Analysis of the Genetic Diversity and Mating System of the Endangered Plant Keteleeria davidiana var. calcarea
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Data Analysis
2.2.1. DNA Extraction and SSR-PCR Amplification
2.2.2. Genetic Diversity Analysis
2.2.3. Genetic Structure and Gene Flow Analysis
2.2.4. Mating System Analysis
3. Results
3.1. Genetic Diversity
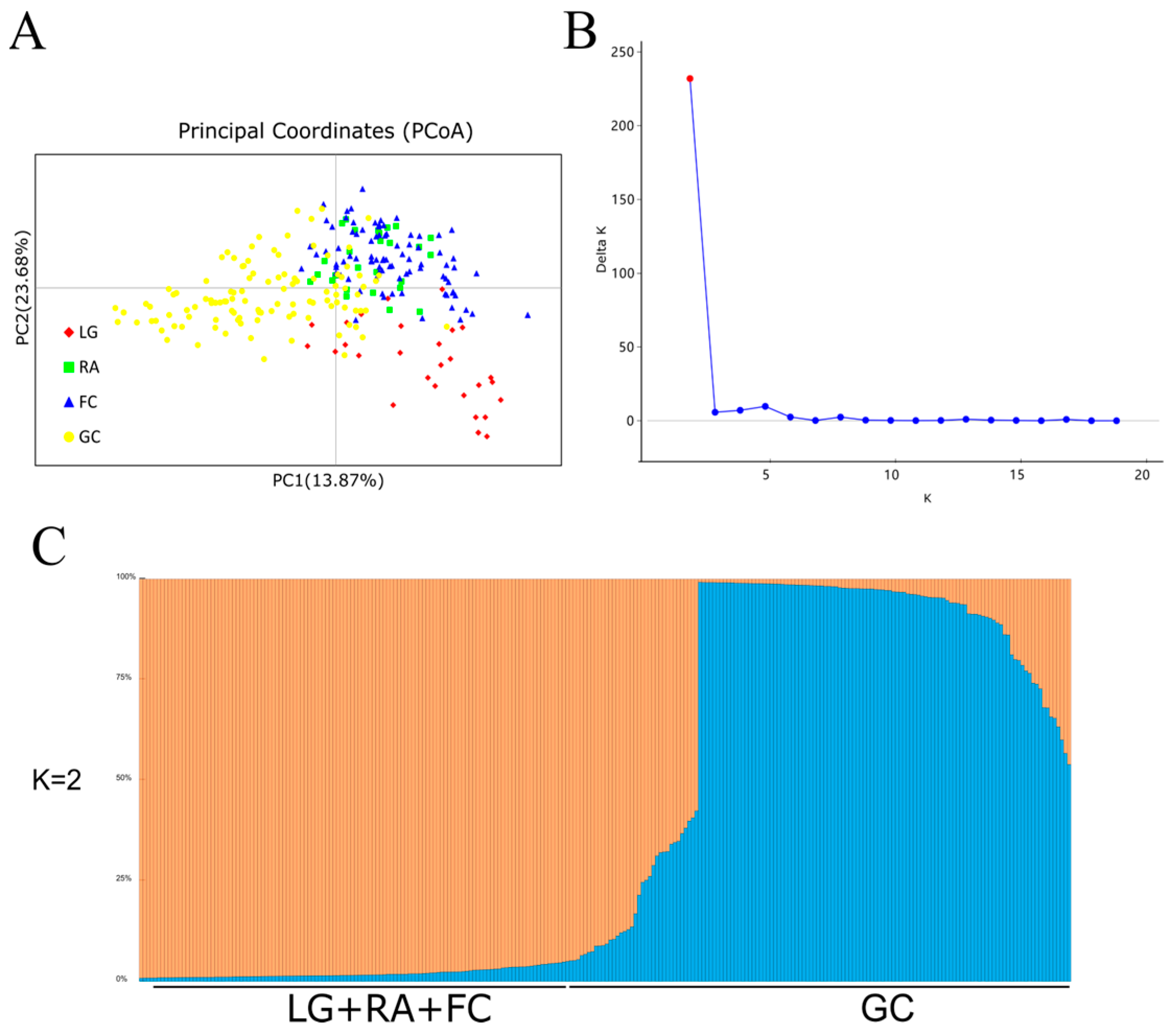
3.2. Genetic Structure
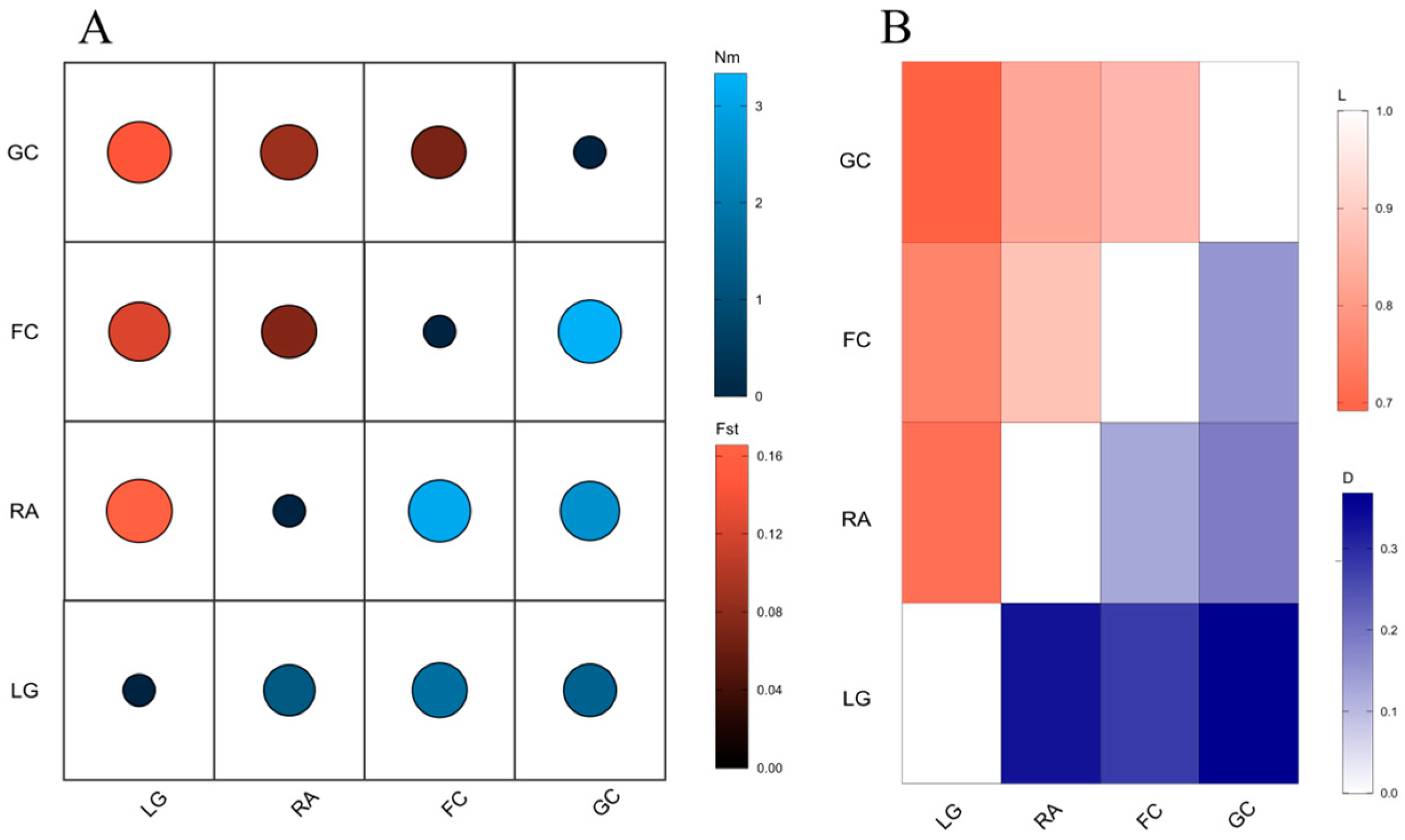
3.3. Genetic Differentiation and Gene Flow
3.4. Mating System
4. Discussion
4.1. Genetic Diversity
4.2. Gene Flow and Gene Structure
4.3. Mating System
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Huang, L. Keteleeria calcarea cheng et lkfua valuable tree for afforesting the limestone hills. Guihaia 1982, 2, 103–104. (In Chinese) [Google Scholar]
- Jiang, B.; Wen, G.; Tang, Y.; Jiang, Q. Effects of different treatments on cuttage cultivation and growth of Keteleeria calcarea. Guihaia 2008, 4, 549–552. (In Chinese) [Google Scholar]
- State Forestry and Grassland Administration of China. National Forestry and Grassland Administration, Ministry of Agriculture and Rural Affairs (No. 15, 2021). List. State Key Prot. Wild Plants 2021, 15, 1–18. (In Chinese) [Google Scholar]
- Lu, Z.; Qin, H.; Jin, X.; Zhang, Z.; Yang, Q.; Hong, D.; Li, D.; Li, K.; Yuan, L.; Zhou, Z.; et al. On the Necessity, Principle, and Process of Updating the List of National Key Protected Wild Plants. Biodivers. Sci. 2021, 29, 1577–1582. [Google Scholar] [CrossRef]
- Jiang, H.; Xie, W.; Chai, S.; Tang, J.; Jiang, Y.; Qin, H.; Wei, X. Seed germination characteristics of Keteleeria calcarea, a precious tree species in karst area. Guihaia 2022, 42, 951–960. (In Chinese) [Google Scholar] [CrossRef]
- Li, J.; Cheng, N.; Shi, Y. The Complete Chloroplast Genome of Keteleeria Davidiana Var. Calcarea (Pinaceae), an Endangered Species Endemic to China. Mitochondrial DNA Part B 2021, 6, 693–695. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Chai, S.; Jiang, Y.; Tang, J.; Chen, Z.; Zou, R.; Wei, X. ISSR analysis on genetic diversity of Keteleeria calcarea. Guihaia 2017, 37, 36–41. (In Chinese) [Google Scholar] [CrossRef]
- Zhou, Q.; Wang, G.; Li, Y. Analysis of the Conservation Status, Genetic Diversity and Population Structure of Endangered Ostrya Rehderiana Resources Using SSR Markers. Forests 2023, 14, 1519. [Google Scholar] [CrossRef]
- Liesebach, H.; Schneck, D. Flowering Behavior of Clones in a Norway Maple (Acer platanoides) Seed Orchard and Mating System Analysis Using Nuclear SSR Markers. Eur. J. For. Res. 2022, 141, 561–569. [Google Scholar] [CrossRef]
- Imai, R.; Tsuda, Y.; Matsumoto, S.; Ebihara, A.; Watano, Y. The Relationship between Mating System and Genetic Diversity in Diploid Sexual Populations of Cyrtomium falcatum in Japan. PLoS ONE 2016, 11, e0163683. [Google Scholar] [CrossRef]
- Yuanyuan, L.; Chaonan, L.; Rong, W.; Shuixing, L.; Shouqian, N.; Jingwen, W.; Xiaoyong, C. Applications of molecular markers in conserving endangered species. Biodivers. Sci. 2020, 28, 367–375. (In Chinese) [Google Scholar] [CrossRef]
- Neel, M.C. Conservation Implications of the Reproductive Ecology of Agalinis acuta (Scrophulariaceae). Am. J. Bot. 2002, 89, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Young, A.; Boyle, T.; Brown, T. The Population Genetic Consequences of Habitat Fragmentation for Plants. Trends Ecol. Evol. 1996, 11, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Seltmann, P.; Cocucci, A.; Renison, D.; Cierjacks, A.; Hensen, I. Mating System, Outcrossing Distance Effects and Pollen Availability in the Wind-Pollinated Treeline Species Polylepis australis BITT. (Rosaceae). Basic Appl. Ecol. 2009, 10, 52–60. [Google Scholar] [CrossRef]
- van Belkum, A.; Scherer, S.; van Alphen, L.; Verbrugh, H. Short-Sequence DNA Repeats in Prokaryotic Genomes. Microbiol. Mol. Biol. Rev. MMBR 1998, 62, 275–293. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Korol, A.B.; Fahima, T.; Beiles, A.; Nevo, E. Microsatellites: Genomic Distribution, Putative Functions and Mutational Mechanisms: A Review. Mol. Ecol. 2002, 11, 2453–2465. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-C.; Korol, A.B.; Fahima, T.; Nevo, E. Microsatellites within Genes: Structure, Function, and Evolution. Mol. Biol. Evol. 2004, 21, 991–1007. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Shrivastava, N.; Padh, H. Advances in Molecular Marker Techniques and Their Applications in Plant Sciences. Plant Cell Rep. 2008, 27, 617–631. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wang, F.; Cui, R.; Liu, X.; Li, X.; Dong, J.; Sun, L.; Qin, S.; Wang, R.; Zheng, P.; et al. Studies on Reproductive Strategies of Vitex negundo L. Var. Heterophylla (Franch.) Rehder (Lamiaceae) Based on Morphological Characteristics and SSR Markers. Ecol. Evol. 2020, 10, 5270–5280. [Google Scholar] [CrossRef]
- Zhang, Z.; Meng, J.; Pan, D.; Yang, C.; Li, Y. Mating System and Progeny Genetic Diversity of Camellia Oleifera ‘Ruan Zhi’. J. For. Res. 2019, 30, 1805–1810. [Google Scholar] [CrossRef]
- Fuchs, E.J.; Cascante-Marín, A.; Madrigal-Brenes, R.; Quesada, M. Genetic Diversity and Phylogeographic Patterns of the Dioecious Palm Chamaedorea tepejilote (Arecaceae) in Costa Rica: The Role of Mountain Ranges and Possible Refugia. AoB PLANTS 2023, 15, plac060. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Chai, S.; Tang, J.; Jiang, Y.; Wei, X. Habitat Condition and Population Structure Characteristics of Keteleeria calcarea Forest in Karst Area of Guangxi. J. Guangxi Acad. Sci. 2020, 36, 56–64. [Google Scholar] [CrossRef]
- Shi, Y.; Lai, I.; Wang, X.; Qin, H.; Chai, S. Development of EST-SSR Primers in Keteleeria calcarea Based on Tran- scriptome Sequencing. Mol. Plant Breed. 2024, 22, 2257–2264. (In Chinese) [Google Scholar]
- Schuelke, M. An Economic Method for the Fluorescent Labeling of PCR Fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J.-M. FSTAT, a Program to Estimate and Test Gene Diversities and Fixation Indices (Version 2.9.3). 2001. Available online: https://www2.unil.ch/popgen/softwares/fstat.htm (accessed on 23 February 2024).
- Rousset, F. Genepop’007: A Complete Re-Implementation of the Genepop Software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research–an Update. Bioinform. Oxf. Engl. 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
- Diniz-Filho, J.A.F.; Soares, T.N.; Lima, J.S.; Dobrovolski, R.; Landeiro, V.L.; de Campos Telles, M.P.; Rangel, T.F.; Bini, L.M. Mantel Test in Population Genetics. Genet. Mol. Biol. 2013, 36, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Ritland, K. Extensions of Models for the Estimation of Mating Systems Using n Independent Loci. Heredity 2002, 88, 221–228. [Google Scholar] [CrossRef]
- Ritland, K. A Series of FORTRAN Computer Programs for Estimating Plant Mating Systems. J. Hered. 1990, 81, 236–237. [Google Scholar] [CrossRef]
- Kaljund, K.; Jaaska, V. No Loss of Genetic Diversity in Small and Isolated Populations of Medicago sativa Subsp. Falcata. Biochem. Syst. Ecol. 2010, 38, 510–520. [Google Scholar] [CrossRef]
- Shih, K.-M.; Chang, C.-T.; Chung, J.-D.; Chiang, Y.-C.; Hwang, S.-Y. Adaptive Genetic Divergence Despite Significant Isolation-by-Distance in Populations of Taiwan Cow-Tail Fir (Keteleeria davidiana Var. Formosana). Front. Plant Sci. 2018, 9, 92. [Google Scholar] [CrossRef]
- Lieu, T.T.; Phong, D.T.; Hien, V.T.T. Genetic diversity among natural populations of Keteleeria evelyniana mast. in central higlands of vietnam using ssr markers. Vietnam J. Sci. Technol. 2018, 56, 275. [Google Scholar] [CrossRef]
- Lanes, É.C.M.; Motoike, S.Y.; Kuki, K.N.; Resende, M.D.V.; Caixeta, E.T. Mating System and Genetic Composition of the Macaw Palm (Acrocomia aculeata): Implications for Breeding and Genetic Conservation Programs. J. Hered. 2016, 107, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Nazareno, A.G.; dos Reis, M.S. At Risk of Population Decline? An Ecological and Genetic Approach to the Threatened Palm Species Butia eriospatha (Arecaceae) of Southern Brazil. J. Hered. 2013, 105, 120–129. [Google Scholar] [CrossRef]
- Hu, X.-S. Mating System as a Barrier to Gene Flow: Mating system and isolating barrier. Evolution 2015, 69, 1158–1177. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Zou, Z.; Hu, X.; Yang, S. Genetic Diversity and Mating System of Two Mangrove Species (Rhizophora apiculata and Avicennia marina) in a Heavily Disturbed Area of China. Diversity 2022, 14, 115. [Google Scholar] [CrossRef]
- Zhu, X.; Zou, R.; Tang, J.; Deng, L.; Wei, X. Genetic Diversity Variation during the Natural Regeneration of Vatica Guangxiensis, an Endangered Tree Species with Extremely Small Populations. Glob. Ecol. Conserv. 2023, 42, e02400. [Google Scholar] [CrossRef]
- Reed, D.H.; Frankham, R. Correlation between Fitness and Genetic Diversity. Conserv. Biol. 2003, 17, 230–237. [Google Scholar] [CrossRef]
- Dillon, R.; Coates, D.; Standish, R.; Monks, L.; Waycott, M. Assessing Plant Translocation Success: Common Metrics Mask High Levels of Inbreeding in a Recently Established Banksia brownii (Proteaceae) Population. Aust. J. Bot. 2023, 71, 79–92. [Google Scholar] [CrossRef]
- Leimu, R.; Mutikainen, P.; Koricheva, J.; Fischer, M. How General Are Positive Relationships between Plant Population Size, Fitness and Genetic Variation? J. Ecol. 2006, 94, 942–952. [Google Scholar] [CrossRef]
- Charlesworth, B.; Charlesworth, D. The Genetic Basis of Inbreeding Depression. Genet. Res. 1999, 74, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Compton, S.G.; Chen, X.-Y. Fragmentation Can Increase Spatial Genetic Structure without Decreasing Pollen-Mediated Gene Flow in a Wind-Pollinated Tree: Spatial genetic structure of castanopsis. Mol. Ecol. 2011, 20, 4421–4432. [Google Scholar] [CrossRef] [PubMed]
- Zong, J.-W.; Zhao, T.-T.; Ma, Q.-H.; Liang, L.-S.; Wang, G.-X. Assessment of Genetic Diversity and Population Genetic Structure of Corylus mandshurica in China Using SSR Markers. PLoS ONE 2015, 10, e0137528. [Google Scholar] [CrossRef] [PubMed]
- Dafni, A.; Tzohari, H.; Ben-Shlomo, R.; Vereecken, N.J.; Ne’eman, G. Flower Colour Polymorphism, Pollination Modes, Breeding System and Gene Flow in Anemone coronaria. Plants 2020, 9, 397. [Google Scholar] [CrossRef] [PubMed]
- Escudero, M.; Vargas, P.; Valcárcel, V.; Luceño, M. Strait of Gibraltar: An Effective Gene-flow Barrier for Wind-pollinated Carex helodes (Cyperaceae) as Revealed by DNA Sequences, AFLP, and Cytogenetic Variation. Am. J. Bot. 2008, 95, 745–755. [Google Scholar] [CrossRef]
- Lenormand, T. Gene Flow and the Limits to Natural Selection. Trends Ecol. Amp Evol. 2002, 17, 183–189. [Google Scholar] [CrossRef]
- Sexton, J.P.; Hangartner, S.B.; Hoffmann, A.A. Genetic Isolation by Environment or Distance: Which Pattern of Gene Flow Is Most Common? Evol. Int. J. Org. Evol. 2014, 68, 1–15. [Google Scholar] [CrossRef]
- BïLgen, B.B.; Kurt, Y.; Kaya, N. Mating System in Natural Populations of Taurus Cedar (Cedrus libani A.Rich.). Turk. J. Agric. For. 2012, 36, 379–387. [Google Scholar] [CrossRef]
- Burczyk, J. Mating System Variation in a Scots Pine Clonal Seed Orchard. Silvae Genet. 1998, 47, 155–158. [Google Scholar]
- Ritland, K. The Genetic-Mating Structure of Subdivided Populations II. Correlated Mating Models. Theor. Popul. Biol. 1988, 34, 320–346. [Google Scholar] [CrossRef]
- Vinson, C.C.; Mangaravite, E.; Sebbenn, A.M.; Lander, T.A. Using Molecular Markers to Investigate Genetic Diversity, Mating System and Gene Flow of Neotropical Trees. Braz. J. Bot. 2018, 41, 481–496. [Google Scholar] [CrossRef]
- Díaz-Hernández, B.G.; Colombo, C.A.; Morales-Marroquín, J.A.; Sanitá-Rodrigues, M.; Azevedo-Filho, J.A.; Zucchi, M.I. Assessing the Genetic Vulnerability of Macaúba Palm [Acrocomia aculeata (Jacq.) Lodd. ex Mart.] through the Mating System and Genetic Diversity of Open-pollinated Progenies. Ann. Appl. Biol. 2023, 184, 238–249. [Google Scholar] [CrossRef]
- Tambarussi, E.V.; Sebbenn, A.M.; Alves-Pereira, A.; Vencovsky, R.; Cambuim, J.; Da Silva, A.; Moraes, M.; De Moraes, M.L.T. Dipteryx alata Vogel (Fabaceae) a Neotropical Tree with High Level of Selfing: Implication for Conservation and Breeding Programs. Ann. For. Res. 2017, 60, 243–261. [Google Scholar] [CrossRef]
- Perrut-Lima, P.; Sebbenn, A.M.; Francisconi, A.F.; Picanço-Rodrigues, D.; Clement, C.R. Genetic Diversity and Mating System of Euterpe precatoria in Three Localities along the Lower Solimões River in Central Amazonia. Silvae Genet. 2023, 72, 81–91. [Google Scholar] [CrossRef]
- Li, F.; Chen, H.; Liu, S.; Zhang, H.; Zhou, Z. Mating Systems of Single Families and Population Genetic Diversity of Endangered Ormosia hosiei in South China. Genes 2022, 13, 2117. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, H.; Yang, J.; Dao, Z.; Sun, W. Conservation Genetics and Potential Geographic Distribution Modeling of Corybas taliensis, a Small “sky Island” Orchid Species in China. BMC Plant Biol. 2024, 24, 11. [Google Scholar] [CrossRef] [PubMed]
- Price, J.H.; Brandvain, Y.; Smith, K.P. Measurements of Lethal and Nonlethal Inbreeding Depression Inform the de Novo Domestication of Silphium integrifolium. Am. J. Bot. 2021, 108, 980–992. [Google Scholar] [CrossRef]
- Andersson, S.; Waldmann, P. Inbreeding Depression in a Rare Plant, Scabiosa canescens (Dipsacaceae). Hereditas 2002, 136, 207–211. [Google Scholar] [CrossRef]
- Lloyd, M.W.; Tumas, H.R.; Neel, M.C. Limited Pollen Dispersal, Small Genetic Neighborhoods, and Bimaternalal Inbreeding in Vallisneria americana. Am. J. Bot. 2018, 105, 227–240. [Google Scholar] [CrossRef]
- Almeida-Rocha, J.M.; Soares, L.A.S.S.; Andrade, E.R.; Gaiotto, F.A.; Cazetta, E. The Impact of Anthropogenic Disturbances on the Genetic Diversity of Terrestrial Species: A Global Meta-Analysis. Mol. Ecol. 2020, 29, 4812–4822. [Google Scholar] [CrossRef] [PubMed]
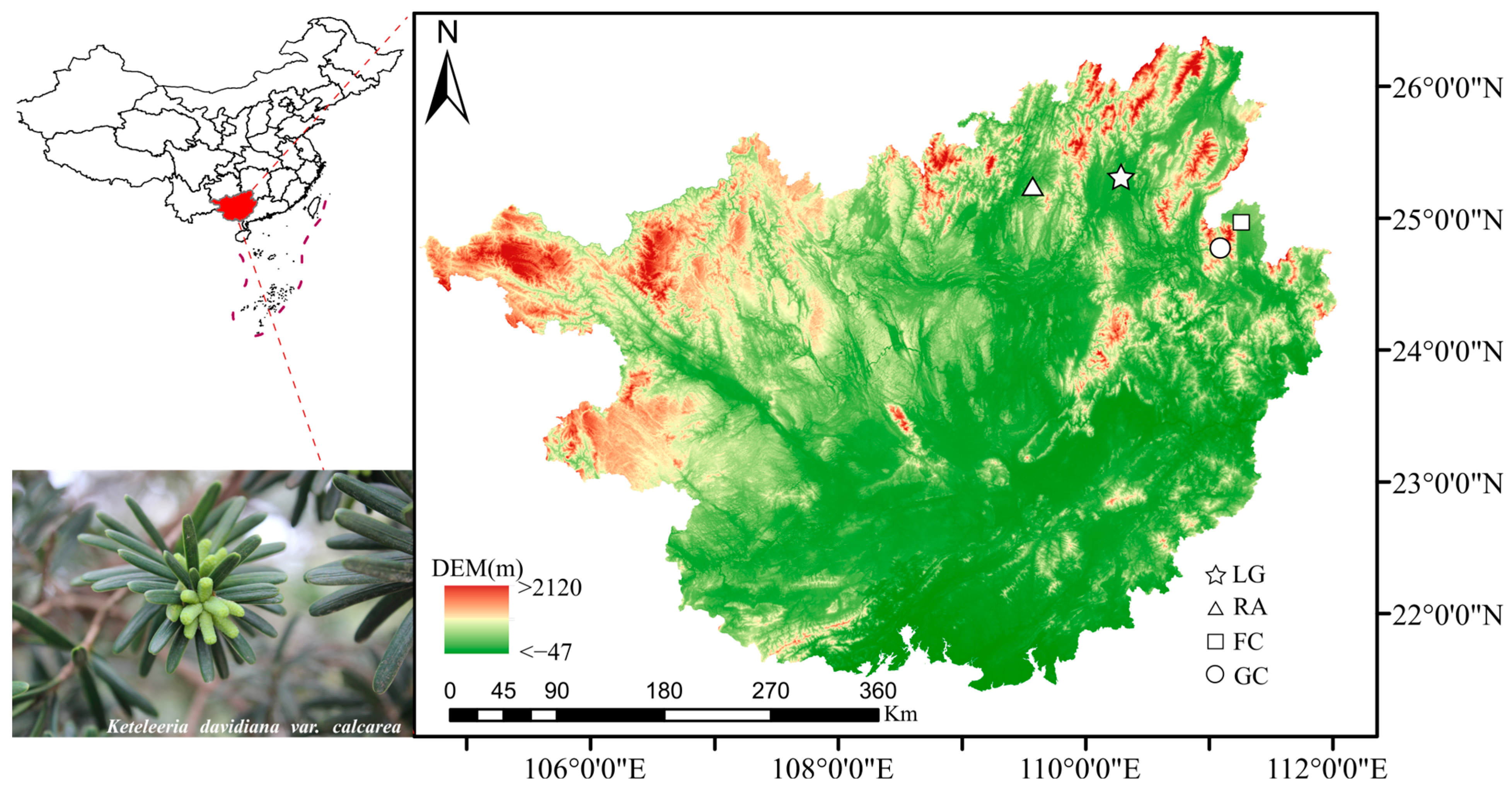
| Population | Elevation (m) | Aspect | Slope (°) | Latitude | Longitude | Maternal Trees | Progeny |
|---|---|---|---|---|---|---|---|
| GC | 670 | WS | 45 | 25°50′26″ | 110°05′47″ | 20 | 98 |
| LG | 160 | WS | 40 | 25°12′47″ | 110°11′51″ | 5 | 22 |
| FC | 334 | ES | 35 | 25°02′24″ | 111°18′47″ | 15 | 73 |
| RA | 480 | WS | 45 | 25°01′46″ | 109°34′50″ | 6 | 21 |
| Primer No. | Repeat Motif | Primer Sequence (5′-3′) | Expected Size (bp) | Tm (°C) |
|---|---|---|---|---|
| P7 | (AGA)8 | F: CCAACATTGCAGTTGACGAC | 132 | 54 |
| R: CATCATCCTCACCACATTGC | ||||
| P49 | (TC)10 | F: TTGGCCAGGTCAGAAATAGG | 171 | 60 |
| R: CCCATTGCCTCAAGAGAGAG | ||||
| P56 | (TA)11 | F: GTCAAAGACAACAGACGCCA | 212 | 59.8 |
| R: CTTGGGGATACAACCAAGGA | ||||
| P60 | (AT)11 | F: GAACCCCCACCTGTACCTCT | 238 | 60.4 |
| R: CGAGTATGCATCCACGTCAA | ||||
| P64 | (TA)11 | F: GCTGCGAAGCTGCTAAAACT | 255 | 60 |
| R: CGGCCTCTCACTTCTGGTAG | ||||
| P130 | (TA)11 | F: CAAATTCTCCCAGAGGAAGC | 234 | 55 |
| R: GGTCAGTGTCCTTCCTCCAA | ||||
| P150 | (TG)12 | F: ATCTCCTTGCTGATTGGGTG | 194 | 55 |
| R: CCTCCTGCAACGGTTATGTT | ||||
| P153 | (GT)10 | F: CCTTTCACACGCACTAGCAA | 267 | 56.5 |
| R: CGCACCTCTTATCCACCACT | ||||
| P156 | (AT)11 | F: ATCCGTATCCGTTTCCGTTT | 110 | 53.5 |
| R: TTTTTGCTGGAGTGTGTTGC |
| Primer | Maternal Trees | Progeny | ||||||
|---|---|---|---|---|---|---|---|---|
| LG | RA | FC | GC | LG | RA | FC | GC | |
| P130 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| P150 | 2 | 1 | 2 | 2 | 2 | 1 | 2 | 2 |
| P153 | 3 | 5 | 3 | 5 | 3 | 6 | 7 | 7 |
| P49 | 3 | 1 | 2 | 3 | 3 | 2 | 3 | 3 |
| P56 | 2 | 2 | 5 | 8 | 2 | 3 | 7 | 11 |
| P60 | 2 | 2 | 3 | 4 | 2 | 3 | 3 | 5 |
| P64 | 6 | 4 | 7 | 10 | 7 | 5 | 6 | 11 |
| P156 | 1 | 3 | 3 | 3 | 2 | 3 | 5 | 4 |
| P7 | 1 | 1 | 2 | 3 | 1 | 2 | 10 | 3 |
| total | 22 | 21 | 29 | 40 | 24 | 27 | 45 | 48 |
| Stage | Population | Ne (SE) | Ho (SE) | He (SE) | uHe (SE) | Ar (SE) | F (SE) | PPL (%) |
|---|---|---|---|---|---|---|---|---|
| Maternal trees | LG | 2.444(0.322) | 0.333(0.042) | 0.387(0.088) | 0.430(0.075) | 2.058(0.032) | 0.106(0.008) | 77.78 |
| RA | 2.333(0.182) | 0.361(0.003) | 0.341(0.046) | 0.383(0.003) | 1.964(0.091) | −0.007(0.000) | 66.67 | |
| FC | 3.222(0.029) | 0.375(0.039) | 0.402(0.037) | 0.418(0.022) | 2.059(0.003) | 0.010(0.001) | 100.00 | |
| GC | 4.444(0.090) | 0.450(0.036) | 0.496(0.098) | 0.509(0.093) | 2.315(0.005) | 0.085(0.002) | 100.00 | |
| average | 3.111(0.355) | 0.380(0.65) | 0.406(0.033) | 0.435(0.033) | 2.099(0.048) | 0.048(0.014) | 86.11 | |
| Progeny | LG | 2.670(0.035) | 0.350(0.091) | 0.390(0.066) | 0.398(0.025) | 1.901(0.009) | 0.120(0.002) | 88.89 |
| RA | 3.000(0.165) | 0.330(0.082) | 0.360(0.014) | 0.365(0.071) | 1.907(0.071) | 0.070(0.000) | 88.89 | |
| FC | 5.000(0.175) | 0.310(0.034) | 0.450(0.039) | 0.456(0.033) | 2.164(0.038) | 0.270(0.000) | 100.00 | |
| GC | 5.330(0.092) | 0.380(0.033) | 0.480(0.064) | 0.488(0.019) | 2.224(0.048) | 0.220(0.007) | 100.00 | |
| average | 4.000(0.316) | 0.340(0.020) | 0.420(0.040) | 0.427(0.067) | 2.049(0.048) | 0.170(0.001) | 94.44 |
| tm (SE) | ts (SE) | tm-ts (SE) | rt (SE) | rp(m) (SE) | rp(s) (SE) | rp(s)-rp(m) (SE) | Nep | δ | |
|---|---|---|---|---|---|---|---|---|---|
| LG | 1.000 (0.000) | 0.918 (0.037) | 0.082 (0.037) | 0.103 (0.000) | 0.064 (0.028) | 0.073 (0.019) | 0.009 (0.015) | 15.384 | 0 |
| RA | 1.134 (0.079) | 0.977 (0.090) | 0.158 (0.090) | −0.443 (0.512) | 0.043 (0.395) | 0.021 (0.293) | 0.001 (0.268) | 23.255 | −0.057 |
| FC | 0.859 (0.046) | 0.603 (0.079) | 0.257 (0.049) | −0.215 (0.318) | 0.584 (0.137) | 0.808 (0.248) | 0.224 (0.165) | 1.712 | 1.029 |
| GC | 0.810 (0.060) | 0.671 (0.061) | 0.140 (0.024) | 0.325 (0.179) | 0.192 (0.080) | 0.095 (0.084) | −0.097 (0.070) | 5.208 | 1.048 |
| Species level | 0.902 (0.025) | 0.606 (0.035) | 0.295 (0.028) | 0.145 (0.114) | 0.490 (0.057) | 0.461 (0.099) | −0.028 (0.069) | 2.040 | 0.967 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, X.; Qin, H.; Jiang, H.; Peng, L.; Yang, Y.; Tang, J.; Zou, R.; Chen, Z.; Wei, X.; Chai, S. Analysis of the Genetic Diversity and Mating System of the Endangered Plant Keteleeria davidiana var. calcarea. Forests 2024, 15, 793. https://doi.org/10.3390/f15050793
Pan X, Qin H, Jiang H, Peng L, Yang Y, Tang J, Zou R, Chen Z, Wei X, Chai S. Analysis of the Genetic Diversity and Mating System of the Endangered Plant Keteleeria davidiana var. calcarea. Forests. 2024; 15(5):793. https://doi.org/10.3390/f15050793
Chicago/Turabian StylePan, Xinfeng, Huizhen Qin, Haidu Jiang, Lihui Peng, Yishan Yang, Jianmin Tang, Rong Zou, Zongyou Chen, Xiao Wei, and Shengfeng Chai. 2024. "Analysis of the Genetic Diversity and Mating System of the Endangered Plant Keteleeria davidiana var. calcarea" Forests 15, no. 5: 793. https://doi.org/10.3390/f15050793
APA StylePan, X., Qin, H., Jiang, H., Peng, L., Yang, Y., Tang, J., Zou, R., Chen, Z., Wei, X., & Chai, S. (2024). Analysis of the Genetic Diversity and Mating System of the Endangered Plant Keteleeria davidiana var. calcarea. Forests, 15(5), 793. https://doi.org/10.3390/f15050793









