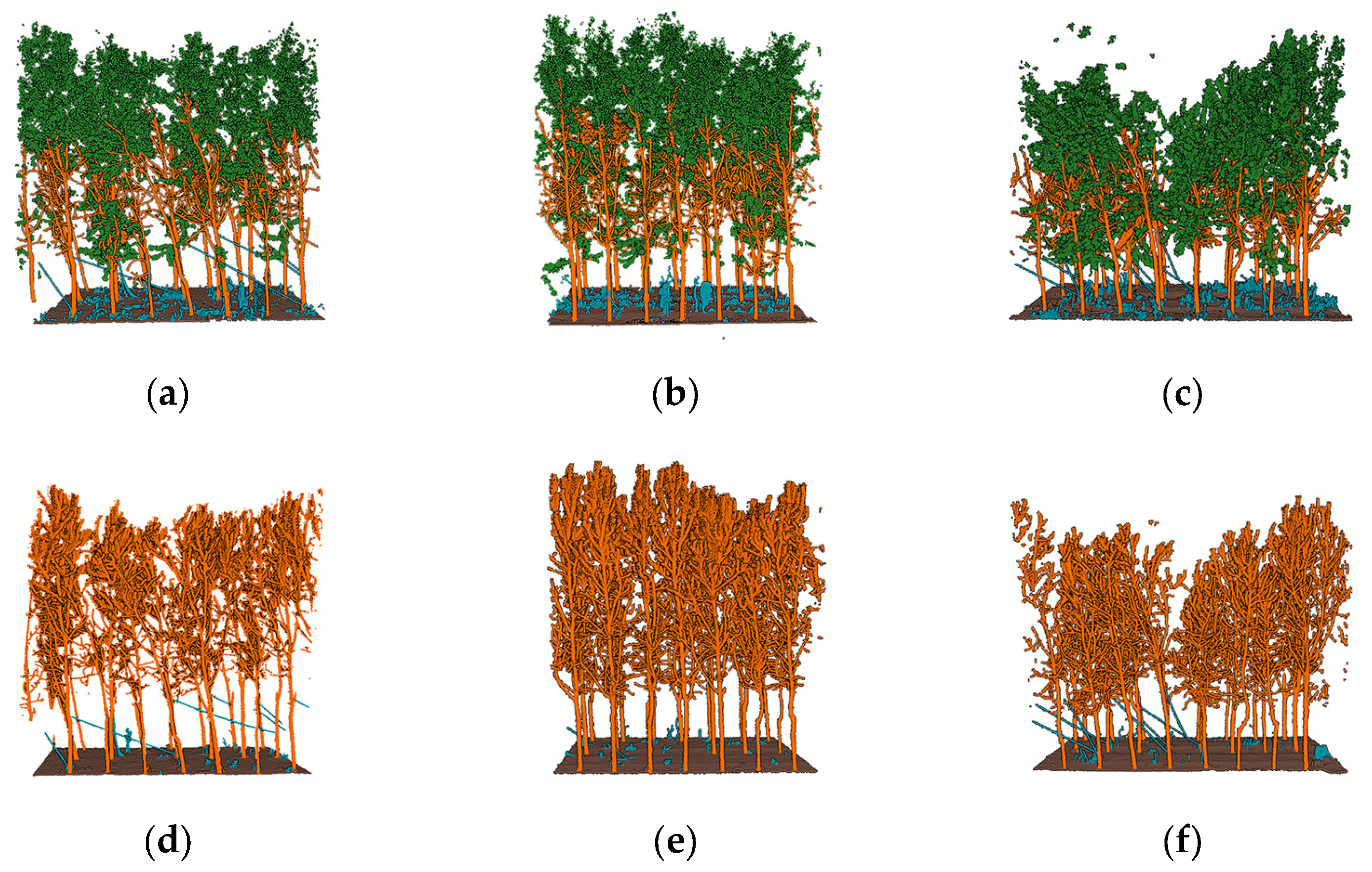
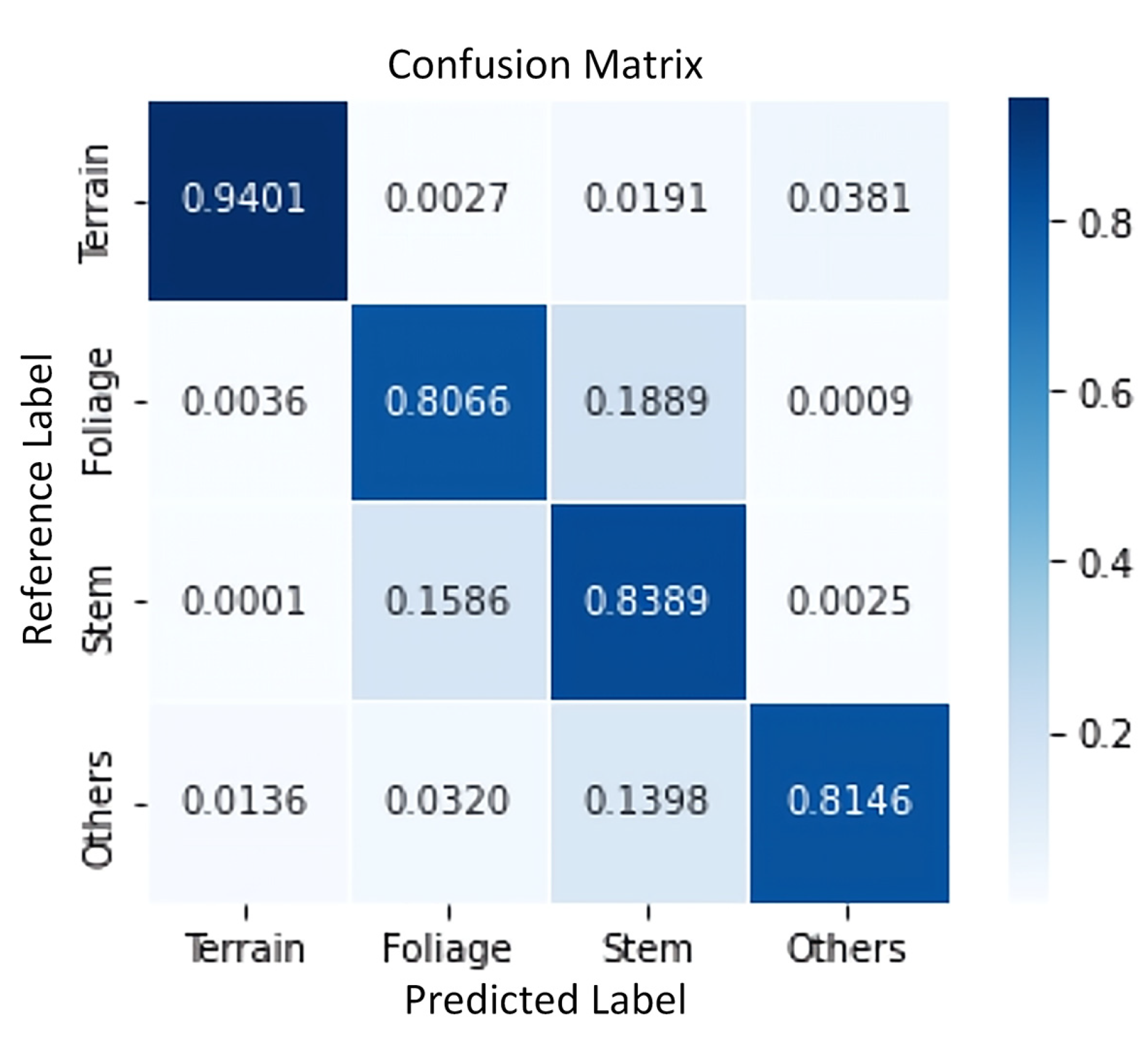
3.1. Semantic Segmentation Results
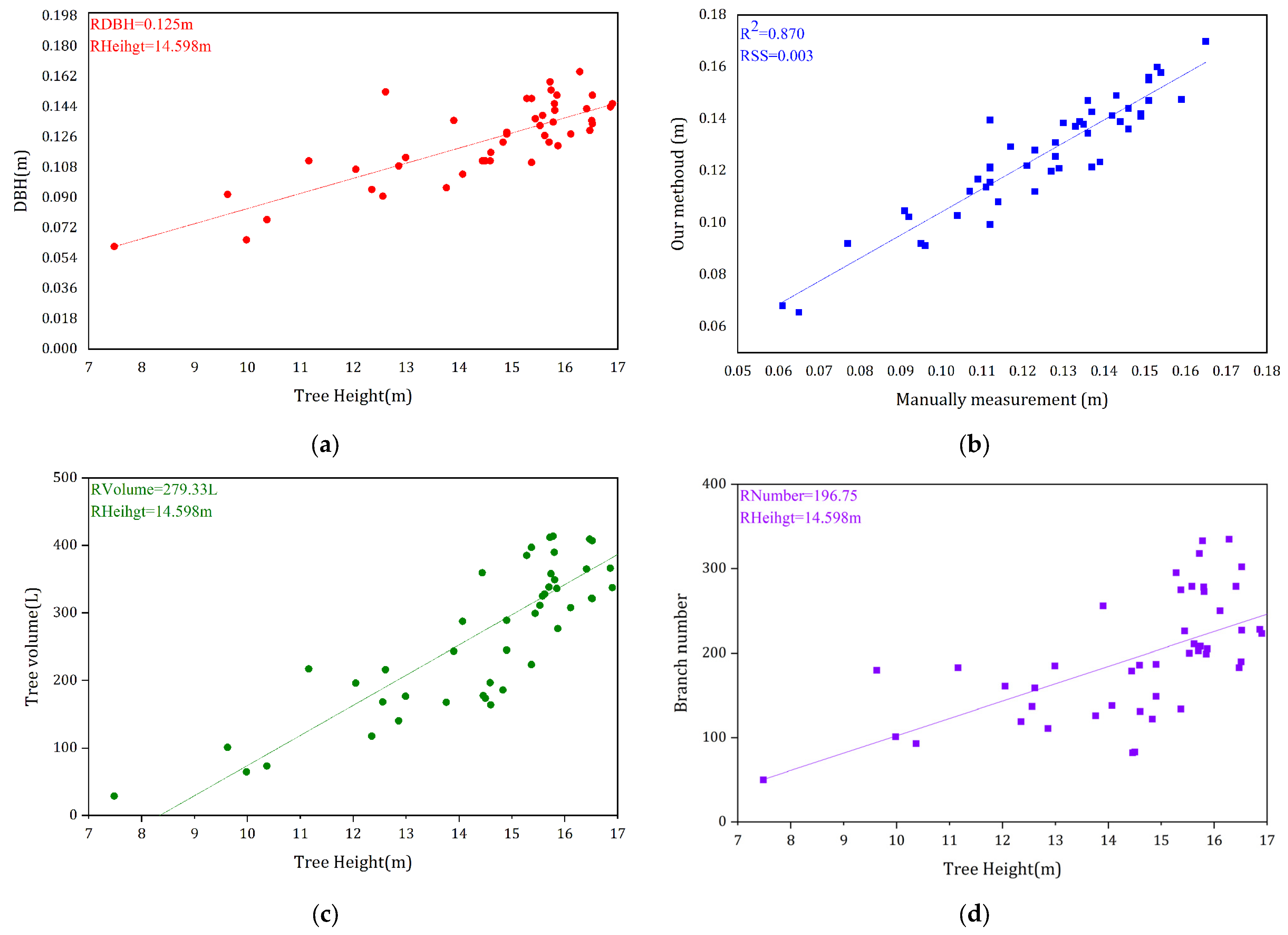
3.2. QSM Model Results and the Extracted Parameters
4. Discussion
4.1. Evaluation of Our Methoud
4.2. Comparison with Similar Methods
4.3. Future Work
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Calders, K.; Origo, N.; Disney, M.; Nightingale, J.; Woodgate, W.; Armston, J.; Lewis, P. Variability and bias in active and passive ground-based measurements of effective plant, wood and leaf area index. Agric. For. Meteorol. 2018, 252, 231–240. [Google Scholar] [CrossRef]
- Calders, K.; Schenkels, T.; Bartholomeus, H.; Armston, J.; Verbesselt, J.; Herold, M. Monitoring spring phenology with high temporal resolution terrestrial LiDAR measurements. Agric. For. Meteorol. 2015, 203, 158–168. [Google Scholar] [CrossRef]
- Weiser, H.; Schäfer, J.; Winiwarter, L.; Krašovec, N.; Fassnacht, F.E.; Höfle, B. Individual tree point clouds and tree measurements from multi-platform laser scanning in German forests. Earth Syst. Sci. Data 2022, 14, 2989–3012. [Google Scholar] [CrossRef]
- Wang, D. Unsupervised semantic and instance segmentation of forest point clouds. ISPRS J. Photogramm. Remote Sens. 2020, 165, 68–97. [Google Scholar] [CrossRef]
- Shen, X.; Huang, Q.; Wang, X.; Li, J.; Xi, B. A Deep Learning-Based Method for Extracting Standing Wood Feature Parameters from Terrestrial Laser Scanning Point Clouds of Artificially Planted Forest. Remote Sens. 2022, 14, 3842. [Google Scholar] [CrossRef]
- Wang, D.; Momo Takoudjou, S.; Casella, E. LeWoS: A universal leaf-wood classification method to facilitate the 3D modelling of large tropical trees using terrestrial LiDAR. Methods Ecol. Evol. 2020, 11, 376–389. [Google Scholar] [CrossRef]
- Tian, Z.; Li, S. Graph-Based Leaf–Wood Separation Method for Individual Trees Using Terrestrial Lidar Point Clouds. IEEE Trans. Geosci. Remote Sens. 2022, 60, 1–11. [Google Scholar] [CrossRef]
- Krishna Moorthy, S.M.; Calders, K.; Vicari, M.B.; Verbeeck, H. Improved Supervised Learning-Based Approach for Leaf and Wood Classification from LiDAR Point Clouds of Forests. IEEE Trans. Geosci. Remote Sens. 2020, 58, 3057–3070. [Google Scholar] [CrossRef]
- Hamraz, H.; Jacobs, N.B.; Contreras, M.A.; Clark, C.H. Deep learning for conifer/deciduous classification of airborne LiDAR 3D point clouds representing individual trees. ISPRS J. Photogramm. Remote Sens. 2019, 158, 219–230. [Google Scholar] [CrossRef]
- Han, T.; Sánchez-Azofeifa, G.A. A Deep Learning Time Series Approach for Leaf and Wood Classification from Terrestrial LiDAR Point Clouds. Remote Sens. 2022, 14, 3157. [Google Scholar] [CrossRef]
- Tan, K.; Zhang, W.; Dong, Z.; Cheng, X.; Cheng, X. Leaf and Wood Separation for Individual Trees Using the Intensity and Density Data of Terrestrial Laser Scanners. IEEE Trans. Geosci. Remote Sens. 2021, 59, 7038–7050. [Google Scholar] [CrossRef]
- Charles, R.Q.; Su, H.; Kaichun, M.; Guibas, L.J. PointNet: Deep Learning on Point Sets for 3D Classification and Segmentation. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 77–85. [Google Scholar]
- Chen, X.; Jiang, K.; Zhu, Y.; Wang, X.; Yun, T. Individual Tree Crown Segmentation Directly from UAV-Borne LiDAR Data Using the PointNet of Deep Learning. Forests 2021, 12, 131. [Google Scholar] [CrossRef]
- Qi, C.R.; Yi, L.; Su, H.; Guibas, L.J. PointNet++: Deep hierarchical feature learning on point sets in a metric space. arXiv 2017, arXiv:1706.02413. [Google Scholar]
- Krisanski, S.; Taskhiri, M.S.; Gonzalez Aracil, S.; Herries, D.; Turner, P. Sensor Agnostic Semantic Segmentation of Structurally Diverse and Complex Forest Point Clouds Using Deep Learning. Remote Sens. 2021, 13, 1413. [Google Scholar] [CrossRef]
- Li, Y.; Bu, R.; Sun, M.; Wu, W.; Di, X.; Chen, B. Pointcnn: Convolution on x-transformed points. In Proceedings of the Advances in Neural Information Processing Systems 31: Annual Conference on Neural Information Processing Systems 2018, NeurIPS 2018, Montréal, QC, Canada, 3–8 December 2018; Volume 31. [Google Scholar]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. TensorFlow: A System for Large-Scale Machine Learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation, OSDI 2016, Savannah, GA, USA, 2–4 November 2016; Volume 16, pp. 265–283. [Google Scholar]
- Krůček, M.; Král, K.; Cushman, K.C.; Missarov, A.; Kellner, J.R. Supervised segmentation of ultra-high-density drone lidar for large-area mapping of individual trees. Remote Sens. 2020, 12, 3260. [Google Scholar]
- Raumonen, P.; Kaasalainen, M.; Åkerblom, M.; Kaasalainen, S.; Kaartinen, H.; Vastaranta, M.; Holopainen, M.; Disney, M.; Lewis, P. Fast Automatic Precision Tree Models from Terrestrial Laser Scanner Data. Remote Sens. 2015, 5, 491–520. [Google Scholar] [CrossRef]
- Raumonen, P.; Casella, E.; Calders, K.; Murphy, S.; Åkerblom, M.; Kaasalainen, M. Massive-Scale Tree Modelling from Tls Data. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci 2015, 2, 189–196. [Google Scholar] [CrossRef]
- Markku, Å.; Raumonen, P.; Kaasalainen, M.; Casella, E. Analysis of Geometric Primitives in Quantitative Structure Models of Tree Stems. Remote Sens. 2015, 7, 4581–4603. [Google Scholar] [CrossRef]
- Calders, K.; Newnham, G.; Burt, A.; Murphy, S.; Raumonen, P.; Herold, M.; Culvenor, D.; Avitabile, V.; Disney, M.; Armston, J.; et al. Nondestructive estimates of above-ground biomass using terrestrial laser scanning. Methods Ecol. Evol. 2014, 6, 198–208. [Google Scholar] [CrossRef]
- Zhao, X.; Li, X.; Hu, W.; Liu, J.; Di, N.; Duan, J.; Li, D.; Liu, Y.; Guo, Y.; Wang, A.; et al. Long-term variation of the sap flow to tree diameter relation in a temperate poplar forest. J. Hydrol. 2023, 618, 129189. [Google Scholar] [CrossRef]
- Cai, J.-X.; Mu, T.-J.; Lai, Y.-K.; Hu, S.-M. LinkNet: 2D-3D linked multi-modal network for online semantic segmentation of RGB-D videos. Computers & Graphics. 2021, 98, 37–47. [Google Scholar]
- Qiu, S.; Anwar, S.; Barnes, N. Semantic Segmentation for Real Point Cloud Scenes via Bilateral Augmentation and Adaptive Fusion. In Proceedings of the IEEECVF Conference on Computer Vision and Pattern Recognition, Virtual, 19–25 June 2021; pp. 1757–1767. [Google Scholar]
- Ye, X.; Li, J.; Huang, H.; Du, L.; Zhang, X. 3D Recurrent Neural Networks with Context Fusion for Point Cloud Semantic Segmentation. In Proceedings of the Computer Vision—ECCV 2018—15th European Conference, Munich, Germany, 8–14 September 2018. [Google Scholar]
- Manduchi, G. Commonalities and differences between MDSplus and HDF5 data systems. Fusion Eng. Des. 2010, 85, 583–590. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Hui, Z.; Jin, S.; Xia, Y.; Wang, L.; Ziggah, Y.Y.; Cheng, P. Wood and leaf separation from terrestrial LiDAR point clouds based on mode points evolution. ISPRS J. Photogramm. Remote Sens. 2021, 178, 219–239. [Google Scholar] [CrossRef]
- Windrim, L.; Bryson, M. Detection, Segmentation, and Model Fitting of Individual Tree Stems from Airborne Laser Scanning of Forests Using Deep Learning. Remote Sens. 2020, 12, 1469. [Google Scholar] [CrossRef]

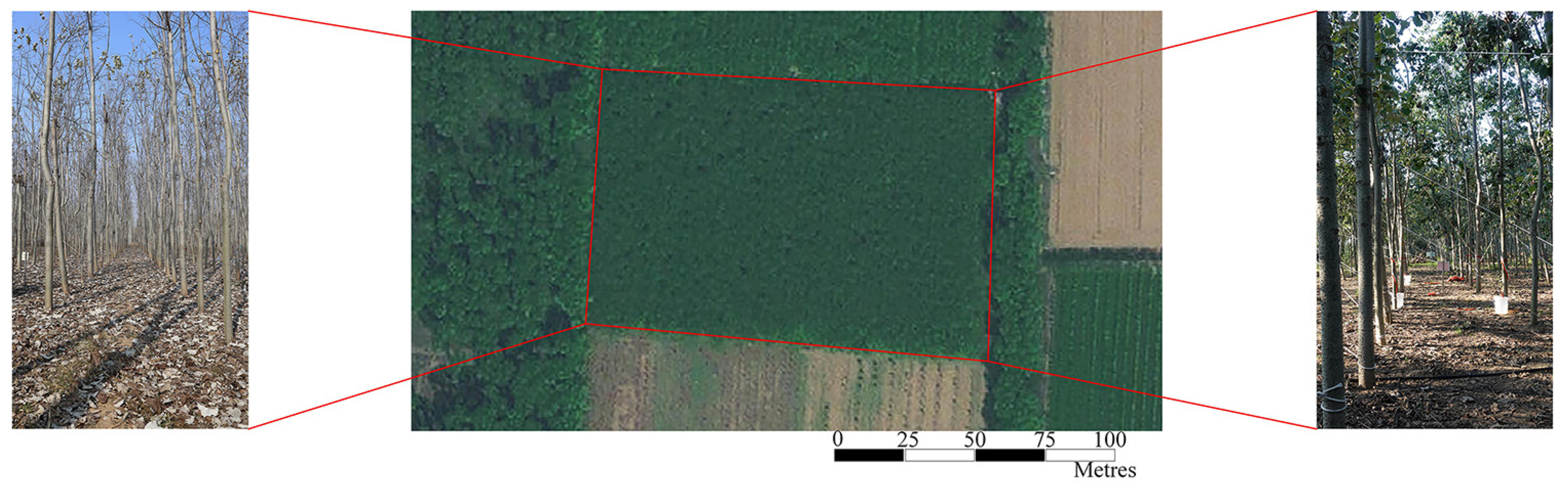


| Data Type | Sensing Method | Forest Type | Training Data | Training Data NT | Training Data Area | Validation Data | Validation Data NT | Validation Data Area | Labeling Time |
|---|---|---|---|---|---|---|---|---|---|
| Leafy data | Terrestrial Laser Scanner (RieglVZ-2000i) | Artificial plantation of Populus tomentosa Carr. | YT1 | 24 | 126.9 m2 | V1 | 23 | 129.6 m2 | 850 min |
| YT2 | 20 | 99.4 m2 | V2 | 20 | 114.4 m2 | ||||
| YT3 | 20 | 99.1 m2 | V3 | 20 | 117.7 m2 | ||||
| YT4 | 20 | 100.1 m2 | - | - | - | ||||
| YT5 | 20 | 108.6 m2 | - | - | - | ||||
| YT6 | 21 | 130.7 m2 | - | - | - | ||||
| YT7 | 20 | 132.2 m2 | - | - | - | ||||
| Leafless data | NT1 | 24 | 120.7 m2 | - | - | - | 105 min | ||
| NT2 | 20 | 97.8 m2 | - | - | - | ||||
| NT3 | 20 | 96.1 m2 | - | - | - | ||||
| NT4 | 19 | 99.7 m2 | - | - | - | ||||
| NT5 | 20 | 113.7 m2 | - | - | - | ||||
| NT6 | 20 | 98.4 m2 | - | - | - | ||||
| NT7 | 20 | 100.1 m2 |
| Data Name | Forest Type | NT | NC | NCET | Area |
|---|---|---|---|---|---|
| Testing data1 | Foliaged Populus tomentosa Carr. | 15 | 1,009,873 | 50,457.5 | 103.699 m2 |
| Testing data2 | 17 | 1,288,463 | 64,728.7 | 96.933 m2 | |
| Testing data3 | 21 | 1,497,158 | 59,627 | 131.647 m2 |
| Training Parameters | Value |
|---|---|
| Basic learning rate | 0.0002 |
| Batch study size | 8 |
| Block size | 50 |
| Epoch | 100 |
| Block point limit | 8200 |
| Testing Data | Training Data | Indicators | Terrain | Foliage | Stem | Other |
|---|---|---|---|---|---|---|
| T1 | YT2-YT7, NT1 | Precision | 0.982 | 0.891 | 0.715 | 0.853 |
| Recall | 0.922 | 0.834 | 0.834 | 0.744 | ||
| Weighted Precision | 0.854 | |||||
| Weighted Recall | 0.844 | |||||
| T2 | Precision | 0.988 | 0.947 | 0.709 | 0.792 | |
| Recall | 0.944 | 0.647 | 0.963 | 0.758 | ||
| Weighted Precision | 0.851 | |||||
| Weighted Recall | 0.816 | |||||
| T3 | Precision | 0.978 | 0.815 | 0.848 | 0.817 | |
| Recall | 0.954 | 0.913 | 0.713 | 0.805 | ||
| Weighted Precision | 0.847 | |||||
| Weighted Recall | 0.845 | |||||
| Training Data | Terrain Recall | Foliage Recall | Stem Recall | Others Recall | Overall Accuracy |
|---|---|---|---|---|---|
| YT1-YT7 | 0.970 | 0.804 | 0.801 | 0.817 | 0.825 |
| YT2-YT7, NT1 | 0.940 | 0.807 | 0.839 | 0.815 | 0.837 |
| YT3-YT7, NT1, NT2 | 0.902 | 0.711 | 0.847 | 0.764 | 0.789 |
| YT4-YT7, NT1-NT3 | 0.883 | 0.654 | 0.841 | 0.742 | 0.760 |
| YT5-YT7, NT1-NT4 | 0.867 | 0.538 | 0.838 | 0.691 | 0.697 |
| Study | Method | Terrain Recall | Foliage Recall | Stem Recall | Others Recall | Overall Accuracy |
|---|---|---|---|---|---|---|
| 4 | Unsupervised Learning | - | - | - | - | 0.888 |
| 8 | Supervised Learning | - | - | - | - | 0.876 |
| 15 | Modified Pointnet++ approach | 0.959 | 0.960 | 0.961 | 0.550 | 0.954 |
| 29 | Mode points evolution | 0.892 | ||||
| 30 | Voxel 3D-FCN | - | 0.971 * | 0.771 * | - | - |
| 0.975 ** | 0.642 ** | |||||
| Voxel 3D-FCN (r) | - | 0.975 * | 0.744 * | - | - | |
| 0.975 ** | 0.703 ** | |||||
| Pointnet | - | 0.976 * | 0.572 * | - | - | |
| 0.932 ** | 0.505 ** | |||||
| Pointnet (r) | - | 0.985 * | 0.727 * | - | - | |
| 0.896 ** | 0.573 ** | |||||
| Ours | PointCNN | 0.940 | 0.807 | 0.839 | 0.815 | 0.837 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, X.; Huang, Q.; Wang, X.; Xi, B. A Method for Extracting the Tree Feature Parameters of Populus tomentosa in the Leafy Stage. Forests 2023, 14, 1757. https://doi.org/10.3390/f14091757
Shen X, Huang Q, Wang X, Xi B. A Method for Extracting the Tree Feature Parameters of Populus tomentosa in the Leafy Stage. Forests. 2023; 14(9):1757. https://doi.org/10.3390/f14091757
Chicago/Turabian StyleShen, Xingyu, Qingqing Huang, Xin Wang, and Benye Xi. 2023. "A Method for Extracting the Tree Feature Parameters of Populus tomentosa in the Leafy Stage" Forests 14, no. 9: 1757. https://doi.org/10.3390/f14091757






