Multi-Omics Analyses Unravel Genetic Relationship of Chinese Coffee Germplasm Resources
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection, DNA Extraction, and RNA Extraction
2.2. SLAF-seq and SNP Analyses
2.3. Transcriptome Sequencing
2.4. Data Analysis
3. Results
3.1. SLAF Sequence
3.2. Transcriptome Sequence Assembly
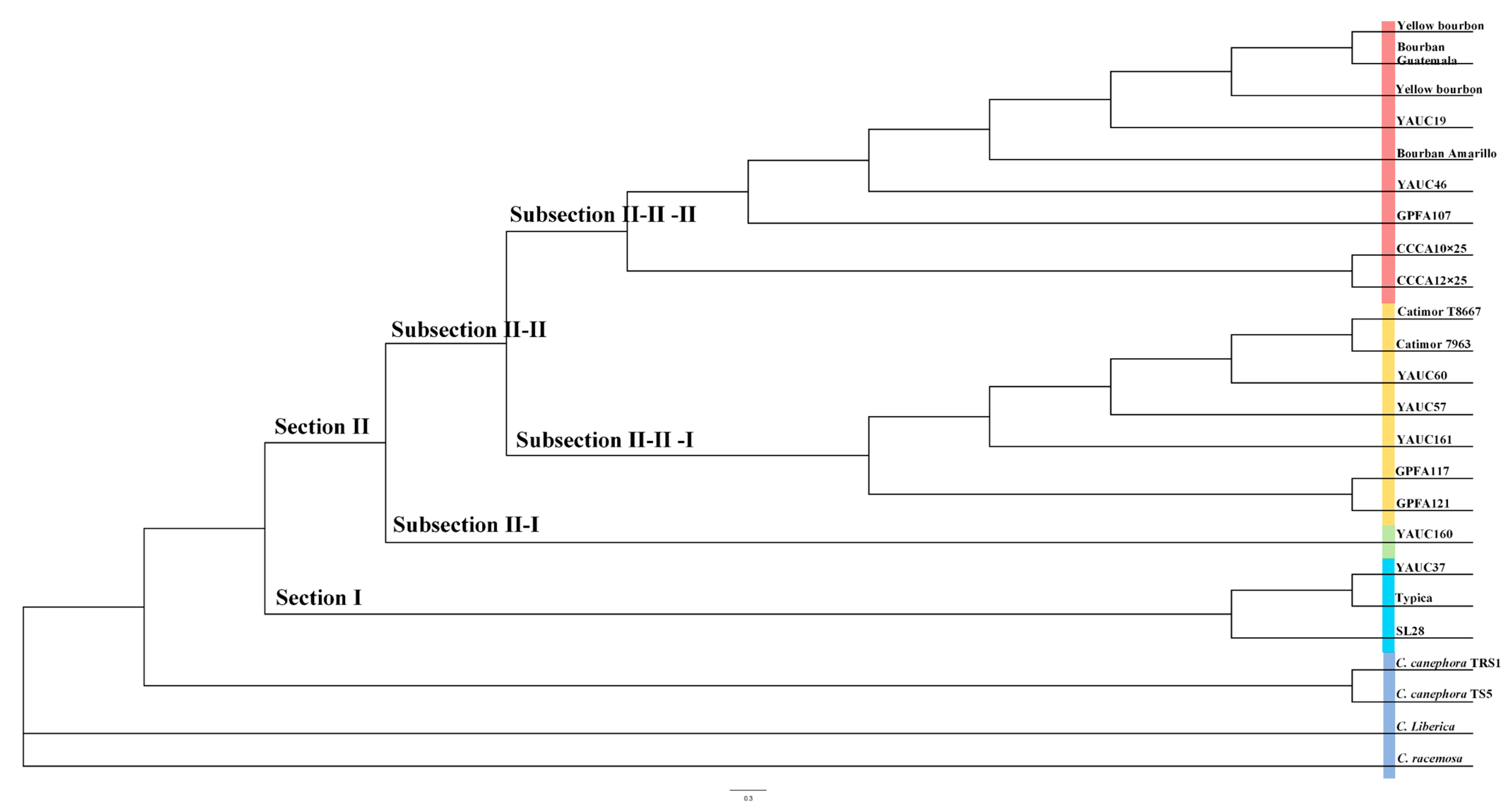
3.3. Phylogenetic Analysis
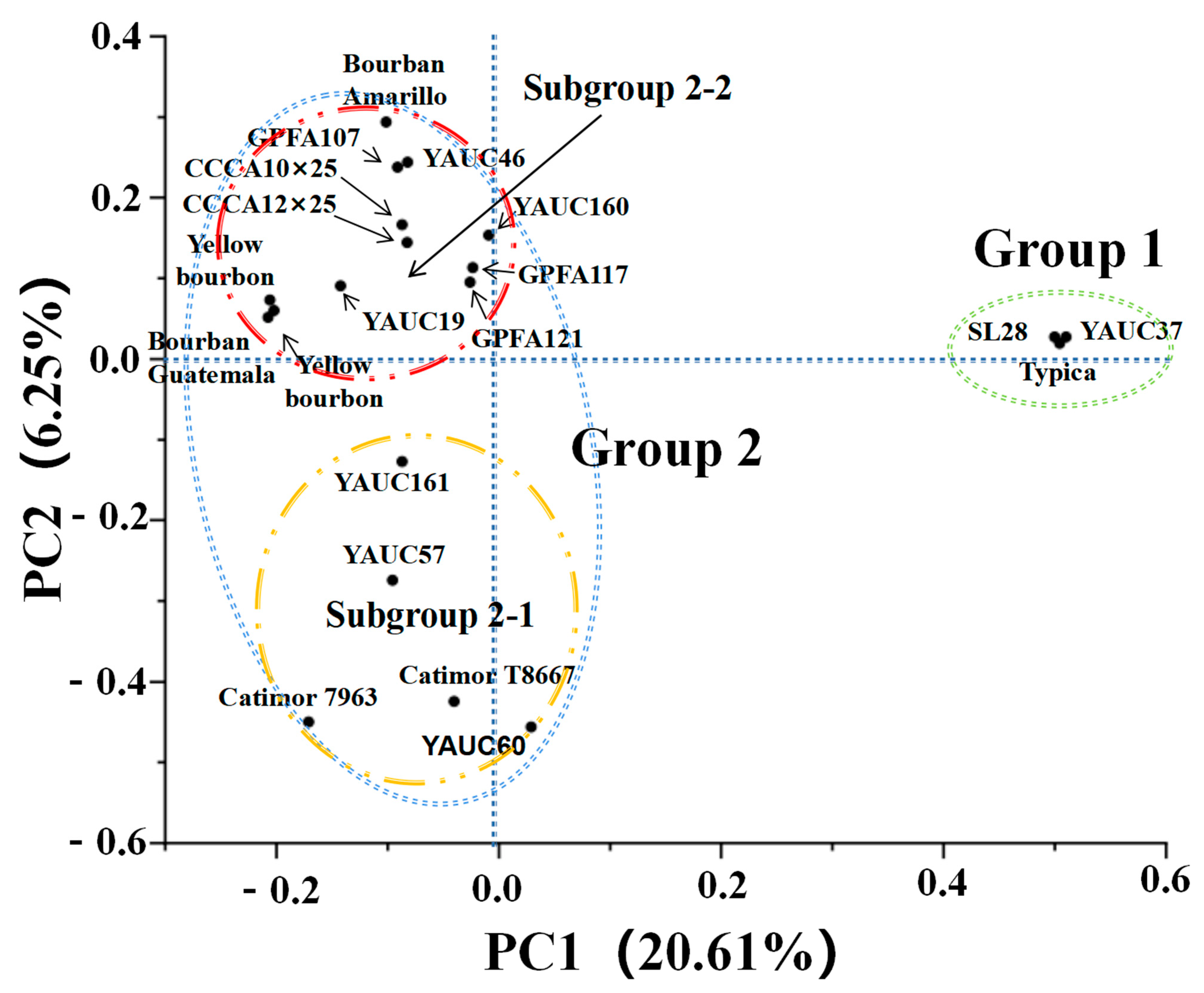
3.4. Principal Component Analysis
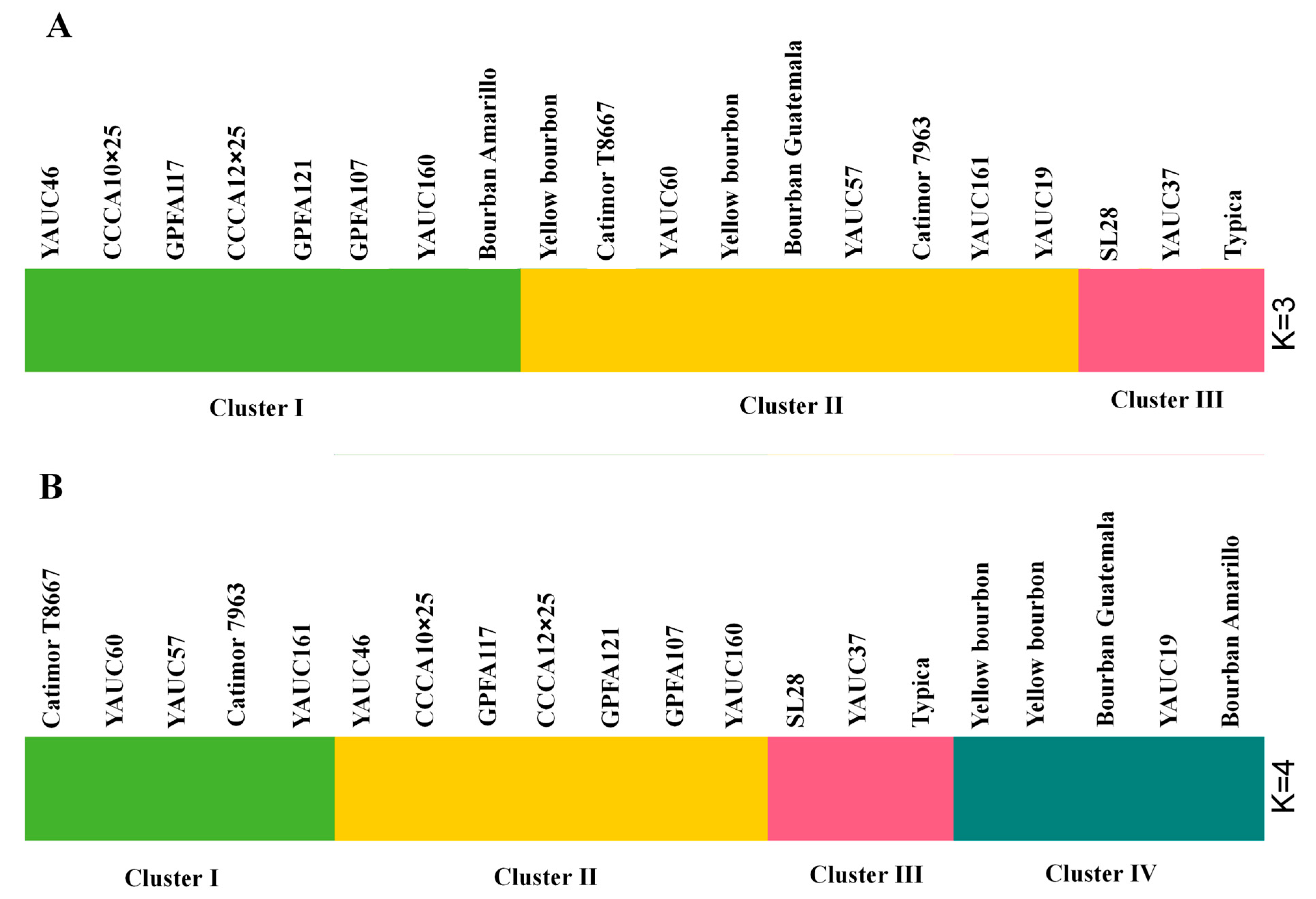
3.5. Genetic Structure Analysis
3.6. Correlation Analysis Using Transcriptome Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Denoeud, F.; Carretero-Paulet, L.; Dereeper, A.; Droc, G.; Guyot, R.; Pietrella, M.; Zheng, C.; Alberti, A.; Anthony, F.; Aprea, G.; et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 2014, 345, 1181–1184. [Google Scholar] [CrossRef]
- DaMatta, F.M.; Rahn, E.; Läderach, P.; Ghini, R.; Ramalho, J.C. Why could the coffee crop endure climate change and global warming to a greater extent than previously estimated? Clim. Chang. 2019, 152, 167–178. [Google Scholar] [CrossRef]
- International Coffee Organization. Coffee Statistics. Available online: http://www.ico.org/trade_statistics.asp (accessed on 10 October 2022).
- Tran, H.T.; Lee, L.S.; Furtado, A.; Smyth, H.; Henry, R.J. Advances in genomics for the improvement of quality in coffee. J. Sci. Food Agric. 2016, 96, 3300–3312. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.P.; Vega, F.E.; Solano, W.; Su, F.Y.; Infante, F.; Meinhardt, L.W. Selecting a core set of nuclear SNP markers for molecular characterization of Arabica cofee (Coffea arabica L.) genetic resources. Conserv. Genet. Resour. 2021, 13, 329–335. [Google Scholar] [CrossRef]
- Davis, A.P.; Tosh, J.; Ruch, N.; Fay, M.F. Growing coffee: Psilanthus (Rubiaceae) subsumed on the basis of molecular and morphological data; implications for the size, morphology, distribution and evolutionary history of Coffea. Bot. J. Linn. Soc. 2011, 167, 357–377. [Google Scholar] [CrossRef]
- International Coffee Organization (ICO). 2023. Available online: http://www.ico.org/prices/po-production.pdf (accessed on 18 August 2023).
- Wang, X.Y.; Zhou, H.; Chen, J.H.; Li, J.H.; Long, Y.Z.; Dong, Y.P. Genetic diversity of coffee germplasms by ISSR markers. Chin. J. Trop. Crops 2019, 40, 300–307. (In Chinese) [Google Scholar]
- Ge, Y.; Zhang, F.; Xie, C.; Qu, P.; Jiang, K.L.; Du, H.B.; Zhao, M.; Lu, Y.F.; Wang, B.T.; Shi, X.D.; et al. Effects of different altitudes on Coffea arabica rhizospheric soil chemical properties and soil microbiota. Agronomy 2023, 13, 471. [Google Scholar] [CrossRef]
- Depecker, J.; Verleysen, L.; Asimonyio, J.A.; Hatangi, Y.; Kambale, J.L.; Mwanga, I.M.; Ebele, T.; Dhed, B.; Bawin, Y.; Staelens, A.; et al. Genetic diversity and structure in wild Robusta coffee (Coffea canephora A. Froehner) populations in Yangambi (DR Congo) and their relation to forest disturbance. Heredity 2023, 130, 145–153. [Google Scholar] [CrossRef]
- Silva, B.S.R.D.; Santana, G.C.; Chaves, C.L.; Androcioli, L.G.; Ferreira, R.V.; Sera, G.H.; Charmetant, P.; Leroy, T.; Pot, D.; Domingues, D.S.; et al. Population structure and genetic relationships between Ethiopian and Brazilian Coffea arabica genotypes revealed by SSR markers. Genetica 2019, 147, 205–216. [Google Scholar] [CrossRef]
- Ogutu, C.; Fang, T.; Yan, L.; Wang, L.; Huang, L.F.; Wang, X.Y.; Ma, B.Q.; Deng, X.B.; Owiti, A.; Nyende, A.; et al. Characterization and utilization of microsatellites in the Coffea canephora genome to assess genetic association between wild species in Kenya and cultivated coffee. Tree Genet. Genomes 2016, 12, 54. [Google Scholar] [CrossRef]
- Montagnon, C.; Mahyoub, A.; Solano, W.; Sheibani, F. Unveiling a unique genetic diversity of cultivated Coffea arabica L. in its main domestication center: Yemen. Genet. Resour. Crop Evol. 2021, 68, 2411–2422. [Google Scholar] [CrossRef]
- Al-Ghamedi, K.; Alaraidh, I.; Afzal, M.; Mahdhi, M.; Al-Faifi, Z.; Oteef MD, Y.; Tounekti, T.; Alghamdi, S.S.; Khemira, H. Assessment of genetic diversity of local coffee populations in southwestern Saudi Arabia using SRAP markers. Agronomy 2023, 13, 302. [Google Scholar] [CrossRef]
- Steiger, D.; Nagai, C.; Moore, P.; Morden, C.; Osgood, R.; Ming, R. AFLP analysis of genetic diversity within and among Coffea arabica cultivars. Theor. Appl. Genet. 2002, 105, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Anthony, F.; Bertrand, B.; Quiros, O.; Wilches, A.; Lashermes, P.; Berthaud, J.; Charrier, A. Genetic diversity of wild coffee (Coffea arabica L.) using molecular markers. Euphytica 2001, 118, 53–65. [Google Scholar] [CrossRef]
- Spinoso-Castillo, J.L.; Escamilla-Prado, E.; Aguilar-Rinco, V.H.; Ramos, V.M.; de los Santos, G.G.; Perez-Rodrıguez, P.; Corona-Torres, T. Genetic diversity of coffee (Coffea spp.) in Mexico evaluated by using DArTseq and SNP markers. Genet. Resour. Crop Evol. 2020, 67, 1795–1806. [Google Scholar] [CrossRef]
- Zaidan, I.R.; Ferreira1, A.; Noia, L.R.; Santos, J.G.; de Arruda, V.C.; do Couto, D.P.; Braz1, R.A.; de Brites Senra, J.F.; Partelli, F.L.; Azevedo, C.F.; et al. Diversity and structure of Coffea canephora from old seminal crops in Espírito Santo, Brazil: Genetic resources for cofee breeding. Tree Genet. Genomes 2023, 19, 19. [Google Scholar] [CrossRef]
- Carvalho, H.F.; Galli, G.; Ferrão, L.F.V.; Nonato, J.V.A.; Padilha, L.; Maluf, M.P.; de Resende, M.F.R., Jr.; Filho, O.G.; Fritsche-Neto, R. The effect of bienniality on genomic prediction of yield in arabica coffee. Euphytica 2020, 216, 101. [Google Scholar] [CrossRef]
- Mekbib, Y.; Tesfaye, K.; Dong, X.; Saina, J.K.; Hu, G.W.; Wang, Q.F. Whole-genome resequencing of Coffea arabica L. (Rubiaceae) genotypes identify SNP and unravels distinct groups showing a strong geographical pattern. BMC Plant Biol. 2022, 22, 69. [Google Scholar] [CrossRef]
- Ge, Y.; Zhang, T.; Wu, B.; Tan, L.; Ma, F.N.; Zou, M.H.; Chen, H.H.; Pei, J.L.; Liu, Y.Z.; Chen, Z.H.; et al. Genome-wide assessment of avocado germplasm determined from specific length amplified fragment sequencing and transcriptomes: Population structure, genetic diversity, identification, and application of race-specific markers. Genes 2019, 3, 215. [Google Scholar] [CrossRef]
- Lyu, Y.Z.; Dong, X.Y.; Huang, L.B.; Zheng, J.W.; He, X.D.; Sun, H.N.; Jiang, Z.P. SLAF-seq uncovers the genetic diversity and adaptation of Chinese elm (Ulmus parvifolia) in eastern China. Forests 2020, 11, 80. [Google Scholar] [CrossRef]
- Li, C.X.; Liu, M.X.; Sun, F.J.; Zhao, X.Y.; He, M.Y.; Li, T.S.; Lu, P.; Xu, Y. Genetic divergence and population structure in weedy and cultivated broomcorn millets (Panicum miliaceum L.) revealed by specific-locus amplified fragment sequencing (SLAF-Seq). Front. Plant Sci. 2021, 12, 688444. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; He, Y.; Iqbal, Y.; Shi, Y.L.; Huang, H.M.; Yi, Z.L. Investigation of genetic relationships within three Miscanthus species using SNP markers identified with SLAF-seq. BMC Genom. 2022, 23, 43. [Google Scholar] [CrossRef]
- Iorizzo, M.; Senalik, D.A.; Grzebelus, D.; Bowman, M.; Cavagnaro, P.F.; Matvienko, M.; Ashrafi, H.; Deynze, A.V.; Simon, P.W. De novo assembly and characterization of the carrot transcriptome reveals novel genes, new markers, and genetic diversity. BMC Genom. 2011, 12, 389. [Google Scholar] [CrossRef] [PubMed]
- Lebot, V.; Melteras, M.; Pilecki, A.; Labouisse, J.-P. Chemometric evaluation of cocoa (Theobroma cacao L.) and coffee (Coffea spp.) germplasm using HPTLC. Genet. Resour. Crop Evol. 2020, 67, 895–911. [Google Scholar] [CrossRef]
- Nadaleti, D.H.S.; de RAbrahão, J.C.; Andrade, V.T.; Andrade, V.T.; Malta, M.R.; Botelho, C.E.; Carvalho, G.R. Sensory quality characterization and selection from a Coffea arabica germplasm collection in Brazil. Euphytica 2022, 218, 35. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeqIllumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- Ge, Y.; Ramchiary, N.; Wang, T.; Liang, C.; Wang, N.; Wang, Z.; Choi, S.R.; Lim, Y.P.; Piao, Z.Y. Development and linkage mapping of unigene-derived microsatellite markers in Brassica rapa L. Breed. Sci. 2011, 61, 160–167. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernysky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 2, 1297–1303. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Mortazavi, A.; Kwan, G.; Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. We checked. There’s no problem. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Hamon, P.; Grover, C.E.; Davis, A.P.; Rakotomalala, J.-J.; Raharimalala, N.E.; Albert, V.A.; Sreenath, H.L.; Stoffelen, P.; Mitchell, S.E.; Couturon, E.; et al. Genotyping-by-sequencing provides the first well-resolved phylogeny for coffee (Coffea) and insights into the evolution of caffeine content in its species GBS coffee phylogeny and the evolution of caffeine content. Mol. Phylogenetics Evol. 2017, 109, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Anagbogu, C.F.; Ilori, C.O.; Bhattacharjee, R.; Olaniyi, O.O.; Beckles, D.M. Gas chromatography-mass spectrometry and single nucleotide polymorphism-genotype-by-sequencing analyses reveal the bean chemical profiles and relatedness of Coffea canephora genotypes in Nigeria. Plants 2019, 8, 425. [Google Scholar] [CrossRef] [PubMed]
- Sant’Ana, G.G.; Pereira, L.F.P.; Pot, D.; Ivamoto, S.T.; Domingues, D.S.; Ferreira, R.V.; Pagiatto, N.F.; da Silva, B.S.R.; Nogueira, L.M.; Kitzberger, D.S.G.; et al. Genome-wide association study reveals candidate genes influencing lipids and diterpenes contents in Coffea arabica L. Sci. Rep. 2018, 8, 465. [Google Scholar] [CrossRef]
- Sousa, T.V.; Caixeta, E.T.; Alkimim, E.R.; Oliveira, A.C.B.; Pereira, A.A.; Sakiyama, N.S.; Zambolim, L.; Resende, M.D.V. Early selection enabled by the implementation of genomic selection in Coffea arabica breeding. Front. Plant Sci. 2019, 9, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Gimase, J.M.; Thagana, W.M.; Omondi, C.O.; Cheserek, J.J.; Gichimu, B.M.; Gichuru, E.K.; Ziyomo, C.; Sneller, C.H. Genome-wide association study identify the genetic loci conferring resistance to coffee berry disease (Colletotrichum kahawae) in Coffea arabica var. Rume Sudan. Euphytica 2020, 216, 86. [Google Scholar] [CrossRef]
- Ji, K.; Zhang, D.; Motilal, L.; Boccara, M.; Lachenaud, P.; Meinhardt, L.W. Genetic diversity and parentage in farmer varieties of cacao (Theobroma cacao L.) from Honduras and Nicaragua as revealed by single nucleotide polymorphism (SNP) markers. Genet. Resour. Crop Evol. 2013, 60, 441–453. [Google Scholar] [CrossRef]
- Wu, G.A.; Prochnik, S.; Jenkins, J.; Salse, J.; Hellsten, U.; Murat, F.; Perrier, X.; Ruiz, M.; Scalabrin, S.; Terol, J.; et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 2014, 32, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.P.; Meinhardt, L.W.; Tan, H.W.; Zhou, L.; Mischke, S.; Zhang, D.P. Varietal identification of tea (Camellia sinensis) using nanofluidic array of single nucleotide polymorphism (SNP) markers. Hortic. Res. 2014, 1, 14035. [Google Scholar] [CrossRef]
- Wang, B.Y.; Tan, H.W.; Fang, W.P.; Meinhardt, L.W.; Mischke, S.; Matsumoto, T.; Zhang, D.P. Developing single nucleotide polymorphism (SNP) markers from transcriptome sequences for identification of longan (Dimocarpus longan) germplasm. Hortic. Res. 2015, 2, 14065. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xiao, Z.D.; Bao, X.L.; Yang, X.Y.; Fang, J.; Xiang, X. Identifying litchi (Litchi chinensis Sonn.) cultivars and their genetic relationships using single nucleotide polymorphism (SNP) markers. PLoS ONE 2015, 10, e0135390. [Google Scholar] [CrossRef]

| Accession Number or Name | Type | Source of Introduction | Species | Fruit Color |
|---|---|---|---|---|
| Yellow Bourbon | C | Taqiri, Myanmar | C. arabica | Yellow |
| Bourbon Guatemala | C | La Aurora, Guatemala | C. arabica | Red |
| Yellow Bourbon | C | Taqiri, Myanmar | C. arabica | Yellow |
| YAUC19 | LS | Yunnan province, China | C. arabica | Red |
| Bourbon Amarillo | C | Campinas, Brasil | C. arabica | Red |
| YAUC46 | LS | Yunnan province, China | C. arabica | Red |
| GPFA107 | C | Nestle R & D Center, Tours, France | C. arabica | Red |
| CCCA10×25 | C | Nestle R & D Center, Tours, France | C. arabica | Red |
| CCCA12×25 | C | Nestle R & D Center, Tours, France | C. arabica | Red |
| Catimor T8667 | C | Coffee Research Center, Mandalay, Myanmar | C. arabica | Red |
| Catimor 7963 | C | Bang Mei Shu, Vietnam | C. arabica | Red |
| YAUC60 | LS | Coffee Research Center, Mandalay, Myanmar | C. arabica | Red |
| YAUC57 | LS | Coffee Research Center, Mandalay, Myanmar | C. arabica | Red |
| YAUC161 | LS | Yunnan province, China | C. arabica | Red |
| GPFA117 | C | Nestle R & D Center, Tours, France | C. arabica | Red |
| GPFA121 | C | Nestle R & D Center, Tours, France | C. arabica | Red |
| YAUC160 | LS | Yunnan province, China | C. arabica | Red |
| YAUC37 | LS | Yunnan province, China | C. arabica | Red |
| Typica | C | Scott Laboratory, Kenya | C. arabica | Red |
| SL28 | C | Scott Laboratory, Kenya | C. arabica | Red |
| C. canephora TRS1 | C | Bang Mei Shu, Vietnam | C. canephora | Red |
| C. canephora TS5 | C | Bang Mei Shu, Vietnam | C. canephora | Red |
| C. liberica | LS | Liberia | C. liberica | Red |
| C. racemosa | LS | Campinas, Brasil | C. racemosa | Red |
| Accessions | Total Reads | GC Percentage (%) | Q30 Percentage (%) | SLAF Numbers | Average Sequencing Depth/x | SNP Numbers | Integrity (%) | Heter Ratio (%) |
|---|---|---|---|---|---|---|---|---|
| Yellow Bourbon | 3,543,006 | 40.36 | 94.99 | 140,363 | 9.19 | 632,143 | 53.28 | 7.04 |
| Bourbon Guatemala | 4,707,446 | 39.87 | 95.32 | 135,758 | 13.19 | 628,483 | 52.97 | 6.83 |
| Yellow Bourbon | 7,210,310 | 38.87 | 96.10 | 154,717 | 16.53 | 782,887 | 65.99 | 8.25 |
| YAUC19 | 6,495,268 | 38.65 | 95.33 | 149,085 | 16.13 | 696,115 | 58.67 | 8.44 |
| Bourbon Amarillo | 6,062,294 | 38.69 | 96.31 | 125,176 | 17.63 | 597,475 | 50.36 | 7.49 |
| YAUC46 | 6,035,786 | 38.81 | 96.29 | 171,026 | 12.73 | 854,496 | 72.02 | 7.74 |
| GPFA107 | 6,215,380 | 39.46 | 96.18 | 159,690 | 14.32 | 748,077 | 63.05 | 10.04 |
| CCCA10×25 | 5,044,688 | 39.47 | 95.93 | 145,016 | 13.71 | 656,741 | 55.35 | 8.28 |
| CCCA12×25 | 4,626,678 | 39.51 | 96.08 | 155,561 | 11.04 | 719,958 | 60.68 | 8.26 |
| Catimor T8667 | 4,695,444 | 39.05 | 96.36 | 167,929 | 10.05 | 835,261 | 70.40 | 11.17 |
| Catimor 7963 | 4,491,620 | 40.91 | 96.12 | 144,938 | 10.79 | 693,483 | 58.45 | 8.21 |
| YAUC60 | 4,691,242 | 39.13 | 96.30 | 164,554 | 9.98 | 836,199 | 70.48 | 9.94 |
| YAUC57 | 5,159,478 | 39.45 | 95.91 | 140,285 | 14.04 | 661,919 | 55.79 | 7.87 |
| YAUC161 | 7,158,700 | 38.49 | 96.28 | 156,314 | 17.23 | 734,970 | 61.95 | 7.71 |
| GPFA117 | 5,229,644 | 39.60 | 96.10 | 148,262 | 13.72 | 694,779 | 58.56 | 10.60 |
| GPFA121 | 5,600,726 | 40.52 | 96.25 | 138,847 | 15.46 | 627,588 | 52.90 | 10.19 |
| YAUC160 | 6,147,466 | 39.39 | 95.51 | 159,478 | 14.58 | 773,980 | 65.24 | 9.29 |
| YAUC37 | 7,230,872 | 38.95 | 95.56 | 167,258 | 15.15 | 903,823 | 76.18 | 10.36 |
| Typica | 5,751,332 | 39.70 | 95.10 | 161,563 | 12.46 | 808,842 | 68.17 | 9.48 |
| SL28 | 4,473,964 | 39.45 | 96.27 | 157,103 | 10.43 | 779,764 | 65.72 | 8.89 |
| C. canephora TRS1 | 3,558,942 | 39.74 | 95.13 | 89,727 | 13.28 | 604,472 | 50.95 | 16.39 |
| C. canephora TS5 | 6,043,846 | 39.44 | 96.21 | 98,025 | 19.59 | 692,389 | 58.36 | 17.49 |
| C. liberica | 4,345,980 | 40.07 | 96.27 | 70,160 | 17.09 | 586,840 | 49.46 | 17.48 |
| C. racemosa | 2,665,424 | 39.48 | 95.69 | 46,234 | 15.18 | 381,571 | 32.16 | 13.89 |
| Accession | Clean Reads | Clean Bases (G) | GC Percentage (%) | Q30 Percentage (%) |
|---|---|---|---|---|
| Yellow Bourbon | 21,550,444 | 6.45 | 45.90 | 95.16 |
| Bourbon Guatemala | 24,555,127 | 7.35 | 44.64 | 95.08 |
| Yellow Bourbon | 23,196,887 | 6.94 | 45.43 | 95.26 |
| YAUC19 | 20,564,708 | 6.16 | 44.86 | 94.74 |
| Bourbon Amarillo | 19,484,349 | 5.83 | 44.58 | 94.92 |
| YAUC46 | 20,604,275 | 6.17 | 44.46 | 95.15 |
| GPFA107 | 24,411,515 | 7.31 | 44.84 | 95.20 |
| CCCA10×25 | 20,656,300 | 6.18 | 45.27 | 95.40 |
| CCCA12×25 | 20,636,047 | 6.17 | 44.55 | 94.98 |
| Catimor T8667 | 19,194,907 | 5.74 | 44.72 | 94.63 |
| Catimor 7963 | 21,505,386 | 6.44 | 45.59 | 95.26 |
| YAUC60 | 20,424,207 | 6.11 | 45.01 | 95.18 |
| YAUC57 | 20,698,969 | 6.19 | 45.09 | 94.86 |
| YAUC161 | 19,810,059 | 5.93 | 44.71 | 94.84 |
| GPFA117 | 21,505,386 | 6.44 | 45.59 | 95.26 |
| GPFA121 | 22,370,461 | 6.70 | 45.29 | 95.09 |
| YAUC160 | 21,662,528 | 6.48 | 44.60 | 94.63 |
| YAUC37 | 22,692,972 | 6.79 | 44.69 | 95.14 |
| Typica | 22,334,079 | 6.68 | 44.36 | 94.55 |
| SL28 | 21,543,184 | 6.45 | 51.09 | 95.28 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, Y.; Wang, B.; Shi, X.; Zhang, Z.; Qi, M.; Du, H.; Qu, P.; Jiang, K.; Chen, Z.; Li, X. Multi-Omics Analyses Unravel Genetic Relationship of Chinese Coffee Germplasm Resources. Forests 2024, 15, 163. https://doi.org/10.3390/f15010163
Ge Y, Wang B, Shi X, Zhang Z, Qi M, Du H, Qu P, Jiang K, Chen Z, Li X. Multi-Omics Analyses Unravel Genetic Relationship of Chinese Coffee Germplasm Resources. Forests. 2024; 15(1):163. https://doi.org/10.3390/f15010163
Chicago/Turabian StyleGe, Yu, Butian Wang, Xuedong Shi, Zhenwei Zhang, Meijun Qi, Huabo Du, Peng Qu, Kuaile Jiang, Zhihua Chen, and Xuejun Li. 2024. "Multi-Omics Analyses Unravel Genetic Relationship of Chinese Coffee Germplasm Resources" Forests 15, no. 1: 163. https://doi.org/10.3390/f15010163
APA StyleGe, Y., Wang, B., Shi, X., Zhang, Z., Qi, M., Du, H., Qu, P., Jiang, K., Chen, Z., & Li, X. (2024). Multi-Omics Analyses Unravel Genetic Relationship of Chinese Coffee Germplasm Resources. Forests, 15(1), 163. https://doi.org/10.3390/f15010163






