1. Introduction
The walnut genus (
Juglans L.) consists of approximately 22 species [
1], with species native to China [
2], namely
J. regia L.,
J. mandshurica Maxium,
J. cathayensis Dode,
J. sigillata Dode, and
J. hopeiensis Hu. Among them, ordinary walnuts are the most widely cultivated species both domestically and internationally and are known as one of four major nuts [
3]. According to the records of leaf fossils and carbonized nuts from the
14C era, ordinary walnuts have a history of 7335 ± 100 years in the Shandong and Hebei provinces of China [
4]. The production of walnuts (including iron walnuts) in China reached approximately 5.4035 million tons in 2021, firmly ranking first in the world [
5]. China, one of the centers for genetic diversity of walnuts, provides important germplasm resources for the cultivation of new walnut varieties both domestically and internationally [
6]. According to the “Records of Chinese Fruit Trees-Walnut Volume”, Chinese farmers and breeders have cultivated over 380 walnut varieties (germplasm resources) through hybridization and other means. The southwest region is the main distribution center of walnut germplasm resources in China, and it has been recognized as having a high level of genetic diversity [
4,
6]. Therefore, exploring scientific and efficient identification methods has positive guiding significance for the identification, protection, and utilization of walnut germplasm resources.
The traditional methods for identifying walnut varieties mainly rely on morphological detection based on appearance features [
7]. However, this method is not only time-consuming and labor-intensive, but also influenced by the external environment. With the advancement of science and technology, research methods represented by molecular markers have been widely applied in the field of walnut variety identification [
8,
9,
10]. At present, molecular marker techniques mainly include restriction fragment length polymorphism (RFLP) [
11], random amplified polymorphism DNA (RAPD) [
12], amplified fragment length polymorphism (AFLP) [
13,
14], simple sequence repeat (SSR) [
15], and single nucleotide polymorphism (SNP) [
16]. Among them, SSR is also called microsatellite DNA, and the core sequence of its tandem repeat is usually 1–6 bp [
17]. Because of its codominance inheritance, locus specificity, and high polymorphism [
18], it has been widely used in the study of genetic diversity and evolutionary relationships of walnuts. The International Union for the Protection of New Varieties of Plants (UPOV) recommends using SSR and SNP markers as the preferred methods for constructing plant DNA fingerprints [
19]. In addition, capillary electrophoresis has gradually replaced traditional polyacrylamide gel electrophoresis (PAGE) due to its advantages of rapidity and automation, and has become the mainstream technology for separating and quantifying PCR products [
20].
In recent years, many scholars have used SSR markers to study the genetic diversity, population genetic structure, and evolutionary relationship of many walnut germplasm resources and progeny [
6,
15,
21]. At present, the approaches to developing SSR markers include searching EST sequences in public databases such as GenBank [
22], genome re-sequencing [
23], transcriptome sequencing [
24], and chloroplast genome sequencing [
25]. Doğan et al. used 25 pairs of RAPD primers, 25 pairs of ISSR primers, and 16 pairs of SSR primers to identify 59 foreign and Turkish walnut varieties. The polymorphism rate of SSR was 99.1%, much higher than those of RAPD (69.1%) and ISSR (71.1%) [
26]. Magige et al. used 31 SSR markers to analyze the genetic structure of 12 walnut populations [
9]. Jin et al. used WJR265, WGA331, and WJR031 (or WJR281, WGA321, WGA032) to fully distinguish 21 walnut materials under test [
27]. Davoodi et al. used WGA001, WGA009, and WGA276 to completely separate 21 high-quality walnut varieties from Iran [
10]. With the continuous increase of new walnut germplasm resources, there is an urgent need to develop highly polymorphic SSR markers to quickly and efficiently distinguish the increasing number of germplasm resources. It is imperative to use well-characterized SSR markers to construct DNA molecular fingerprints. This not only has guiding significance for the rapid identification of walnut resources and the establishment of walnut DNA fingerprint databases, but also contributes to the screening of walnut core germplasm and the development of germplasm innovation work.
Based on the genome re-sequencing results of Baokexiang, this study used eight different walnut samples as materials and selected 14 polymorphic SSR markers from 60 pairs of primers. These SSR markers were used to conduct molecular identification of 47 walnut germplasm via capillary electrophoresis detection, analyzing the genetic diversity, genetic relationship, population genetic structure, and principal coordinate analysis among different germplasm. This study seeks to achieve several goals, including molecularly identifying walnut germplasm resources, examining genetic diversity and population structure, creating a fingerprint map, and conducting association analysis between SSR markers and walnut traits. Overall, this study will provide a reference for future breeding of walnut germplasm with excellent traits and lay a theoretical foundation for screening walnut resources with excellent traits.
3. Results
3.1. Distribution Characteristics of SSR Loci in Baokexiang
Based on the genomic re-sequencing results of Baokexiang (
Table 3 and
Table 4), a total of 3,509,660 sequences with a total length of 529,208,302 bp were detected, and a total of 943,186 SSR loci were screened. Among them, dinucleotide repeat locus accounted for the largest proportion (350,229, 37.13%), followed by single nucleotide repeat (26.06%), tetranucleotide repeat (18.86%), trinucleotide repeat (12.54%), pentanucleotide repeat (3.40%), and hexanucleotide repeat (2.01%), respectively (
Table S2). In dinucleotide repeats, the majority (66,349, 18.94%) of SSR loci repeated six times. As the number of repeats increased, the frequency of distributed repeat bases gradually decreased, and similar phenomena were also observed in other repeat base types. Analysis of base repeat types (
Table S2) showed that A/T base types accounted for the highest proportion (233,978, 24.81%) in the microsatellite sites of Baokexiang, followed by AA/TT (139,372, 14.78%), AT/AT (118,186, 12.53%), and AAA/TTT (87,664, 9.29%).
3.2. Development and Genetic Diversity Analysis of SSR Primers in Walnuts
By conducting PCR pre-experiments on eight walnut varieties, including Baokexiang, Jinghong 1, Jinghong 2, Black walnut, Jingyi 2, Xiaoxiaoqiu, Yangbipao, and Changshan 99-11, 60 pairs of primers were designed to amplify bands, accounting for 100% of the total. However, only 23 pairs of primers amplified bands without primer dimers, accounting for 38.33%. The amplification results of SSR primers within walnut species, black walnut, and hickory showed that these 23 pairs of SSR primers could amplify the target product bands within walnut species, black walnut, and hickory, accounting for 100%. That was to say, these 23 pairs of SSR primers had good generality. However, the polymorphism of Jr18, Jr19, Jr24, Jr25, and Jr51 was poor and would not be considered in subsequent analysis. Therefore, the remaining 18 pairs of SSR primers with good specificity and clear bands were selected for capillary electrophoresis detection. The genetic diversity of walnut germplasm is shown in
Table 5. The variation range of Na in experimental samples with 18 pairs of polymorphic primers ranged from 2 to 6, with an average value of 3.556. The variation range of Ne was 1.882~4.741, with an average value of 2.611. The variation range of MAF was 0.250~0.688, with an average value of 0.538. The variation range of Ho ranged from 0 to 0.875, with an average value of 0.167. The variation range of He ranged from 0.469 to 0.789, with an average value of 0.590. The variation range of uHe was 0.500~0.842, with an average value of 0.629. The variation range of PIC was 0.359~0.757, with an average value of 0.526. The variation range of I ranged from 0.662 to 1.646, with an average value of 1.048. The variation range of GD was 0.456~0.789, with an average value of 0.590. The variation range of Fst ranged from −0.043 to 1.000, with an average value of 0.762. However, Jr09, Jr22, and Jr48 amplified more hetero-bands besides the target site, which were not suitable for constructing the fingerprint of 47 walnut resources. In addition, Jr60 was not considered in the construction of 47 walnut resource fingerprints due to the low MAF value. Other SSR markers had rich polymorphism, and eight experimental samples tested had high heterozygosity, which was more suitable for molecular markers in subsequent walnut populations.
3.3. Cluster Analysis
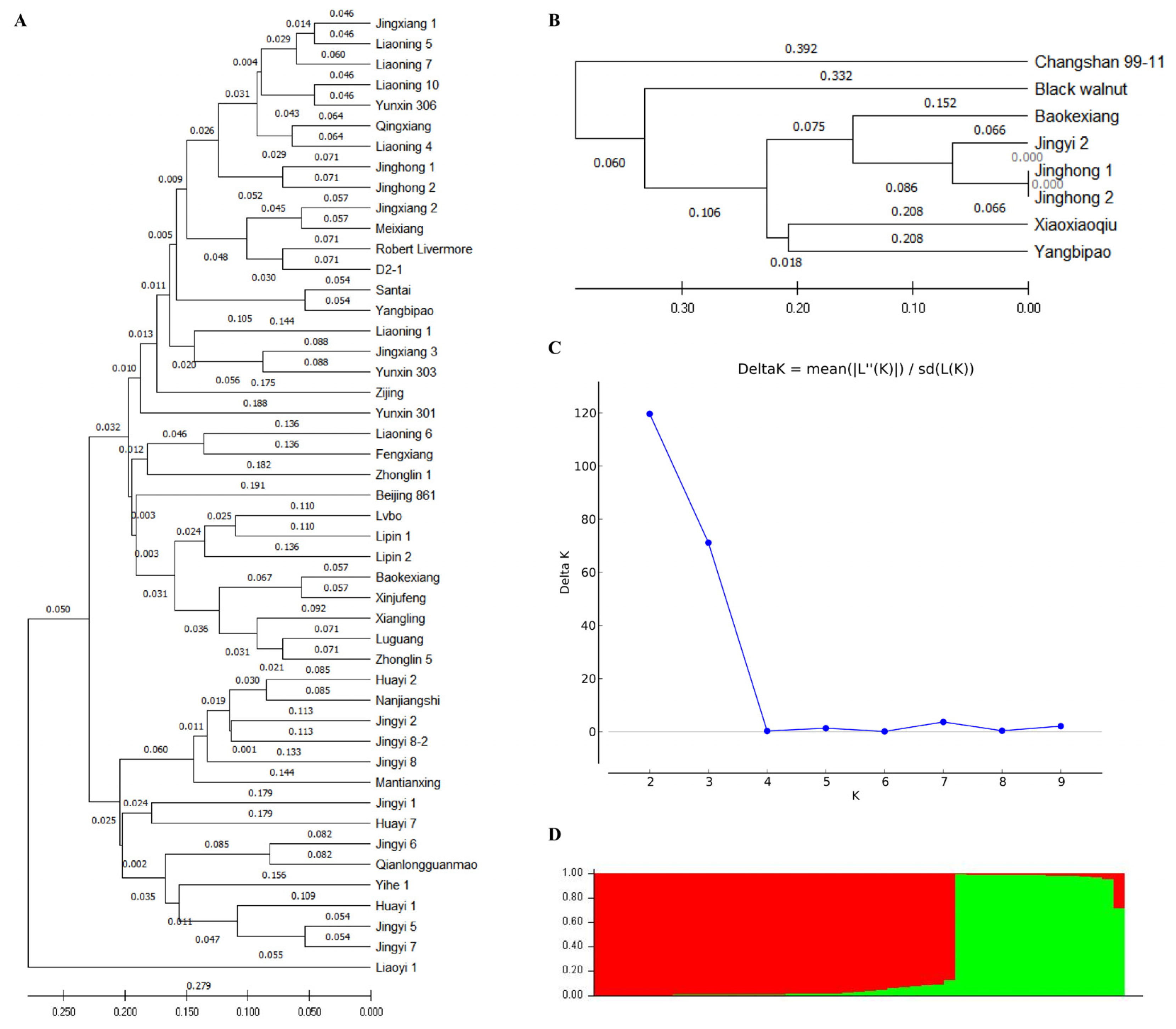
Based on the amplification of 18 SSR markers to eight walnut varieties, the genetic distance between eight samples was calculated. The results showed that Jr05 + Jr29 + Jr35 + Jr50 + Jr55 + Jr60 could effectively classify these eight walnut varieties (
Figure 1B). The genetic distance between Changshan 99-11 and the other seven samples of walnuts was large, which meant these could be separated. Within the walnut genus, black walnut had a distant genetic relationship with other samples, so it was clustered on different branches from the other six samples. In addition, both Jinghong 1 and Jinghong 2 were descendants of
J. regia ‘Robert Livermore’. Jingyi 2 was a hybrid offspring of
J. regia and
J. mandshurica. The above three belonged to the ecological type of Beijing walnut. Baokexiang was the offspring of Xinjiang’s early fruiting walnut, but these four samples were all of the northern walnut ecological type. Therefore, Jinghong 1, Jinghong 2, and Jingyi 2 first gathered together and then formed a group with Baokexiang.
3.4. Genetic Diversity
Capillary electrophoresis was performed on 47 walnut resources, and a total of 64 allele loci were obtained from 14 SSR markers (
Table 6). The variation range of Na ranged from 2 (Jr52) to 9 (Jr45), with an average of 4.571. The variation range of MAF ranged from 0.468 (Jr15) to 0.947 (Jr52), with an average of 0.687. The variation range of Ho ranged from 0 (Jr05, Jr11, and Jr12) to 0.681 (Jr55), with an average of 0.202. The variation range of He was 0.101 (Jr52) to 0.731 (Jr45), with an average of 0.456. The variation range of uHe ranged from 0.102 (Jr52) to 0.739 (Jr45), with an average of 0.461. Among them, He of all SSR markers was higher than Ho. The variation range of GD ranged from 0.101 (Jr52) to 0.731 (Jr45), with an average value of 0.458. The variation range of I ranged from 0.208 (Jr52) to 1.723 (Jr45), with an average value of 0.897. The variation range of PIC ranged from 0.096 (Jr52) to 0.711 (Jr45), with an average value of 0.422. Among them, there were 2 SSR markers with low polymorphism (<0.25), 6 SSR markers with moderate polymorphism (0.25~0.5), and 6 SSR markers with high polymorphism (>0.5). The variation range of Fst ranged from −0.312 (Jr53) to 1.000 (Jr05, Jr11, Jr12, and Jr50), with an average value of 0.581. In summary, the 14 selected SSR markers exhibited rich polymorphism, and the tested walnut resources exhibited high heterozygosity and rich genetic diversity.
3.5. Genetic Relationships and Clustering Characteristics of 47 Walnut Resources
To understand the genetic relationships between members of the walnut population, cluster analysis was conducted on 47 walnut resources based on their genetic similarity coefficients (
Figure 1A). The first branch could be further divided into two sub-branches. The first sub-branch of branch 1 consisted of ordinary walnuts, iron walnuts, and their hybrid offspring, including Jingxiang 1, Liaoning 5, Liaoni ng 7, Liaoning 10, Yunxin 306, Qingxiang, Liaoning 4, Jinghong 1, Jinghong 2, Jingxiang 2, Meixiang, Robert Livermore, D2-1, Santai, Yangbipao, Liaoning 1, Jingxiang 3, Yunxin 303, Zijing, Yunxin 301, Liaoning 6, Fengxiang, Zhonglin 1, Beijing 861, Lvbo, Lipin 1, Lipin 2, Baokexiang, Xinjufeng, Xiangling, Luguang, and Zhonglin 5. The second sub-branch of branch 1 was composed of
J. hopeiensis, including Huayi 2, Nanjiangshi, Jingyi 2, Jingyi 8-2, Jingyi 8, Mantianxing, Jingyi 1, Huayi 7, Jingyi 6, Qianlongguanmao, Yihe 1, Huayi 1, Jingyi 5, and Jingyi 7. The second branch only had one member, Liaoyi 1, which was the actual offspring of
J. cordiformis in the
J. mandshurica group.
3.6. Population Genetic Structure Analysis and Principal Coordinate Analysis of 47 Walnut Cultivars
To further evaluate the genetic characteristics of these 47 walnut varieties, population genetic structure analysis and principal coordinate analysis were conducted for walnut populations (
Figure 1C,D and
Figure 2). When K = 2, ΔK had a maximum value, indicating that 47 walnut resources could be best divided into two groups (
Table S3). Group I consisted of 32 varieties, including Santai, Yangbipao, Yunxin 301, Yunxin 303, Yunxin 306, Liaoning 1, Liaoning 4, Liaoning 5, Liaoning 6, Liaoning 7, Liaoning 10, Jingxiang 1, Jingxiang 2, Jingxiang 3, Baokexiang, Beijing 861, Lvbo, Fengxiang, Qingxiang, Meixiang, Luguang, Xinjufeng, Xiangling, Lipin 1, Lipin 2, Zhonglin 1, Zhonglin 5, Zijing, Robert Livermore, Jinghong 1, Jinghong 2, and D2-1. Among them, the Qi values of all varieties in group I were greater than 0.8. In addition, group I included two iron walnuts, three hybrid offspring of ordinary walnuts and iron walnuts, and 20 ordinary walnuts. Group II consisted of 15 varieties, namely Liaoyi 1, Jingyi 1, Jingyi 2, Jingyi 5, Jingyi 6, Jingyi 7, Jingyi 8, Jingyi 8-2, Huayi 1, Huayi 2, Huayi 7, Yihe 1, Qianlongguanmao, Nanjiangshi, and Mantianxing. Among them, the Qi value of 14 varieties in group II was greater than 0.8, while the Qi value of Liaoyi 1 was less than 0.8, which was 0.712. In addition, group II was composed of both
J. hopeiensis and
J. mandshurica. The genetic sources within each group were single, and the similarity between varieties was high. Therefore, Group I and II were divided into ordinary walnuts and
J. hopeiensis, respectively.
PCoA analysis (
Figure 2) showed that 47 walnut resources could be roughly divided into two groups. Group I was mainly composed of ordinary walnuts and iron waluts, including Santai, Yangbipao, Yunxin 301, Yunxin 303, Yunxin 306, Liaoning 1, Liaoning 4, Liaoning 5, Liaoning 6, Liaoning 7, Liaoning 10, Jingxiang 1, Jingxiang 2, Jingxiang 3, Baokexiang, Beijing 861, Lvbo, Fengxiang, Qingxiang, Meixiang, Luguang, Xinjufeng, Xiangling, Lipin 1, Lipin 2, Zhonglin 1, Zhonglin 5, Zijing, Robert Livermore, Jinghong 1, Jinghong 2, and D2-1. Group II was mainly composed of
J. hopeiensis, including Jingyi 1, Jingyi 2, Jingyi 5, Jingyi 6, Jingyi 7, Jingyi 8, Jingyi 8-2, Huayi 1, Huayi 2, Huayi 7, Yihe 1, Qianlongguanmao, Nanjiangshi, and Mantianxing, all of which were
J. hopeiensis. However, Liaoyi 1 had a certain distance from both Group I and Group II, which was consistent with the results of the cluster analysis. Therefore, Group I and Group II were divided into ordinary walnuts and
J. hopeiensis, respectively.
3.7. The Identification Ability of SSR Markers and the Construction of DNA Fingerprints
To evaluate the fingerprint recognition ability of these 14 SSR markers, PI and Pisibs were calculated separately (
Table 6). The PI variation range of each molecular marker was 0.092 (Jr45) to 0.814 (Jr52), with an average value of 0.368. Assuming that all marker loci were independently separated, the probability of two random individuals having identical multi-locus genotypes among 14 SSR markers was estimated to be 6.4 × 10
−8. Usually, Pisibs was defined as the upper limit of PI, with a variation range of 0.408 (Jr45) to 0.903 (Jr53) for the 14 SSR markers, with an average value of 0.614. In addition, the combined Pisibs was 7.2 × 10
−4.
Based on the data of gene locus combinations, the recognition ability of 14 random combinations of SSR markers was calculated (
Table 6) to evaluate the number of markers required to fully distinguish 47 walnut varieties. The SSR markers with strong identification ability included Jr29, Jr40, Jr45, and Jr56, which could identify five, five, five, and three different varieties, respectively (
Table 7). Jr29 could identify Yunxin 301, Fengxiang, Baokexiang, Liaoyi 1, and Xinjufeng. Jr40 could identify Liaoyi 1, Liaoning 1, Huayi 1, Robert Livermore, and Zijing. Jr45 could identify Robert Livermore, Jingyi 6, Jingyi 8, Huayi 7, and Mantianxing. Jr56 could distinguish between Jingyi 5, Huayi 7, and Mantianxing. Furthermore, Jr11, Jr12, Jr35, Jr44, Jr50, Jr52, Jr53, and Jr55 could only identify one variety, namely Yunxin 301, Lipin 1, Beijing 861, Liaoning 6, Yihe 1, Yunxin 306, Yunxin 303, and Mantianxing, respectively. However, Jr05 and Jr15 could not independently identify any walnut varieties.
When five SSR markers were combined, the PI value approached 0, and 47 walnut varieties could be independently identified (
Figure 3,
Table S4). Based on the principle of core marker selection and genotype data results, five SSR markers were selected as core markers. Due to its high Na, Ne, He, GD, I, and PIC values, as well as lower PI and Pisibs values, Jr45 was selected as one of the core markers. This marker could separate five varieties independently from all varieties. When the second SSR marker (Jr40) was added, 24 out of 47 varieties could be identified, accounting for 51.06%. When the third SSR marker (Jr29) was added, 32 varieties could be identified, accounting for 68.09%. When the fourth SSR marker (Jr35) was added, 43 varieties could be identified, accounting for 91.49%. When adding the fifth SSR marker (Jr11), 47 walnut varieties could be completely distinguished.
The allele sizes and frequencies of the five core markers Jr45, Jr40, Jr29, Jr35, and Jr11 are shown in
Figure 4. Jr45 had nine alleles, with the highest allele frequency of 200 bp (0.479) and the lowest allele frequency of 272 bp (0.021). Jr40 had eight alleles, with the highest frequency of 138 bp (0.500) and the lowest frequency of 146, 154, and 158 bp (0.032). Jr29 had seven alleles, with the highest frequency of 195 bp (0.574), and the lowest frequency of 204 and 228 bp (0.011). Jr35 had four alleles, with the highest allele frequency of 136 bp (0.174) and the lowest allele frequency of 127 bp (0.087). Jr11 had five alleles, with the highest allele frequency of 155 bp (0.596) and the lowest allele frequency of 147 bp (0.021). The amplified alleles of 47 walnut varieties were arranged in the order of core markers Jr45, Jr40, Jr29, Jr35, and Jr11 to obtain a unique genotype. The DNA fingerprint data of 47 walnut genetic resources were visualized in the form of heat maps (
Figure 5).
3.8. Correlation Analysis between SSR Markers and Phenotypic Traits of Walnuts
Based on the correlation analysis of 14 SSR markers with 14 morphological traits of walnuts (
Table 8 and
Table S5), four SSR markers (Jr35, Jr50, Jr52, and Jr53) were significantly associated with phenotypic traits of walnuts (
p < 0.05). Jr35 was only significantly associated with dry weight of nuts. Jr50 was significantly associated with green fruit weight, fresh fruit weight, fresh weight of seed kernel, fresh weight of seed meat, and nut kernel rate. Jr52 was significantly associated with green skin rate and fruiting rate (
p < 0.05). Jr53 was significantly correlated with fresh weight of seed kernel, fresh fruit kernel rate, fresh weight of seed meat, dry weight of seed kernel, dry weight of seed meat, dry weight of nut, and nut kernel rate (
p < 0.01), and significantly correlated with green fruit weight, fresh fruit weight, green skin weight, green skin rate, fruiting rate, fresh weight of seed coat, and dry weight of seed coat (
p < 0.05).
There were significant differences in the dry weight of nuts among different genotypes of nuts in Jr35, with AB (124, 136) > AC (124, 139) > CD (127, 139) > CC (139, 139) > AA (124, 124) > BB (136, 136). There were significant differences in green fruit weight, fresh fruit weight, fresh weight of seed kernel, fresh weight of seed meat, and nut kernel rate among different genotypes of Jr50, with AA (170, 170) > BB (165, 165). There were significant differences in the green skin rate and fruiting rate among different genotypes of Jr52, with AB (130, 140) > BB (130, 130) > AA (140, 140), AA (140, 140) > BB (130, 130) > AB (130, 140), respectively. There were significant differences in the green fruit weight, fresh fruit weight, green skin weight, green skin rate, fruiting rate, fresh weight of seed kernel, fresh seed kernel rate, fresh weight of seed meat, fresh weight of seed coat, dry weight of seed kernel, dry weight of seed meat, dry weight of seed coat, dry weight of nut, and nut kernel rate among different genotypes of Jr53, with AA (140, 140) > BB (175, 175).