Genome-Wide Analysis of Homologous E6-AP Carboxyl-Terminal E3 Ubiquitin Ligase Gene Family in Populus trichocarpa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of HECT Genes in P. trichocarpa
2.2. Chromosomal Location
2.3. Phylogenetic Analysis and Sequence Analysis
2.4. Promoter cis-Element Analysis
2.5. Protein Interaction Network Analysis
2.6. Homology Model of HECT Genes
2.7. Tissue-Specific Expression Analyses
2.8. Plant Materials, Drought Stress, and Salt Treatment
2.9. RNA Extraction and Real-Time Quantitative PCR (qRT-PCR)
2.10. Statistical Analysis
3. Results
3.1. Identification of HECT Genes in P. trichocarpa
3.2. Chromosomal Locations
3.3. Sequence Alignment, Phylogenetic Analysis, and Domain Assessment
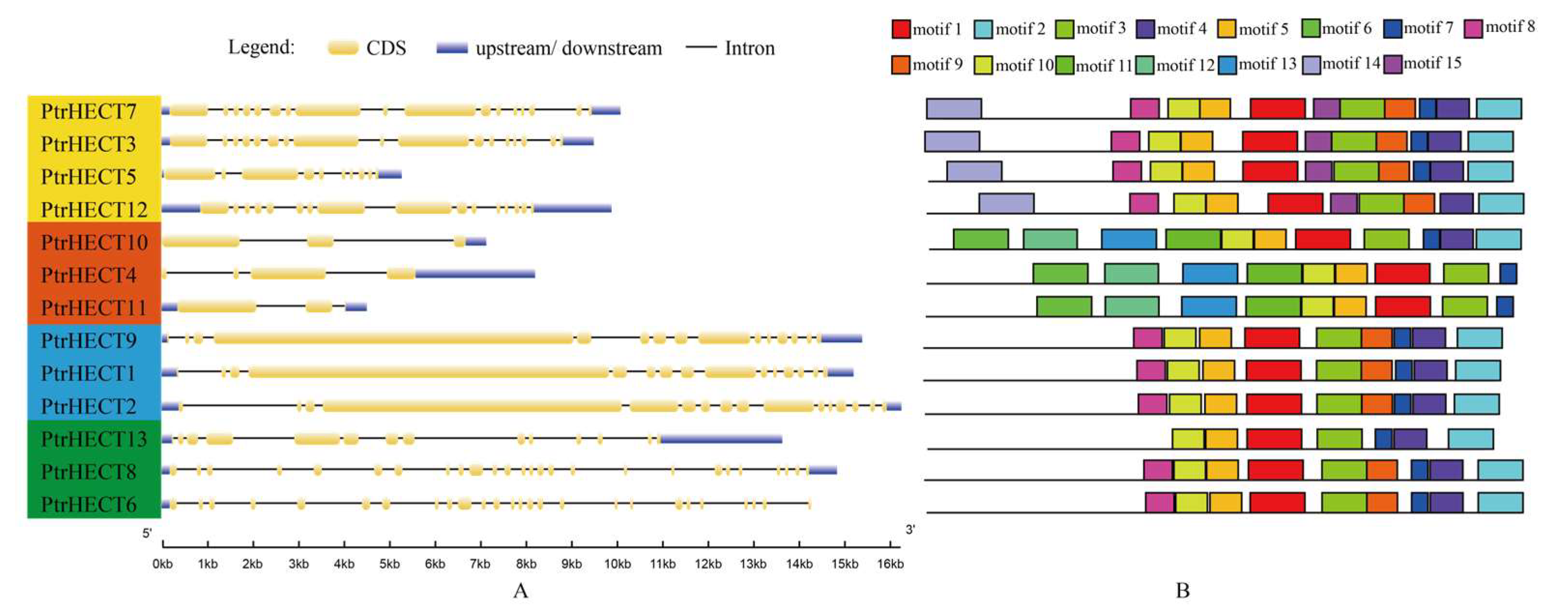
3.4. Gene Structure and Conserved Motif
3.5. Promoter cis-Element Analysis
3.6. Analysis of the HECT Protein Interaction Network in P. trichocarpa
3.7. Homology Modeling and Structure Validation of Modeled Proteins
3.8. Expression Profiles of HECT Genes in P. trichocarpa
3.9. Expression Pattern of PtrHECT Genes under Drought Stress
3.10. Expression Pattern of PtrHECT Genes under Salinity
4. Discussion
4.1. The HECT Genes Family in Populus Trichocarpa
4.2. The Promoter cis-Elements of PtrHECT
4.3. Transcript Profiles of HECT Genes under Drought Stress and Salinity
4.4. The Predicted Interaction Network of PtrHECT Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shu, K.; Yang, W. E3 Ubiquitin Ligases: Ubiquitous Actors in Plant Development and Abiotic Stress Responses. Plant Cell Physiol. 2017, 58, 1461–1476. [Google Scholar] [CrossRef] [PubMed]
- Mazzucotelli, E.; Belloni, S.; Marone, D.; De Leonardis, A.M.; Guerra, D.; Fonzo, N.; Cattivelli, L.; Mastrangelo, A.M. The E3 ubiquitin ligase gene family in plants: Regulation by degradation. Curr. Genom. 2006, 7, 509–522. [Google Scholar] [CrossRef] [PubMed]
- Lyzenga, W.J.; Stone, S.L. Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 2012, 63, 599–616. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Y.; Hellmann, H. Plant E3 Ligases: Flexible Enzymes in a Sessile World. Mol. Plant 2013, 6, 1388–1404. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Saxena, H.; Negi, H. Genome-wide analysis of HECT E3 ubiquitin ligase gene family in Solanum lycopersicum. Sci. Rep. 2021, 11, 15891. [Google Scholar] [CrossRef]
- Wang, Z.; Orosa-Puente, B.; Nomoto, M.; Grey, H.; Potuschak, T.; Matsuura, T.; Mori, I.C.; Tada, Y.; Genschik, P.; Spoel, S.H. Proteasome-associated ubiquitin ligase relays target plant hormone-specific transcriptional activators. Sci. Adv. 2022, 8, eabn4466. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L. The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front. Plant Sci. 2014, 5, 135. [Google Scholar] [CrossRef]
- Kim, H.T.; Kim, K.P.; Lledias, F.; Kisselev, A.F.; Scaglione, K.M.; Skowyra, D.; Gygi, S.P.; Goldberg, A.L. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 2007, 282, 17375–17386. [Google Scholar] [CrossRef]
- Wang, M.; Pickart, C.M. Different HECT domain ubiquitin ligases employ distinct mechanisms of polyubiquitin chain synthesis. Embo J. 2005, 24, 4324–4333. [Google Scholar] [CrossRef]
- Scheffner, M.; Nuber, U.; Huibregtse, J.M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 1995, 373, 81–83. [Google Scholar] [CrossRef]
- Guzmán, P. ATLs and BTLs, plant-specific and general eukaryotic structurally related E3 ubiquitin ligases. Plant Sci. 2014, 215, 69–75. [Google Scholar] [CrossRef]
- Duplan, V.; Rivas, S. E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front. Plant Sci. 2014, 5, 42. [Google Scholar] [CrossRef] [PubMed]
- Yee, D.; Goring, D.R. The diversity of plant U-box E3 ubiquitin ligases: From upstream activators to downstream target substrates. J. Exp. Bot. 2009, 60, 1109–1121. [Google Scholar] [CrossRef]
- Wang, Z.; Spoel, S.H. HECT ubiquitin ligases as accessory proteins of the plant proteasome. Essays Biochem. 2022, 66, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Cheng, D.M.; Peng, J.M.; Pickart, C.M. Molecular determinants of polyubiquitin linkage selection by an HECT ubiquitin ligase. Embo J. 2006, 25, 1710–1719. [Google Scholar] [CrossRef] [PubMed]
- Marín, I. Evolution of Plant HECT Ubiquitin Ligases. PLoS ONE 2013, 8, e68536. [Google Scholar] [CrossRef] [PubMed]
- Huibregtse, J.M.; Scheffner, M.; Beaudenon, S.; Howley, P.M. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 1995, 92, 2563–2567. [Google Scholar] [CrossRef] [PubMed]
- Furniss, J.J.; Grey, H.; Wang, Z.; Nomoto, M.; Jackson, L.; Tada, Y.; Spoel, S.H. Proteasome-associated HECT-type ubiquitin ligase activity is required for plant immunity. PLoS Pathog. 2018, 14, e1007447. [Google Scholar] [CrossRef] [PubMed]
- Grau-Bové, X.; Sebé-Pedrós, A.; Ruiz-Trillo, I. A Genomic Survey of HECT Ubiquitin Ligases in Eukaryotes Reveals Independent Expansions of the HECT System in Several Lineages. Genome Biol. Evol. 2013, 5, 833–847. [Google Scholar] [CrossRef]
- Downes, B.P.; Stupar, R.M.; Gingerich, D.J.; Vierstra, R.D. The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis:: UPL3 has a specific role in trichome development. Plant J. 2003, 35, 729–742. [Google Scholar] [CrossRef]
- Lan, W.; Qiu, S.; Ren, Y.; Miao, Y. Expression Profile and Function of HECT E3 sin Arabidopsis thaliana. Acta Bot. Boreali-Occident. Sin. 2017, 37, 2112–2119. [Google Scholar]
- Patra, B.; Pattanaik, S.; Yuan, L. Ubiquitin protein ligase 3 mediates the proteasomal degradation of GLABROUS 3 and ENHANCER OF GLABROUS 3, regulators of trichome development and flavonoid biosynthesis in Arabidopsis. Plant J. 2013, 74, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Lan, W.; Ma, W.B.; Zheng, S.; Qiu, Y.H.; Zhang, H.; Lu, H.S.; Zhang, Y.; Miao, Y. Ubiquitome profiling reveals a regulatory pattern of UPL3 with UBP12 on metabolic-leaf senescence. Life Sci. Alliance 2022, 5, e202201492. [Google Scholar] [CrossRef] [PubMed]
- El Refy, A.; Perazza, D.; Zekraoui, L.; Valay, J.G.; Bechtold, N.; Brown, S.; Hülskamp, M.; Herzog, M.; Bonneville, J.M. The Arabidopsis KAKTUS gene encodes a HECT protein and controls the number of endoreduplication cycles. Mol. Genet. Genom. 2004, 270, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.W.; Yang, T.; Liu, J.; Zhao, M.D.; Wang, J.L. Genome-wide identification and evolution of HECT genes in wheat. PeerJ 2020, 8, e10457. [Google Scholar] [CrossRef]
- Lan, W.; Miao, Y. New Aspects of HECT-E3 Ligases in Cell Senescence and Cell Death of Plants. Plants 2019, 8, 483. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Wang, C.; Rahman, S.U.; Wang, Y.; Wang, A.; Tao, S. Genome-Wide Identification and Evolution of HECT Genes in Soybean. Int. J. Mol. Sci. 2015, 16, 8517–8535. [Google Scholar] [CrossRef]
- Alam, I.; Cui, D.-L.; Batool, K.; Yang, Y.-Q.; Lu, Y.-H. Comprehensive Genomic Survey, Characterization and Expression Analysis of the HECT Gene Family in Brassica rapa L. and Brassica oleracea L. Genes 2019, 10, 400. [Google Scholar] [CrossRef]
- Miller, C.; Wells, R.; McKenzie, N.; Trick, M.; Ball, J.; Fatihi, A.; Dubreucq, B.; Chardot, T.; Lepiniec, L.; Bevan, M.W. Variation in Expression of the HECT E3 Ligase UPL3 Modulates LEC2 Levels, Seed Size, and Crop Yields in Brassica napus. Plant Cell 2019, 31, 2370–2385. [Google Scholar] [CrossRef]
- Xu, J.; Xing, S.; Cui, H.; Chen, X.; Wang, X. Genome-wide identification and characterization of the apple (Malus domestica) HECT ubiquitin-protein ligase family and expression analysis of their responsiveness to abiotic stresses. Mol. Genet. Genom. 2016, 291, 635–646. [Google Scholar] [CrossRef]
- Tuskan, G.A.; DiFazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Leng, X.; Yang, J.; Zhang, M.; Zeng, M.; Xu, X.; Wang, F.; Li, C. Comprehensive analysis of AHL gene family and their expression under drought stress and ABA treatment in Populus trichocarpa. PeerJ 2021, 9, e10932. [Google Scholar] [CrossRef] [PubMed]
- Tao, R.; Liu, Y.Q.; Jing, W.P. Response and Regulatory Network Analysis of Roots and Stems to Abiotic Stress in Populus trichocarpa. Forests 2022, 13, 1300. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.Q.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Benkert, P.; Künzli, M.; Schwede, T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009, 37, W510–W514. [Google Scholar] [CrossRef]
- Borrill, P.; Ramirez-Gonzalez, R.; Uauy, C. expVIP: A Customizable RNA-seq Data Analysis and Visualization Platform. Plant Physiol. 2016, 170, 2172–2186. [Google Scholar] [CrossRef]
- Phillips, G.C.; Garda, M. Plant tissue culture media and practices: An overview. In Vitro Cell. Dev. Biol. Plant 2019, 55, 242–257. [Google Scholar] [CrossRef]
- Li, Q.; Yeh, T.-F.; Yang, C.; Song, J.; Chen, Z.-Z.; Sederoff, R.R.; Chiang, V.L. Populus trichocarpa. Methods Mol. Biol. 2015, 1224, 357–363. [Google Scholar] [CrossRef]
- Huang, L.; Hua, K.; Xu, R.; Zeng, D.; Wang, R.; Dong, G.; Zhang, G.; Lu, X.; Fang, N.; Wang, D.; et al. The LARGE2-APO1/APO2 regulatory module controls panicle size and grain number in rice. Plant Cell 2021, 33, 1212–1228. [Google Scholar] [CrossRef]
- Li, Y.; Zhai, L.; Fan, J.; Ren, J.; Gong, W.; Wang, X.; Huang, J. Genome-wide identification, phylogenetic and expression analysis of the maize HECT E3 ubiquitin ligase genes. Genetica 2019, 147, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Janda, T.; Szalai, G.; Pál, M. Salicylic Acid Signalling in Plants. Int. J. Mol. Sci. 2020, 21, 2655. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.; Ding, Y. Stories of Salicylic Acid: A Plant Defense Hormone. Trends Plant Sci. 2020, 25, 549–565. [Google Scholar] [CrossRef] [PubMed]
- Malcheska, F.; Ahmad, A.; Batool, S.; Müller, H.M.; Ludwig-Mueller, J.; Kreuzwieser, J.; Randewig, D.; Hänsch, R.; Mendel, R.R.; Hell, R.; et al. Drought-Enhanced Xylem Sap Sulfate Closes Stomata by Affecting ALMT12 and Guard Cell ABA Synthesis. Plant Physiol. 2017, 174, 798–814. [Google Scholar] [CrossRef]
- Yoshida, T.; Fernie, A.R. Hormonal regulation of plant primary metabolism under drought. J. Exp. Bot. 2023, erad358. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Z.; Leng, X.; Xu, X.M.; Li, C.H. Comprehensive Analysis of the TIFY Gene Family and Its Expression Profiles under Phytohormone Treatment and Abiotic Stresses in Roots of Populus trichocarpa. Forests 2020, 11, 315. [Google Scholar] [CrossRef]
- Wang, X.; Gomes, M.M.; Bailly, C.; Nambara, E.; Corbineau, F. Role of ethylene and proteolytic N-degron pathway in the regulation of Arabidopsis seed dormancy. J. Integr. Plant Biol. 2021, 63, 2110–2122. [Google Scholar] [CrossRef]
- Miao, Y.; Zentgraf, U. A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J. 2010, 63, 179–188. [Google Scholar] [CrossRef]









| Gene Name | Gene ID | Protein Length (a.a.) | Molecular Weight (Da) | Theoretical pI | GRAVY | The Predicted Location of PtrHECT Proteins |
|---|---|---|---|---|---|---|
| PtrHECT1 | Potri.001G368600 | 3756 | 411,293.51 | 4.95 | −0.213 | Nuclear |
| PtrHECT2 | Potri.002G110500 | 3667 | 405,141.71 | 5.1 | −0.281 | Plas |
| PtrHECT3 | Potri.004G174700 | 1877 | 201,786.1 | 5.97 | −0.308 | Nuclear |
| PtrHECT4 | Potri.006G011700 | 840 | 953,69.61 | 6.34 | −0.129 | Nuclear |
| PtrHECT5 | Potri.006G132000 | 1074 | 119,539.13 | 5.15 | −0.164 | Nuclear |
| PtrHECT6 | Potri.008G101300 | 1033 | 117,999.93 | 6.75 | −0.161 | Cytoplasmic |
| PtrHECT7 | Potri.009G134300 | 1895 | 203,638.89 | 5.62 | −0.315 | Nuclear |
| PtrHECT8 | Potri.010G150000 | 1032 | 118,051.54 | 6.41 | −0.178 | Cytoplasmic |
| PtrHECT9 | Potri.011G094100 | 3749 | 411,209.5 | 4.96 | −0.214 | Nuclear |
| PtrHECT10 | Potri.016G012900 | 853 | 97,311.88 | 6.24 | −0.18 | Nuclear |
| PtrHECT11 | Potri.016G059800 | 782 | 88,833.73 | 5.95 | −0.192 | Nuclear |
| PtrHECT12 | Potri.016G085200 | 1512 | 169,298.31 | 5.31 | −0.11 | E.R._plas |
| PtrHECT13 | Potri.016G096500 | 1173 | 132,025.06 | 7.88 | −0.105 | Chloroplast |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Y.; Li, H.; Li, Y.; Zhao, H.; Yang, D.; Chen, A.; Yang, J. Genome-Wide Analysis of Homologous E6-AP Carboxyl-Terminal E3 Ubiquitin Ligase Gene Family in Populus trichocarpa. Forests 2024, 15, 169. https://doi.org/10.3390/f15010169
Fu Y, Li H, Li Y, Zhao H, Yang D, Chen A, Yang J. Genome-Wide Analysis of Homologous E6-AP Carboxyl-Terminal E3 Ubiquitin Ligase Gene Family in Populus trichocarpa. Forests. 2024; 15(1):169. https://doi.org/10.3390/f15010169
Chicago/Turabian StyleFu, Yanrui, Hui Li, Yaqi Li, Haoqin Zhao, Da Yang, Aihua Chen, and Jingli Yang. 2024. "Genome-Wide Analysis of Homologous E6-AP Carboxyl-Terminal E3 Ubiquitin Ligase Gene Family in Populus trichocarpa" Forests 15, no. 1: 169. https://doi.org/10.3390/f15010169





