Abstract
Soil salinity affects approximately 20% of the world’s arable land, presenting a significant challenge for studying the mechanisms by which plants adapt to saline environments. Cyclocarya paliurus, an invaluable research model due to its ecological and medicinal significance, is primarily concentrated in central and southern China. Nevertheless, Cyclocarya paliurus faces challenges from environmental factors such as soil salinization, which adversely impacts its growth, subsequently affecting the yield and quality of its bioactive compounds. The NAC gene family, a critical group of plant-specific transcription factors, plays pivotal roles in responding to abiotic stresses. However, there has not yet been any studies on NAC genes under salt stress in Cyclocarya paliurus. In this study, we identified 132 NAC genes within the Cyclocarya paliurus genome. Our analysis of the conserved structures and gene organization revealed a high degree of conservation in the proteins of the CpNAC gene family. Cis-element analysis unveiled the participation of these genes in a variety of biological processes, including light responses, phytohormone responses, cell cycle responses, and abiotic stress responses. Under salt stress conditions, the expression of 35 CpNAC genes changed significantly, indicating a response to salt treatment. Furthermore, we provided additional evidence for the identification of the NAC gene family and revealed their potential positive regulatory role in signal transduction by conducting a transcriptional activation activity analysis of CpNAC132(D) and CpNAC040, which are homologous to Arabidopsis thaliana NAC062/91 and NAC103, respectively. This research not only advances our comprehension of the salt stress adaptation in Cyclocarya paliurus but also provides robust support for future investigations into plant responses to environmental stress and the cultivation of salt-tolerant crops.
1. Introduction
With ongoing global climate change and increasing human activities, soil salinization has become an increasingly serious challenge facing global agriculture [1]. This environmental change poses a significant threat not only to agricultural production but also to global food security. In this context, exploring the growth mechanisms of plants in saline stress environments and studying how to improve plant adaptation to this adversity has become an important research direction in the fields of plant biology and agricultural science [2,3,4]. Saline stress, characterized by soil salinity levels exceeding the plant’s tolerance limit, has severely detrimental effects on plant growth and development. It is estimated that approximately 20% of the world’s arable land is affected by soil salinization, highlighting the seriousness of this issue. Cyclocarya paliurus, also known as the sweet tea tree, or the immortal tea tree, is a deciduous tree native to China. It belongs to the Juglandaceae family and is primarily found in central and southern China, including the Guizhou, Hubei, and Hunan provinces [5]. Cyclocarya paliurus has garnered attention due to its rich bioactive compounds such as its flavonoids, polysaccharides, and triterpenes, which are traditionally used in Chinese medicine to treat various ailments [6]. Additionally, Cyclocarya paliurus holds significant value in various fields, including ecology, agriculture, energy, and fundamental science [7]. Therefore, studying the response of Cyclocarya paliurus to saline stress not only aids in addressing salinity issues and promoting sustainable agriculture and ecological restoration but also deepens our understanding of the molecular mechanisms of plant responses to adversity.
Transcription factors (TFs) are key regulatory elements in many physiological and biological processes within cells and organisms. They coordinate the control of various signaling pathways, including the regulation of both biotic and abiotic stress pathways, by upregulating or downregulating the expression of target genes. This has profound implications for crop improvement and protein evolution [8]. In the study of plant responses to salt stress, TFs play a pivotal role in helping plants adapt to saline environments by modulating the plants’ physiological and molecular responses. For instance, in Arabidopsis thaliana, the bZIP family transcription factors, AtbZIP24 and AtbZIP53, enhance salt tolerance by regulating osmotic pressure and ion balance [9,10]. In Cyclocarya paliurus, the transcription factors CpWRKY19 and CpWRKY26 alleviate the negative effects of salt stress. Within the Oryza sativa MYB family, OsMYB2 and OsMYB4 play a positive role in maintaining the ion balance and antioxidant defense within cells, thereby enhancing salt tolerance [11,12]. In Glycine max, GmSALT3 and GmSALT1 primarily influence the genes involved in ion balance, osmotic balance regulation, and antioxidant defense to improve salt stress tolerance [13].
The NAC family is a crucial group of transcription factors specific to plants, with its members playing key roles in multiple physiological processes, including secondary growth regulation, the response to abiotic stress, the development of meristems, and hormone signal transduction [14,15,16]. For example, the SND1 gene plays a significant role in cellulose synthesis and secondary cell wall formation. In the context of abiotic stress responses, genes such as GmNAC81 and ZmNAC074 are involved in regulating endoplasmic reticulum (ER) stress-related genes, promoting cell survival, and participating in the ER stress response [17,18]. NbNAC089, on the other hand, regulates the ER stress pathway by mediating programmed cell death (PCD), contributing to plant responses to abiotic stress [19]. In terms of hormone signal transduction, genes like ANAC019, ANAC055, and ANAC072 (RD26) participate in regulating the jasmonic acid-mediated stress responses in Cyclocarya paliurus [20,21,22]. The regulation of these hormone signaling pathways has far-reaching effects on plants’ growth and environmental adaptation.
Salt stress is a detrimental environmental factor known to impede plant growth and development significantly [23]. To circumvent the deleterious effects of such stress, plants have evolved complex regulatory mechanisms that include the modulation of gene expression [3,24,25]. Notably, the impact of salt stress on the NAC gene family in Cyclocarya paliurus, a species of considerable ecological significance, remains underexplored. Therefore, addressing this knowledge gap, this study conducted a comprehensive genome-wide identification and expression analysis of the NAC family members in Cyclocarya paliurus. By analyzing the expression patterns of these genes under salt stress conditions, we aim to uncover the functions of NAC genes in Cyclocarya paliurus and their roles in salt stress responses. The results of this study will provide new insights into the regulatory mechanisms of NAC genes under salt stress, lay the foundation for further functional studies of NAC genes, and offer important theoretical support for promoting the genetic improvement of Cyclocarya paliurus in terms of its growth, development, and stress resistance.
2. Materials and Methods
2.1. Gene Identification
In this study, the plant material we utilized was Cyclocarya paliurus (Figure S1), and we used a systematic approach to identify and validate the NAC genes in the Cyclocarya paliurus genome. First, we obtained genomic data on Cyclocarya paliurus from the website of the Chinese National Center for Genome Data (CNCB) (https://ngdc.cncb.ac.cn, accessed on 5 September 2023). Subsequently, we successfully performed a preliminary identification of NAC genes using a Hidden Markov Model (HMM), while core NAC sequence identification was provided by the Pfam protein family database. The Hidden Markov Model (HMM) we used was based on the NAC structural domain PF02365, obtained from the Pfam database. Specifically, we used the hmmscan command from the HMMER toolkit, which is a widely used sequence database search tool for identifying the protein structural domains in sequences [26].
In the process of selecting candidate NAC genes, we used several key metrics and threshold criteria. Initially, we ensured the accuracy and reliability of the search results by setting a threshold of less than 1 × 10−5 for the E-value (expected value), a statistical measure that assesses the likelihood of finding a match by chance in a database search. This stringent E-value threshold was chosen to minimize the false-positive rate and to ensure that only highly related sequences were considered NAC candidate genes. In addition, we manually checked and validated the sequences to eliminate any potential false positives. For all sequences screened by E-value thresholding, we further checked their sequence homology and structural domain identity to confirm their NAC gene identity. We validated the annotations using RNA sequencing (RNA-seq) data, organized and classified these gene models, and experimentally validated some of them using polymerase chain reaction (PCR) amplification and sequencing techniques to finally confirm a set of reliable NAC gene models.
Additionally, this study collected additional information regarding the identified NAC proteins, including their sequence length, molecular weight, isoelectric point, and predicted subcellular localization. The physicochemical properties of CpNAC proteins were calculated using the ProtParam tool in ExPASy [27]. Furthermore, protein subcellular localization prediction was conducted using CELLO v.2.5 [28]. The collection of these data not only provided us with a deeper understanding of the basic characteristics of NAC proteins but also laid a solid foundation for subsequent functional studies.
2.2. Element and Gene Structure Analysis of CpNAC Genes
In this study, we utilized the MEME web tool (https://meme-suite.org/meme/tools/meme, accessed on 17 September 2023) to predict the conserved motifs of the Cyclocarya paliurus NAC protein family (CpNAC) [29]. MEME is an efficient bioinformatics tool designed specifically for identifying and analyzing recurring conserved patterns within protein sequences, which is crucial for understanding the functional and structural characteristics of proteins.
Furthermore, we employed TBtools software (version 1.098) to generate structural diagrams of CpNAC genes, including the distribution of their exons and introns [30]. TBtools is a powerful bioinformatics software that provides an intuitive representation of gene’s structural composition, including the arrangement of their exons and introns, facilitating a deeper understanding of gene organization.
TBtools software was also used to display results related to the phylogenetic tree and conserved motifs and gene structure analyses of CpNAC genes. Constructing the phylogenetic tree aids in uncovering the evolutionary relationships between members of the CpNAC gene family, while the analysis of gene structure and conserved motifs further contributes to our understanding of the functions and evolutionary history of these genes. By integrating these analytical results into our work, we can gain a more comprehensive understanding of the characteristics of the CpNAC gene family and their significance in the biology of Cyclocarya paliurus.
2.3. Systematic Evolutionary Analysis of CpNAC Genes
In this study, to explore the phylogenetic relationships within the NAC gene family in Cyclocarya paliurus, we initially performed sequence alignment using ClustalX software (version 2.0). This step aimed to precisely compare and analyze the sequence similarities and differences between different NAC genes. Through this alignment, we could gain a better understanding of the relationships between these genes during the evolutionary process.
Subsequently, we utilized MEGA 7.0 software to construct a phylogenetic tree based on the Maximum Likelihood (ML) method. Maximum Likelihood is a powerful statistical approach used to estimate the evolutionary history of species or genes. Through this method, we could generate a phylogenetic tree that reflects the evolutionary relationships between members of the NAC gene family.
Finally, we classified the genes based on their similarity to homologous genes in Arabidopsis thaliana. Arabidopsis thaliana, as a model plant, has extensively studied genomic information, providing a valuable reference point for comparing and classifying the NAC genes in Cyclocarya paliurus. Through this homology-based classification approach, we could gain a deeper understanding of the functional and evolutionary characteristics of the NAC gene family in Cyclocarya paliurus.
2.4. Analysis of cis-Acting Elements in CpNAC Genes
To gain a deeper understanding of the regulatory mechanisms of CpNAC genes, this study employed the PlantCARE online analysis platform (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 29 September 2023). Through this tool, we conducted a detailed analysis of the promoter regions of CpNAC genes, aiming to identify critical data related to cis-regulatory elements. This step is crucial because cis-regulatory elements play a central role in gene expression regulation, and the analysis’ results will provide us with a deeper insight into the regulatory network of CpNAC genes.
2.5. Chromosome Distribution, Gene Duplication, and Collinearity Analysis of CpNAC Genes
In this study, we utilized various bioinformatics tools to analyze the CpNAC genes in Cyclocarya paliurus. Firstly, using the Circos tool, we successfully mapped these genes to specific chromosomes of Cyclocarya paliurus [31]. This step is crucial for understanding the distribution pattern of genes across chromosomes and their potential genetic regulatory mechanisms. Subsequently, we employed MCScanX software (version 2.0) with its default parameter settings to detect gene duplication events [32]. This analysis helps reveal the evolutionary history of the expansion of the CpNAC gene family. Finally, we utilized dual synteny plotter of TBtools (https://github.com/CJ-Chen/Tbtools, accessed on 8 October 2023) to generate synteny analysis plots [33]. These plots provide an intuitive representation of the relationships between homologous NAC genes, offering us important insights into their evolutionary relationships.
2.6. Expression Profiling and Validation Analysis under Salt Stress
In this study, we utilized the Transcript per Kilobase Million (TPM) metric to quantify transcript abundance. This metric standardizes the mapped read counts for each gene, allowing us to compare gene expression levels across different treatment conditions. Using this approach, we extracted expression profiles from the transcriptome sequencing data of Cyclocarya paliurus under two different treatment conditions, including a control group (CK) and a 0.4% NaCl (68.4 mM) salt stress treatment group [34,35].
To visualize the gene expression patterns of Cyclocarya paliurus under salt stress conditions, we generated a heatmap using TBtools software (version 1.098) (https://github.com/CJ-Chen/TBtools, accessed on 21 October 2023). This graphical representation allows for a clear comparison of gene expression differences between normal and salt stress conditions, providing valuable insights into the mechanisms underlying the response of Cyclocarya paliurus to salt stress.
Furthermore, we conducted gene expression counting, utilizing the count values to validate whether genes exhibit expression changes under salt stress conditions and to assess the genes’ relevance to the salt stress response.
2.7. Histochemical Staining
In this study, we prepare an X-Gal staining solution by dissolving X-Gal powder in dimethylformamide (DMF) to make a stock solution (typically 20 mg/mL). Dilute the stock solution with Z buffer to achieve a final concentration of 1 mg/mL X-Gal containing 1 μm β-mercaptoethanol and 1% Triton X-100. Subject yeast cells to repeated freeze–thaw cycles using liquid nitrogen to facilitate X-Gal entry. Subsequently, incubate the permeabilized yeast cells with the X-Gal staining solution at 37 °C for a suitable duration (usually 1–24 h) to allow β-galactosidase-mediated hydrolysis of X-Gal and the formation of a blue precipitate. Following the incubation period, wash the yeast cells with buffer to remove excess X-Gal solution.
2.8. Transcriptional Activation Activity Assay in Yeast
In this study, we focused on two genes, CpNAC132 and CpNAC040, which possess typical NAC transcription factor structures. To assess the transcriptional activation ability of NAC family members, we selected CpNAC132 and CpNAC040 as our subjects of study. Specifically, we constructed a variant of NAC132(D) lacking its transmembrane domain (with an amino acid range of 1–461) and a full-length CpNAC040 (with an amino acid range of 1–434), each cloned into the pGBKT7 vector containing the GAL4 system DNA-binding domain (DNA-BD). Subsequently, these constructs were transplanted into the Y187 yeast strain for functional analysis. As an experimental control, we also transformed the empty pGBKT7 vector.
In the transcriptional activation assay setup, we designed specific primers for the amplification of CpNAC132 and CpNAC040 segments. The amplification of CpNAC132 used the following forward primer sequence: 5′-CGGAATTCATGGCTATCTCGTTGGATTC-3′, and the following reverse primer sequence: 3′-GAAGATCTGTGAAGGAAGATCCTAGCTC-5′. For CpNAC040, the corresponding forward primer sequence was 5′-CGGAATTCATGGGAAAACATCGCGG-3′, and the reverse primer sequence was 3′-CGGGATCCTGGAGTCTGGGAAAAGC-5′.
A reporter gene (e.g., LacZ) is placed under the control of a promoter region that is responsive to a related transcription factor. When a transcription factor binds to a recognized sequence in the promoter, it activates the transcription of the reporter gene, resulting in a measurable product (e.g., the β-galactosidase activity of LacZ) [36]. We cultured transformed yeast strains on histidine-deficient media and observed yeast growth only when the CpNAC protein activated the expression of the His gene. X-Gal serves as a substrate for the β-galactosidase enzyme, which produces a blue-colored product in the presence of the enzyme, so that we can directly assess the transcriptional activation activity of the CpNAC protein.
2.9. Statistical Analysis
In this study, we performed statistical analysis using SPSS software (version 22). In the SPSS software, a one-way ANOVA test revealed a significant difference between the groups. Subsequently, we employed Tukey’s Honestly Significant Difference (HSD) post hoc test for multiple comparisons to further analyze the significant differences between the groups.
3. Results
3.1. Identification and Physicochemical Characteristics of CpNAC Genes
In this study, we conducted a comprehensive identification of the NAC gene family within the Cyclocarya paliurus genome. A total of 132 CpNAC genes were identified, and they were named CpNAC1 to CpNAC132 based on their positions on the chromosomes (Figure S2). These genes encoded proteins of varying lengths, ranging from 160 amino acids to 547 amino acids, with an average length of approximately 342 amino acids. For instance, CpNAC009 encoded the shortest protein, which had only 160 amino acids, while CpNAC121 encoded the longest protein, consisting of 547 amino acids.
Further analysis revealed that the open reading frame (ORF) lengths of CpNAC genes varied between 483 bp (CpNAC009) and 1644 bp (CpNAC121). Consequently, the predicted molecular weights of these proteins ranged from 18,316.99 Da (CpNAC009) to 61,526.79 Da (CpNAC121). Additionally, the predicted isoelectric points (pI) of the CpNAC proteins ranged from 4.54 (CpNAC052) to 9.43 (CpNAC053).
In the subcellular localization prediction analysis, we found that, except for the protein encoded by CpNAC111, which was predicted to localize to the cell membrane, the localization of the other 131 CpNAC proteins varied. Two were predicted to localize in the vacuoles, three in the cytoplasm, and eight on plastids, while the majority (118) were predicted to localize within the cell nucleus (Table S1). These detailed physicochemical characteristics and localization data provide a solid foundation for further investigating the functions and mechanisms of the CpNAC gene family in Cyclocarya paliurus.
3.2. Motif and Structural Analyses of CpNAC Genes
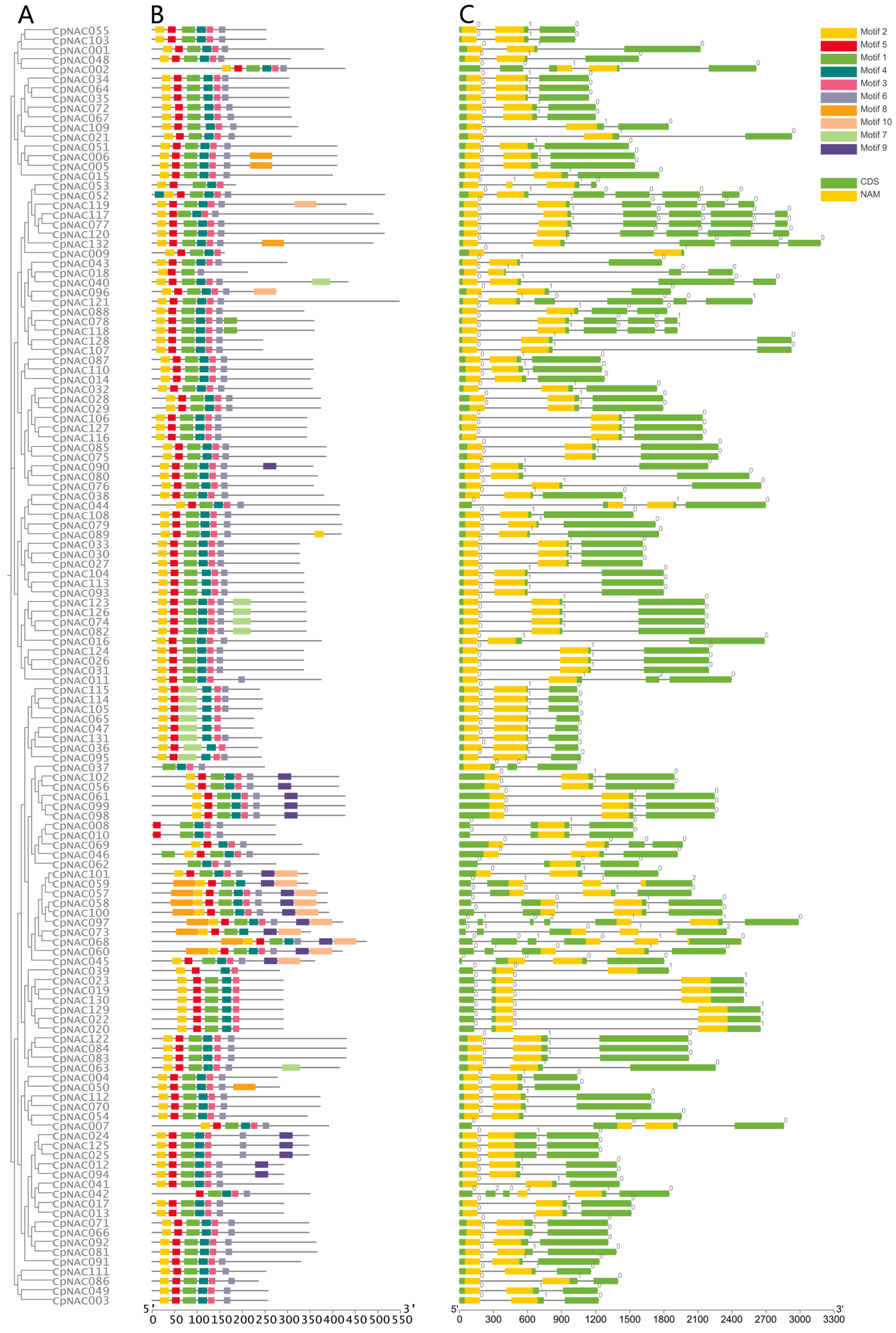
To study the interrelationships among the 132 CpNAC genes in Cyclocarya paliurus, we utilized the Maximum Likelihood (ML) method to construct a phylogenetic tree, aiming to reveal the evolutionary connections between these genes. Additionally, we conducted a detailed investigation of the conserved protein motif structures and gene structures (exons–introns) of the CpNAC genes (Figure 1). We identified a total of 10 conserved protein motifs in the CpNAC gene family and obtained motif lengths ranging from 15 to 50 amino acids (Figure S3). The analysis of the conserved protein motifs indicated that the 132 CpNAC proteins contain at least three conserved motifs, with motifs 1, 2, 3, 4, 5, and 6 appearing most frequently. For instance, the CpNAC097 protein contains up to nine motifs, while the CpNAC037 protein only has four (Figure 1B). The gene structure analysis revealed a similarity in the number of exons–introns within genes of the same clade, with the number of introns in CpNAC genes varying between two and six, with each gene containing at least two introns. Notably, the CpNAC097 gene has the most introns, amounting to six (Figure 1C). Integrating the results of the phylogenetic relationships and conserved motifs and gene structure analyses, we discovered a high level of conservation in the proteins of the CpNAC gene family [37,38]. This finding suggests that genes within the same group may have similar functions. However, further research is needed to more accurately determine the specific functions of these genes.
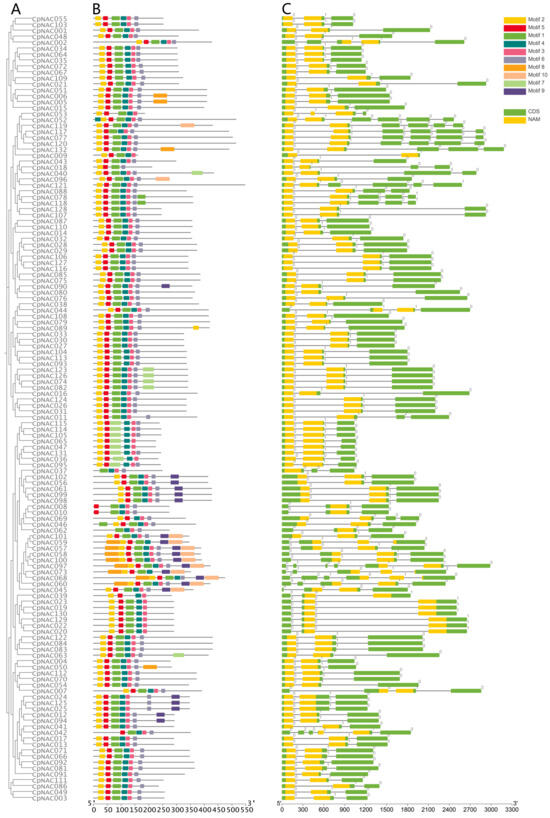
Figure 1.
Phylogenetic relationships, structure of conserved protein motifs, and gene structure of the C. paliurus NAC genes. (A) Phylogenetic tree was constructed from the full-length sequence of a C. paliurus NAC protein. (B) Structural formulae that constitute the C. paliurus NAC protein. Numbers indicate the stage of the corresponding intron. (C) Exon and intron structures of C. paliurus’ NAC structural genes. Green boxes indicate exons and black lines indicate introns. Yellow boxes highlight NAC structural domains. Numbers indicate the stage of the corresponding intron.
3.3. Systematic Evolutionary Analysis of CpNAC Genes
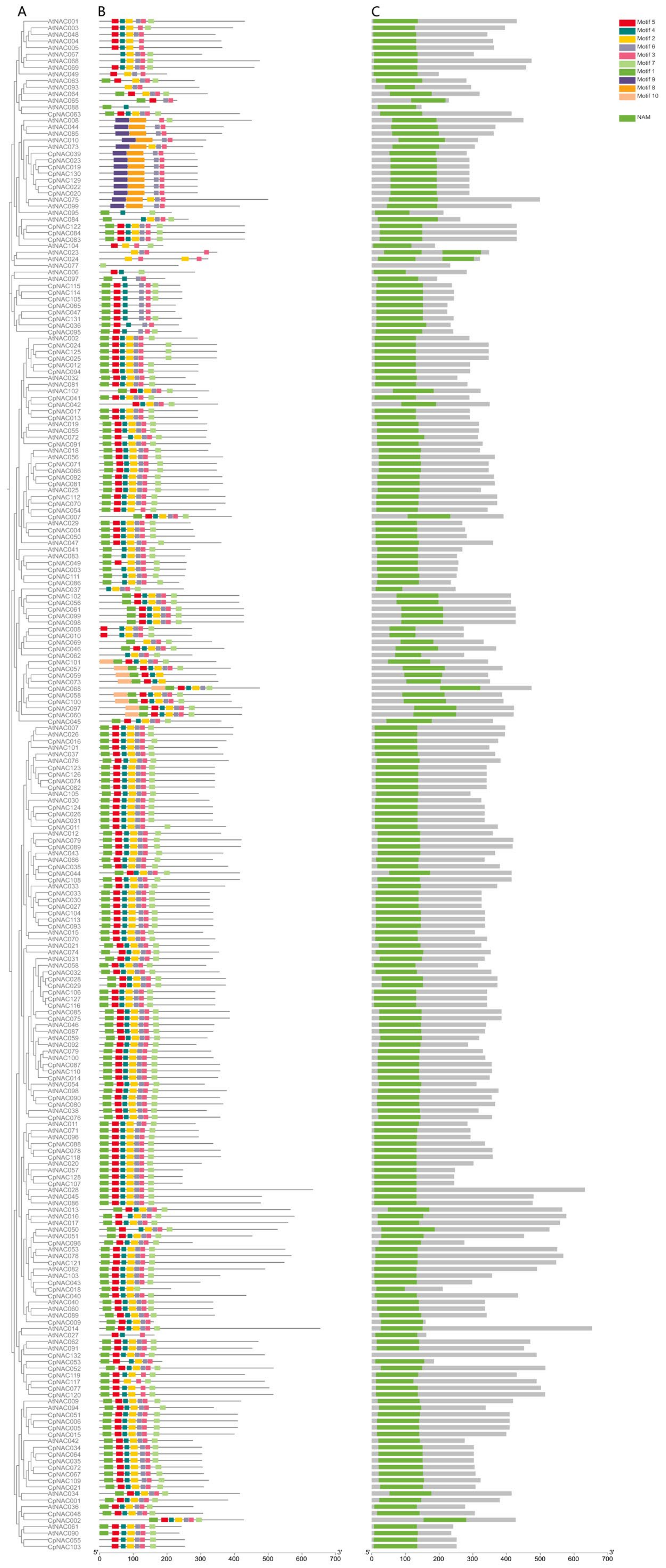
To study the differences in the NAC family between Cyclocarya paliurus (CpNAC) and Arabidopsis thaliana (AtNAC), we constructed a rooted Maximum Likelihood (ML) phylogenetic tree to reveal the similarities and inherent diversity between the NAC gene sequences of Cyclocarya paliurus and Arabidopsis thaliana (Figure 2). The results of the phylogenetic tree revealed the similarities in the NAC gene sequences between Cyclocarya paliurus and Arabidopsis thaliana, while also showcasing the diversity of CpNAC genes and proteins. This discovery not only deepens our understanding of the evolutionary relationships of the NAC gene family in different plant species but also provides a new perspective and direction for the further exploration of plant gene functions. Through this phylogenetic analysis, we are able to more comprehensively understand the interrelationships among these NAC genes, as well as their position and role in the evolutionary history of plant gene families.
3.4. Analysis of cis-Elements in CpNAC Genes
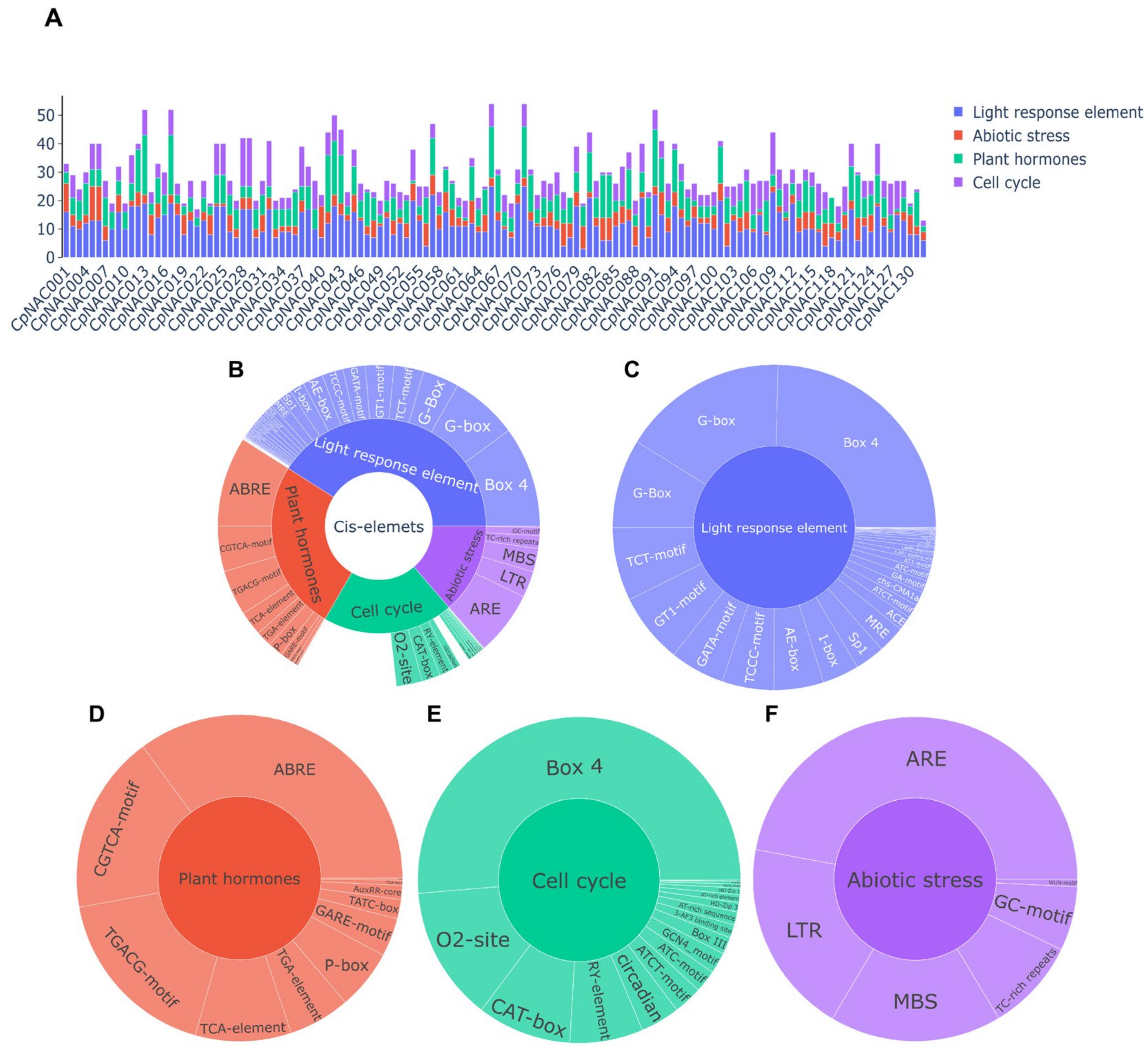
The regulation of gene expression is primarily achieved through cis-acting elements, and understanding how these elements are regulated by cellular signaling pathways is crucial for revealing the functions of these genes in different organisms and developmental processes [39]. Cis-acting elements include promoters, enhancers, silencers, insulators, etc., and they play an important role in regulating gene expression, cell differentiation, and tissue specialization [40,41,42]. They are also key subjects of study in genetics and molecular biology. We utilized the PlantCARE database to identify promoter regions. Specifically, we defined the putative promoter region as the 2000 base pairs upstream of the coding sequence (CDS) of the CpNAC genes and conducted an analysis of four major classes of commonly occurring cis-acting elements: light-responsive elements, plant hormone response elements, cell cycle-related elements, and abiotic stress response elements (Figure 3A). Within the CpNAC gene family, most cis-acting elements were found to overlap, except for eight unique cis-acting elements that do not overlap (Table S2). This overlap of cis-acting elements suggests potential co-regulation among these genes, indicating their involvement in similar biological processes or responses to stress. Conversely, the presence of unique, non-overlapping cis-acting elements in certain CpNAC family genes implies that these genes may have distinct functions or be responsive to specific environmental stimuli, distinct from those affecting other members of the family. The number of light-responsive elements is the greatest, followed by plant hormone response elements and cell cycle response elements, while the number of abiotic stress response elements is relatively smaller (Figure 3B). Of the light-responsive cis-elements, we identified various types, including Box 4, G-box, TCT-motif, GT1-motif, GATA-motif, and more. Box 4 is the most abundant light-responsive cis-element, while Pc-CMA2a is the least (Figure 3C). In terms of plant hormone response elements, there are ABRE, CGTCA-motif, TGACG-motif, etc., among which ABRE is the most abundant, while SARE is the least (Figure 3D). Cell cycle-related cis-elements include Box 4, O2-site, CAT-box, etc., with Box 4 being the most abundant and NON-box the least (Figure 3E). In terms of abiotic stress response elements, there are ARE, LTR, MBS, etc., among which ARE is the most abundant and WUN-motif the least (Figure 3F). These findings suggest that the CpNAC gene family plays a significant role in light responses, plant hormone signaling, cell cycle regulation, and responses to abiotic stress. Therefore, the transcriptional diversity of the CpNAC genes in different cis-regulatory elements indicates the need for further functional studies to reveal their specific mechanisms in plants’ growth, development, and stress responses.
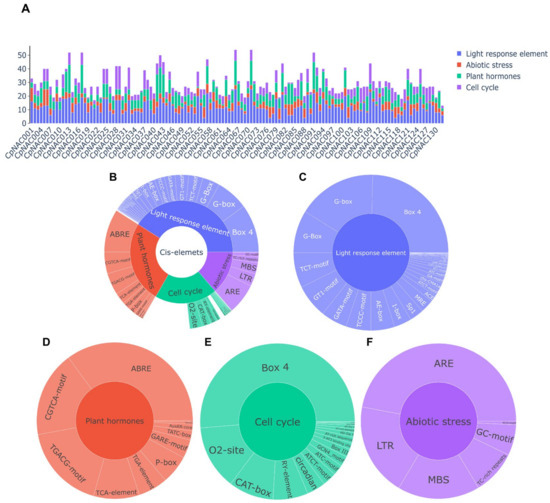
Figure 3.
Cis-regulatory elements in the promoter region of a CpNAC gene. Various cis-elements in the CpNAC gene are depicted in the figure, with different colors indicating different cis-elements. (A) The cis-element in the 132 CpNAC genes are represented as bar graphs; (B) the proportion of each type of cis-element in the CpNAC gene is represented as a pie chart; (C) light response cis-regulatory elements; (D) plant hormone cis-regulatory elements; (E) cell cycle cis-regulatory elements; and (F) abiotic stress cis-regulatory elements.
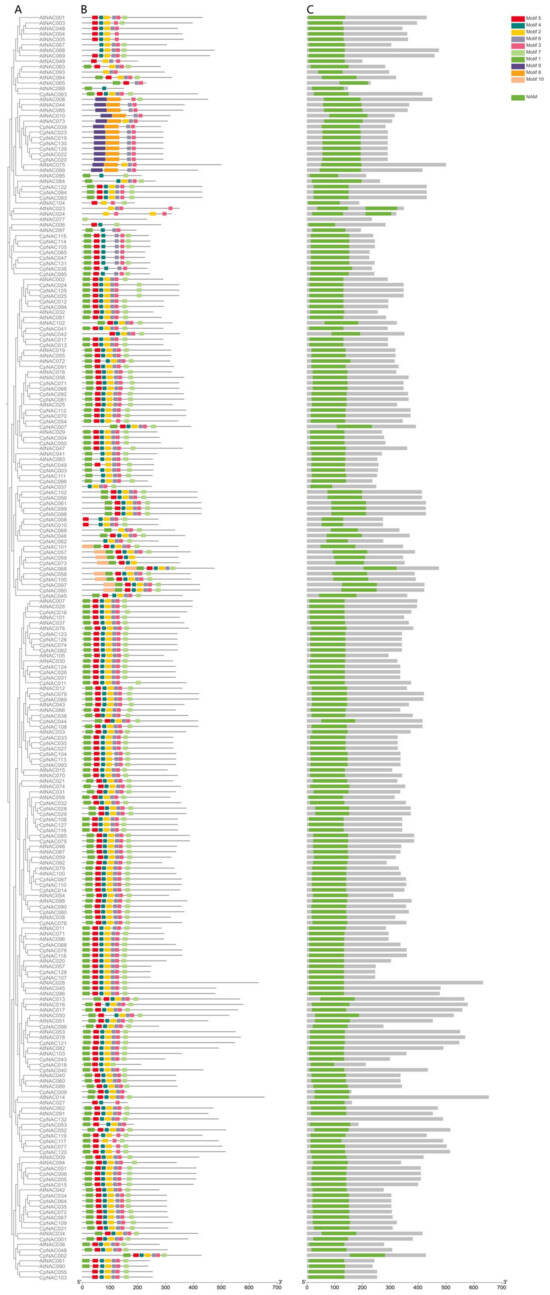
Figure 2.
Phylogenetic relationships, conserved protein structures and gene structures of the NAC genes of C. paliurus and A. thaliana. (A) Rooted Maximum likelihood (ML) phylogenetic tree based on NAC protein sequence comparison between C. paliurus (CpNAC) and A. thaliana (AtNAC). (B) Structural formulae of the NAC proteins of C. paliurus and A. thaliana. (C) Exon structures of C. paliurus and A. thaliana’s NAC structural genes. Gray boxes indicate exons and green boxes highlight NAC structural domains.
Figure 2.
Phylogenetic relationships, conserved protein structures and gene structures of the NAC genes of C. paliurus and A. thaliana. (A) Rooted Maximum likelihood (ML) phylogenetic tree based on NAC protein sequence comparison between C. paliurus (CpNAC) and A. thaliana (AtNAC). (B) Structural formulae of the NAC proteins of C. paliurus and A. thaliana. (C) Exon structures of C. paliurus and A. thaliana’s NAC structural genes. Gray boxes indicate exons and green boxes highlight NAC structural domains.
3.5. Chromosome Localization and Collinearity Analysis of CpNAC Genes
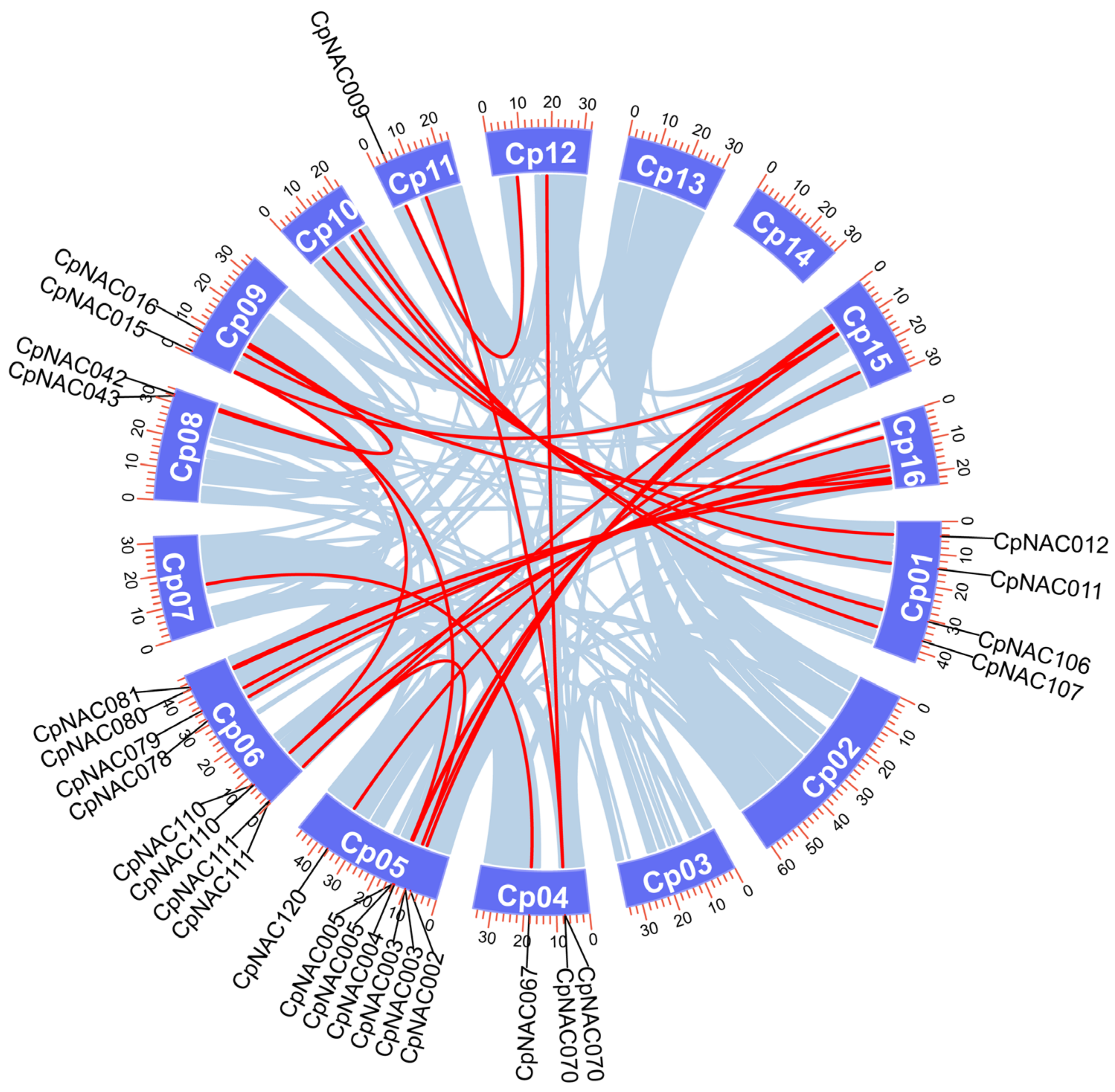
In this study, we conducted a systematic analysis of the distribution of the CpNAC gene family across chromosomes based on a detailed annotation of the Cyclocarya paliurus genome’s DNA sequences. The results indicate that these 132 CpNAC genes are unevenly distributed across the 15 chromosomes of Cyclocarya paliurus (Figure S2). Specifically, chromosome 12 has the highest number of CpNAC genes, a total of 16. Following closely are chromosomes 1 and 9, which possess 14 and 13 CpNAC genes, respectively. Chromosome 10 contains 12 CpNAC genes, while chromosomes 2, 3, and 6 each have 10 CpNAC genes. Chromosomes 4 and 5 have nine CpNAC genes each, while chromosome 16 has seven CpNAC genes. Chromosome 15 has six CpNAC genes, and chromosomes 7 and 8 each have five CpNAC genes. Chromosome 11 contains four CpNAC genes, while chromosome 13 has the fewest CpNAC genes, with only two. This distribution pattern provides crucial information for a deeper understanding of the function and regulatory mechanisms of the CpNAC gene family, laying the foundation for investigating the roles of CpNAC genes in the growth, development, and stress responses of Cyclocarya paliurus.
Furthermore, gene duplication analysis revealed the presence of 27 pairs of homologous genes among the 132 CpNAC genes (Figure 4). These homologous gene pairs are located on different chromosomes, indicating that they are formed through segmental duplication rather than tandem duplication. It is noteworthy that there is one gene duplication pair on chromosome 7, while no duplication pairs were found on chromosomes 2, 3, 13, and 14. These findings suggest that CpNAC genes have undergone segmental duplication events during their long evolutionary history, which has significant implications for the diversity and evolutionary history of the gene family, playing a crucial role in the adaptive evolution of the species.
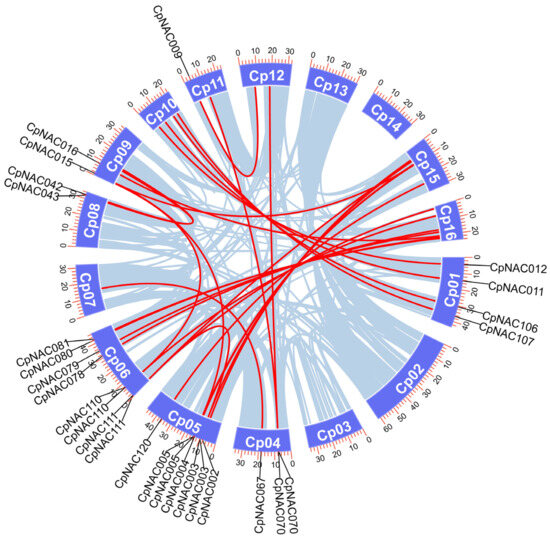
Figure 4.
Schematic diagram of the chromosomal distribution and interchromosomal relationships of the C. paliurus NAC genes. Gray lines indicate all homologous blocks in the C. paliurus genome and red lines indicate duplicated NAC gene pairs.
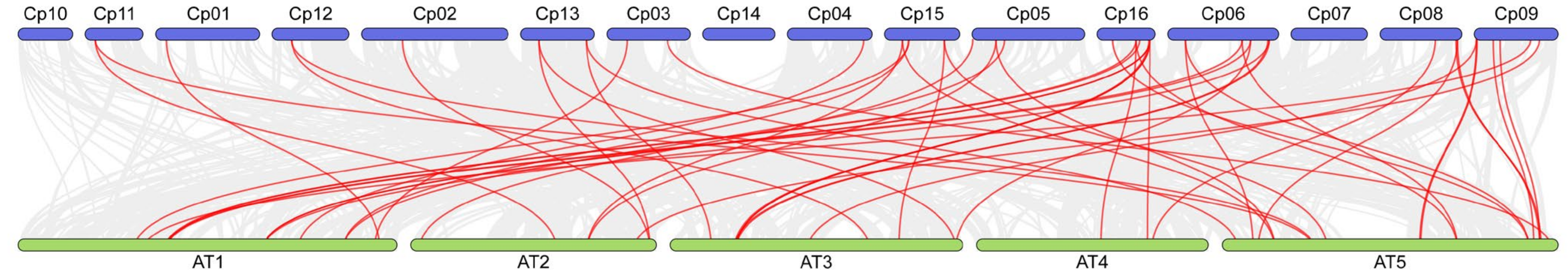
Finally, we conducted a comprehensive comparative analysis of the NAC genes in Cyclocarya paliurus and Arabidopsis thaliana. In both species, a total of 52 pairs of NAC genes were identified. Covariance analysis indicates a high covariance between the NAC genes of Cyclocarya paliurus and Arabidopsis thaliana, suggesting a conservation of these genes between the two species (Figure 5). Despite the presence of some duplicated or missing segments in the NAC genes, these results imply that the NAC gene families in Cyclocarya paliurus and Arabidopsis thaliana may have originated from a common ancestor, although further research is needed to confirm this hypothesis.
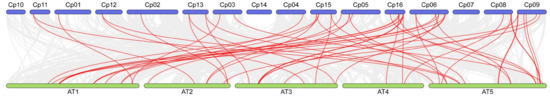
Figure 5.
Synteny analysis of NAC genes between C. paliurus and A. thaliana. Gray lines in the background indicate co-localized blocks in the genomes of C. paliurus and A. thaliana, while red lines highlight homologous NAC gene pairs. Species names prefixed with “Cp” and “At” indicate Cyclocarya paliurus and Arabidopsis thaliana, respectively.
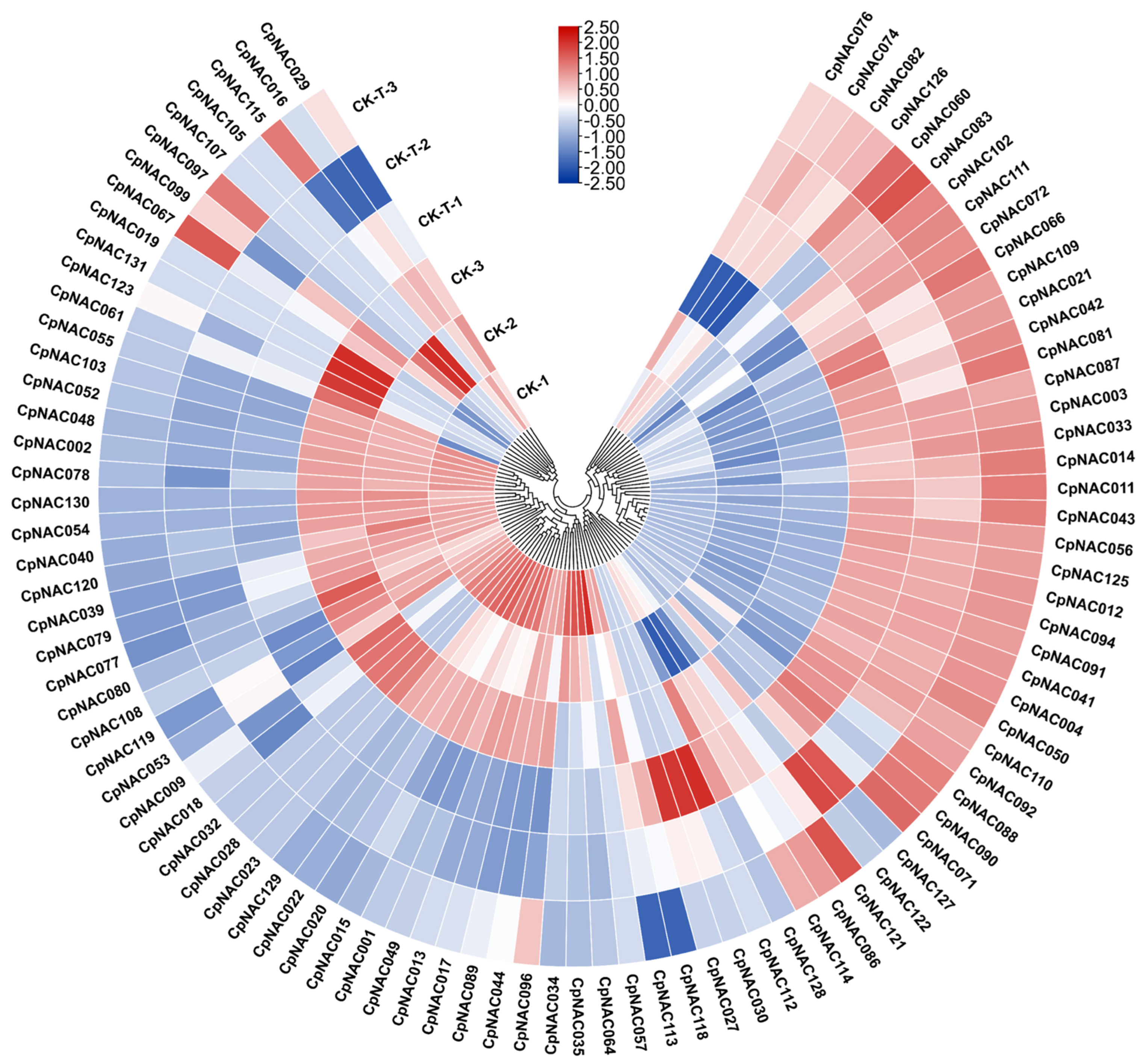
3.6. Salt Stress Induces a Response from the NAC Genes in Cyclocarya paliurus
In this study, we characterized the response of the CpNAC gene family to salt stress, offering novel insights into their potential regulatory roles under this adverse condition. We methodically assessed the expression patterns of CpNAC genes under salt stress by comparing transcriptomic data from control and salt-stressed samples. In this process, the expression of only 93 out of the 132 CpNAC genes were detected (Figure 6). Further investigation showed that 15 out of the 93 CpNAC genes were significant upregulated by salt stress (log2 fold change > 1, p < 0.05), and they were CpNAC033, CpNAC056, CpNAC011, CpNAC091, CpNAC004, CpNAC012, CpNAC094, CpNAC125, CpNAC110, CpNAC041, CpNAC102, CpNAC050, CpNAC087, CpNAC071, and CpNAC111 (Table S3); 20 out of 93 CpNAC genes were significant downregulated by salt stress (log2 fold change < −1, p < 0.05), and they were CpNAC015, CpNAC052, CpNAC129, CpNAC077, CpNAC120, CpNAC108, CpNAC103, CpNAC055, CpNAC002, CpNAC048, CpNAC119, CpNAC040, CpNAC044, CpNAC089, CpNAC079, CpNAC039, CpNAC130, CpNAC028, CpNAC032, and CpNAC080 (Table S3). Additionally, we found that, under salt stress conditions, approximately 14% of genes in the entire genome exhibited significant expression changes (log2 fold change > 1 or <−1, p < 0.05) compared to the control group (Table S4). Specifically, about 16% of the genes in the NAC gene family were significantly upregulated under salt stress conditions, while the proportion of upregulated genes among all other genes was only 5% (log2 fold change > 1, p < 0.05). Similarly, approximately 22% of the genes in the NAC gene family were significantly downregulated, compared to only 8% of genes being downregulated among all other genes (log2 fold change < −1, p < 0.05). These results indicate that, under salt stress conditions, a significant portion of the genome is affected, with notably higher responsiveness in the NAC gene family compared to the rest of the plant’s genes. The upregulation and downregulation of NAC family members’ expression under salt stress suggest that these genes play a crucial role in the plant’s response to salt stress, potentially contributing to enhanced salt tolerance or adaptation.
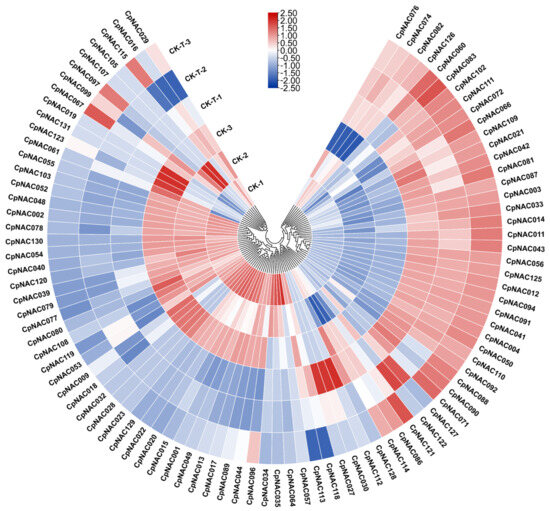
Figure 6.
Clustering expression analysis of the CpNAC genes under salt stress treatments. CK (control) and 0.4% NaCl (68.4 mM) represent two different treatments. The transcript abundance levels were normalized and hierarchically clustered using log2(FPKM + 1) comparisons between genes from different treatments, with expression values presented on a color scale. Gene expression differences are quantified as log2 fold changes with the statistical significance set at p < 0.05. “Up” indicates a log2 fold change > 1 (upregulation in salt treatment relative to control); “Down” indicates a log2 fold change < −1 (downregulation in salt treatment relative to control).
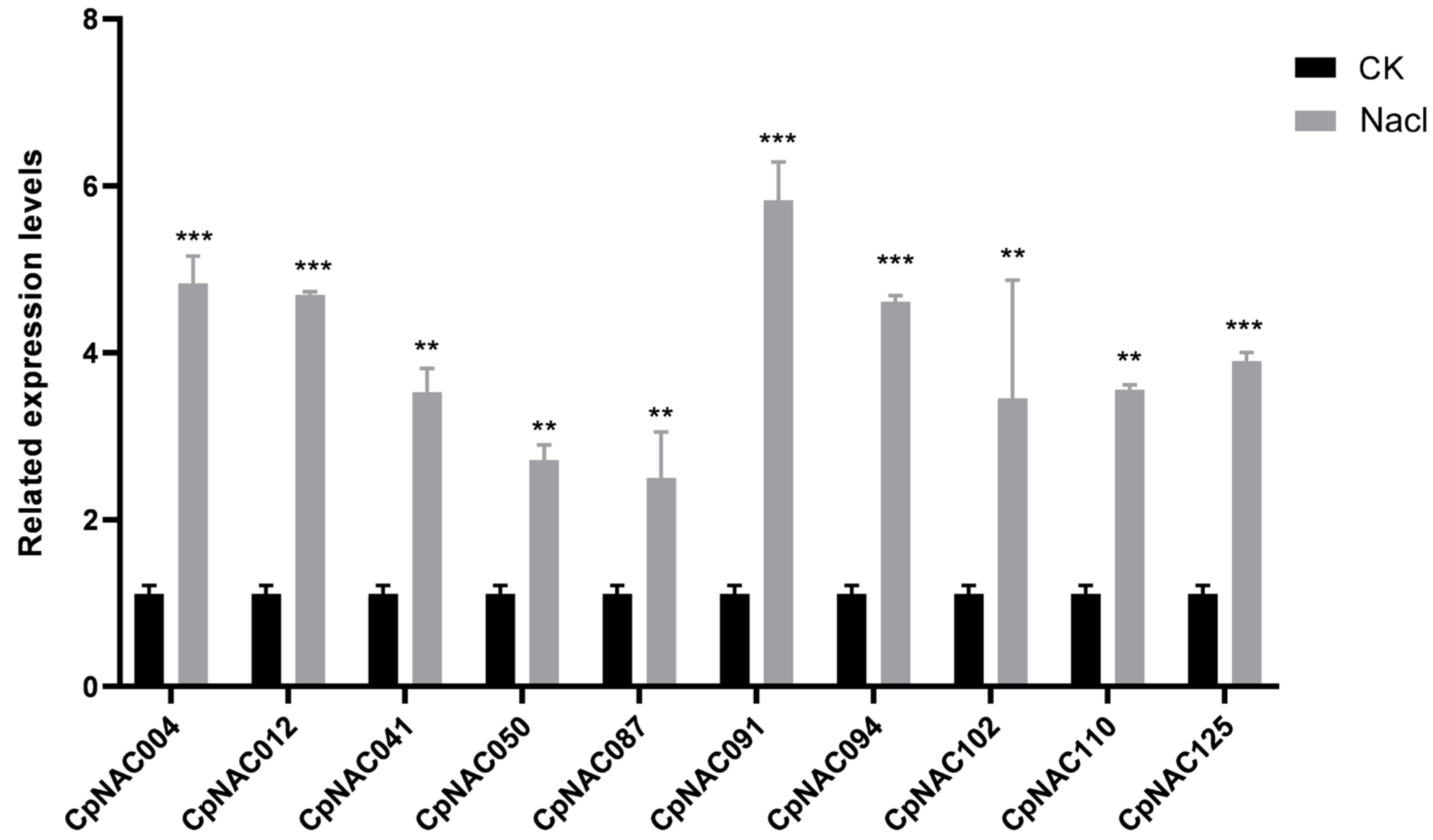
To verify these observations, we conducted gene expression counting to analyze the expression patterns of 10 randomly selected NAC genes, which were CpNAC004, CpNAC012, CpNAC041, CpNAC050, CpNAC087, CpNAC091, CpNAC094, CpNAC102, CpNAC110, and CpNAC125. Among these, CpNAC004, CpNAC012, CpNAC091, and CpNAC094 were identified to have significantly higher expressions under salt stress conditions. The trend of upregulation observed in these genes aligns with their overall transcriptomic profile (Figure 7). This observation underlines the inducible nature of NAC gene expression in response to salt stress, suggesting systematic upregulation as an adaptive strategy. Collectively, these results provide a foundational understanding of the dynamic response of the CpNAC gene family to salt stress in Cyclocarya paliurus. This study not only enhances our comprehension of the molecular mechanisms employed by Cyclocarya paliurus in salt stress adaptation but also sets the stage for the future functional characterization of these genes, potentially contributing to the development of stress-resilient plant varieties.
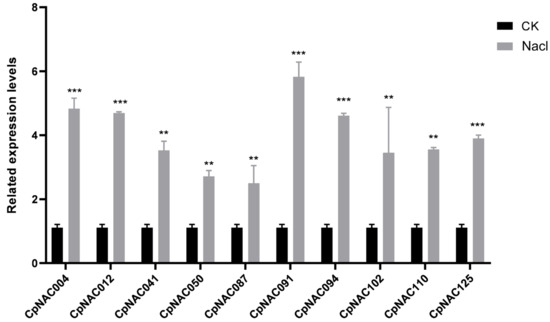
Figure 7.
Expression profiles of 10 candidate CpNAC genes under salt stress treatments, presented as error bar graphs. Significant differences identified using Duncan’s test (p < 0.05), conducted using SPSS v.22 after an analysis of variance, are indicated with asterisks. “**” indicates p < 0.01, “***” indicates p < 0.001.
3.7. The Transcriptional Activation Activity Analysis of CpNAC132 (D) and CpNAC040
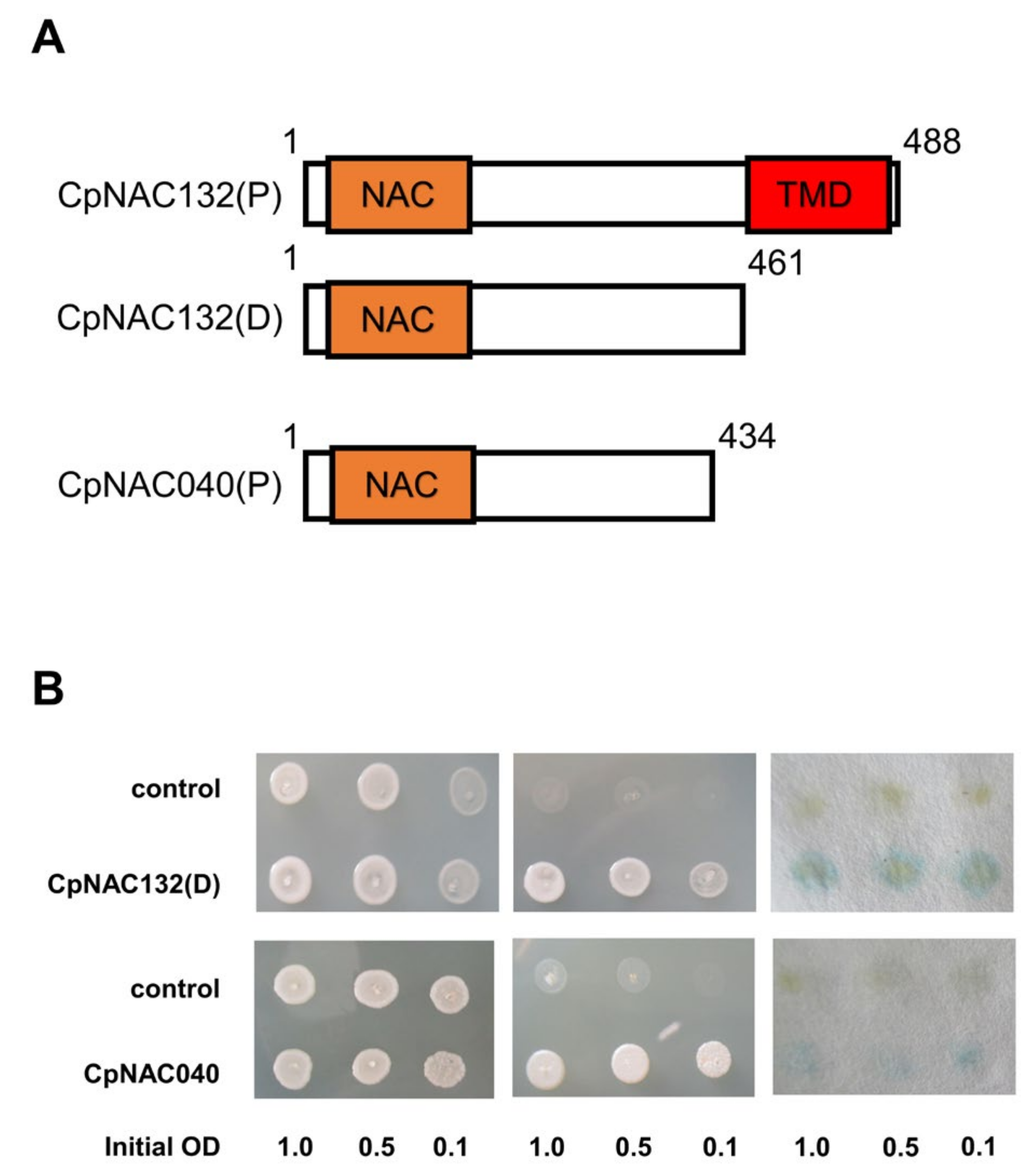
The NAC gene family includes both membrane-bound proteins and nuclear proteins. Currently, most reported NAC family genes possess transcriptional activation activity [43,44,45], such as nuclear proteins (NAC027, NAC039, NAC040) and membrane-bound proteins with their transmembrane domain removed (NAC009, NAC062, NAC132). In our preliminary analysis, there were 118 nuclear proteins and 14 membrane proteins. Among them, CpNAC040 is a homologue of AtNAC103, which has been shown to have transcriptional activation activity, and CpNAC132 is a homologue of AtNAC062/091, which has transcriptional activation activity after the removal of its transmembrane domain. CpNAC132 is a full-length protein with a transmembrane domain (TMD) located after its NAC domain, between amino acids 461 and 488, while CpNAC040 is a truncated protein lacking a transmembrane domain. (Figure 8A). To verify the transcriptional activation activity of a membrane protein (CpNAC132) and a nuclear protein (CpNAC040), we conducted transcriptional activation assays in yeast cells. The experimental results showed that, in the absence of TRP medium, both the control group and the experimental group of yeast colonies were able to grow. However, in the absence of TRP-His medium, the growth of yeast colonies in the control group was inhibited, while the growth of yeast colonies in the experimental group was not inhibited (Figure 8B). This result confirmed the transcriptional activation activity of CpNAC132(D) and CpNAC040 proteins, providing insights into their potential functions in gene regulation. In our transcriptional activation assay, yeast cells containing CpNAC132 and CpNAC040 turned blue in the presence of X-Gal, confirming their ability to activate the LacZ reporter gene. This color reaction evidence is consistent with our molecular research results, confirming the role of CpNAC132 and CpNAC040 as transcriptional activation factors. In summary, our transcriptional activation activity experiments directly validate whether the CpNAC132(D) and CpNAC040 proteins possess the ability to activate gene expression, a critical step in understanding the intracellular mechanisms of these proteins. This provides direction for future research into their specific roles in plants’ development and responses to environmental stress.
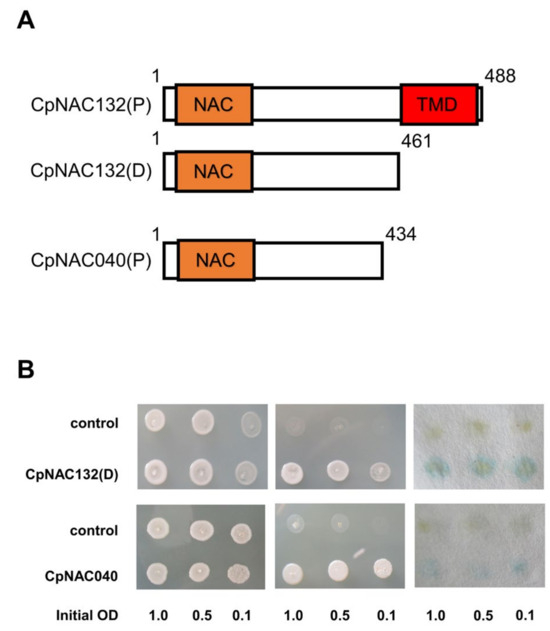
Figure 8.
Transcriptional activation assay of CpNAC132(D) and CpNAC040. (A) Full-length unspliced CpNAC132(P), CpNAC040(P), and truncated CpNAC132(D); scale bar represents 50 μm. (B) Analysis employed to determine the transcriptional activation activity of CpNAC132(D) and CpNAC040 in yeast cells, utilizing a series of yeast cell dilutions in the assessment.
4. Discussion
For a long time, traditional Chinese medicine has widely employed Cyclocarya paliurus, a plant renowned for its therapeutic effects, in the treatment of various diseases. Cyclocarya paliurus contains numerous bioactive compounds, such as flavonoids, polysaccharides, triterpenoid acids, and essential trace elements for the human body. These compounds give Cyclocarya paliurus a wide range of physiological and biological activities, making it an important resource in the field of herbal medicine. Cyclocarya paliurus is commonly used in the treatment of conditions such as hyperlipidemia and coronary heart disease due to its unique medicinal properties. Studies have shown that Cyclocarya paliurus leaves can effectively reduce cholesterol levels and lipid levels, which is beneficial for cardiovascular health [46]. Furthermore, Cyclocarya paliurus leaves also have the ability to lower blood glucose levels, providing a potential natural therapy for controlling high blood sugar in diabetic patients [47]. Further research has revealed that the polysaccharide components in Cyclocarya paliurus possess antioxidant properties, which can alleviate oxidative stress in the liver and kidneys [48]. Since oxidative stress can potentially damage cells and tissues and lead to various diseases, the protective role of Cyclocarya paliurus polysaccharides is particularly important. These research findings highlight the potential health benefits of Cyclocarya paliurus as an herbal remedy in various areas, but a deeper understanding of its specific therapeutic and health-promoting functions requires further research.
Despite the significant medicinal value of Cyclocarya paliurus, it still faces challenges posed by environmental stressors. Among them, salt stress has a detrimental effect on the growth of Cyclocarya paliurus, subsequently affecting the yield and quality of its bioactive compounds. Salt stress has been shown to affect multiple stages of the plant cell cycle, particularly the G1/S and G2/M transitions [49]. Under salt stress conditions, plant cells often exhibit cell cycle delay, primarily because cells require more time to repair the DNA damage caused by salt stress and to replenish the energy and resources needed to enter the next cell cycle phase. Additionally, salt stress can induce an excessive production of reactive oxygen species (ROS) within plant cells, including superoxide anions (O2−), hydrogen peroxide (H2O2), and hydroxyl radicals (OH−). While ROS may participate in signal transduction processes at low concentrations, excessive ROS production can cause structural cellular damage, lipid peroxidation, and DNA damage, thereby affecting plant growth and development [50]. However, plants have evolved a series of protective mechanisms to cope with external threats, and the NAC gene family, in particular, plays a crucial role in them. Therefore, knowledge of the NAC gene family is essential for the conservation and maximization of the medicinal properties of this valuable medicinal plant.
The NAC gene family is a large transcription factor group widely present in all plant species. This study focuses on an in-depth investigation of the NAC gene family in Cyclocarya paliurus. In the genome of Cyclocarya paliurus, we identified 132 NAC genes, and they were named based on their chromosome positions, and labeled as CpNAC1 to CpNAC132. Through the analysis of the conserved motifs in these CpNAC proteins, we found that these NAC proteins contain at least three conserved motifs, indicating significant conservation in their protein structures. This finding is consistent with previous research on the NAC genes in other plant species. Notably, most of the conserved motifs are located at the N-terminus of CpNAC proteins, which is considered an essential component of NAC proteins [51]. This result aligns with previous findings in studies on potato, emphasizing the importance of the N-terminal region in NAC proteins [52]. Additionally, we observed variations in the number of introns among genes in the CpNAC gene family, which ranged from 2 to 6 introns. These differences also showed similarities with genes within the same phylogenetic lineage, further indicating that the genetic composition of the NAC genes in Cyclocarya paliurus is similar to that of other plant species reported previously [53].
The analysis of the NAC cis-regulatory elements in Cyclocarya paliurus revealed the presence of various responsive elements, including Box 4, ABRE, O2-site, ARE, and other cis-elements. This suggests that the NAC genes in Cyclocarya paliurus may play important roles in light responses, plant hormone signaling, cell cycle regulation, and responses to abiotic stress. For example, the ABRE cis-regulatory element has been shown to regulate the expression of dehydration and salt-responsive genes in both Cyclocarya paliurus and Arabidopsis thaliana [54]. Furthermore, we conducted a repeat analysis of the CpNAC genes and found 27 pairs of CpNAC genes with segmental duplications, but no tandem duplicated gene pairs were identified. This suggests a close relationship between the NAC genes in Arabidopsis thaliana and Cyclocarya paliurus, with the possibility that they share a common ancestor and have retained their respective functions. We also delved into the regulatory mechanisms of the NAC genes in Cyclocarya paliurus in response to salt stress. Our research results indicate that the regulation of NAC genes can enhance the salt tolerance of Cyclocarya paliurus and may improve the quality of its secondary products. Therefore, in-depth studies on NAC genes are crucial for addressing the challenges of cultivating Cyclocarya paliurus, a valuable resource, under unfavorable conditions.
Under salt stress treatment conditions, we observed varying levels of expression in the CpNAC genes. Specifically, after eliminating the genes with an undetectable expression, a comparison between the control group and the experimental group revealed changes in the expression of 35 genes. Moreover, the expression patterns of 10 randomly selected NAC genes analyzed in our gene expression counting were consistent with the transcriptome data. These findings suggest that these CpNAC genes play a significant role in the plant’s response to salt stress. Through further validation using yeast experiments, it was observed that yeast cells containing CpNAC132 (D) and CpNAC040 proteins exhibited a blue reaction in the presence of X-Gal, confirming their ability to activate the LacZ reporter gene. This color reaction result is consistent with our molecular research findings and confirms the roles of the CpNAC132 (D) and CpNAC040 proteins as transcriptional activation factors, providing experimental support for a deeper understanding of their functions.
These findings provide important guidance for further research on the genetic improvement of Cyclocarya paliurus, especially in enhancing its environmental resistance and agronomic traits. Additionally, investigating protein–protein interactions helps us to gain a deeper understanding of the molecular mechanisms underlying the physiological and biochemical processes of plants during their growth, development, and responses to environmental stress. This provides new clues and entry points for future research, which may further elucidate the crucial roles of the NAC proteins in plants. This not only contributes to a deeper understanding of plant biology but also offers additional solutions for agricultural and ecosystem management.
5. Conclusions
In this study, based on published Cyclocarya paliurus genome data, we successfully identified 132 CpNAC genes. Through a detailed analysis of each gene, we revealed the high structural conservation of the CpNAC gene family, which are involved in various biological processes including light responses, phytohormone responses, cell cycle regulation, and abiotic stress responses. Additionally, an NAC gene co-linearity analysis with the model plant Arabidopsis thaliana suggests that they may have originated from a common ancestor. Under salt stress conditions, the changes in the expression of 35 CpNAC genes highlight their significant role in Cyclocarya paliurus’s adaptation to salt stress environments. Furthermore, analysis of the transcriptional activation activity of the CpNAC132(D) and CpNAC040 proteins in yeast cells confirms the existence of a positive regulatory role of the CpNAC gene as a transcription factor in signal transduction. Overall, this study not only enhances our understanding of Cyclocarya paliurus’s adaptation to salt stress but also provides strong support for future research into plant responses to environmental stress and the development of salt-tolerant crops. These findings are significant for a deeper understanding of the role of plant gene families in environmental adaptability and for their future application in agricultural biotechnology.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/f15030479/s1, Supplementary Figure S1: a visual image of Cyclocarya paliurus. Supplementary Figure S2: chromosomal positions of CpNAC genes in Cyclocarya paliurus. Supplementary Figure S3: conserved motifs in the NAC protein sequence of Cyclocarya paliurus. Supplementary Table S1: list of the 132 CpNAC genes identified in this study. Supplementary Table S2: list of the eight independent cis-elements identified in this study. Supplementary Table S3: the expression of 35 CpNAC genes was significantly changed under salt stress conditions. Supplementary Table S4: list of genes significantly changed under salt stress conditions.
Author Contributions
Z.Y. (Zhengting Yang) and K.L. designed the research. Z.Y. (Ziwei Yang), Y.A., Q.Y., N.Z., X.L., F.H., Y.Z. and M.T. performed the study. Z.Y. (Ziwei Yang), Y.A., Q.Y. and F.H. analyzed the data. Z.Y. (Ziwei Yang), Y.A., Q.Y. and F.H. wrote the manuscript, Z.Y. (Zhengting Yang) revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This project was funded by National Natural Science Foundation of China (grant number 32360074), Guizhou Provincial Natural Science Foundation of Department of Education [2022]077, the Joint Fund of the National Natural Science Foundation of China and the Karst Science Research Center of Guizhou Province (grant number U1812401).
Data Availability Statement
All data are available upon reasonable request.
Conflicts of Interest
The author Hui Yu was employed by the company Taixing HSCT Chemical Corporation. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Leung, H.; Chan, L.; Law, C.; Li, M.; Lam, H. Twenty years of mining salt tolerance genes in soybean. Mol. Breed. 2023, 43, 45. [Google Scholar] [CrossRef]
- Shultana, R.; Zuan, A.T.K.; Naher, U.A.; Islam, A.K.M.M.; Rana, M.M.; Rashid, M.H.; Irin, I.J.; Islam, S.S.; Rim, A.A.; Hasan, A.K. The PGPR Mechanisms of Salt Stress Adaptation and Plant Growth Promotion. Agronomy 2022, 12, 2266. [Google Scholar] [CrossRef]
- Fatima, T.; Mishra, I.; Verma, R.; Arora, N.K. Mechanisms of halotolerant plant growth promoting Alcaligenes sp. involved in salt tolerance and enhancement of the growth of rice under salinity stress. 3 Biotech 2020, 10, 361. [Google Scholar] [CrossRef]
- Balasubramaniam, T.; Shen, G.; Esmaeili, N.; Zhang, H. Plants’ Response Mechanisms to Salinity Stress. Plants 2023, 12, 2253. [Google Scholar] [CrossRef]
- Du, Z.; Ren, Z.; Yu, B.; Zhu, J.; Li, J. Impacts of climate change on the global distribution of Cyclocarya paliurus. Biologia 2023, 78, 41–53. [Google Scholar] [CrossRef]
- Sun, Y.; Ho, C.; Liu, Y.; Zhan, S.; Wu, Z.; Zheng, X.; Zhang, X. The Modulatory Effect of Cyclocarya paliurus Flavonoids on Intestinal Microbiota and Hypothalamus Clock Genes in a Circadian Rhythm Disorder Mouse Model. Nutrients 2022, 14, 2308. [Google Scholar] [CrossRef]
- Yang, W.; Zhuang, J.; Tian, Y.; Wan, S.; Ding, S.; Zhang, M.; Fang, S. Technical Scheme for Cutting Seedlings of Cyclocarya paliurus under Intelligent Control of Environmental Factors. Sustainability 2023, 15, 10690. [Google Scholar] [CrossRef]
- Song, L.; Li, W.; Chen, X. Transcription factor is not just a transcription factor. Trends Plant Sci. 2022, 27, 1087–1089. [Google Scholar] [CrossRef]
- Yang, O.; Popova, O.V.; Süthoff, U.; Lüking, I.; Dietz, K.; Golldack, D. The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 2009, 436, 45–55. [Google Scholar] [CrossRef]
- Lee, S.; Yang, S.H.; Berberich, T.; Miyazaki, A.; Kusano, T. Characterization of AtbZIP2, AtbZIP11 and AtbZIP53 from the group S basic region-leucine zipper family in Arabidopsis thaliana. Plant Biotechnol.-Nar. 2006, 23, 249–258. [Google Scholar] [CrossRef][Green Version]
- Taghipour, A.M.; Tarang, A.; Zare, N.; Pourebrahim, M.; Seighalani, R.; Selakjani, M.G. Expression analysis of five critical transcription factors (TFs)‘OsbHLH148, OsbZIP72, OsMYB2, OsNAC6 and TRAB1’ in response to drought stress in contrasting Iranian rice genotypes. Plant Omics 2016, 9, 327–333. [Google Scholar] [CrossRef]
- Aydin, G. Oryza sativa Osmyb4 geni aktarılmış transgenik patateste Osmyb4 gen ifadesinin tuzluluk toleransına etkisi. Anadolu J. Agric. Sci. 2020, 35, 1308–8769. [Google Scholar] [CrossRef][Green Version]
- Qu, Y.; Guan, R.; Yu, L.; Berkowitz, O.; David, R.; Whelan, J.; Ford, M.; Wege, S.; Qiu, L.; Gilliham, M. Enhanced reactive oxygen detoxification occurs in salt-stressed soybean roots expressing GmSALT3. Physiol. Plant. 2022, 174, e13709. [Google Scholar] [CrossRef]
- Dong, X.; Jiang, Y.; Yang, Y.; Xiao, Z.; Bai, X.; Gao, J.; Tan, S.; Hur, Y.; Hao, S.; He, F. Identification and Expression Analysis of the NAC Gene Family in Coffea canephora. Agronomy 2019, 9, 670. [Google Scholar] [CrossRef]
- Liu, S.; Guan, Y.; Weng, Y.; Liao, B.; Tong, L.; Hao, Z.; Chen, J.; Shi, J.; Cheng, T. Genome-wide identification of the NAC gene family and its functional analysis in Liriodendron. BMC Plant Biol. 2023, 23, 415. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hu, F.; Yao, Z.; Zhao, X.; Chu, G.; Ye, J. Comprehensive genomic characterisation of the NAC transcription factor family and its response to drought stress in Eucommia ulmoides. Peerj 2023, 11, e16298. [Google Scholar] [CrossRef]
- Pimenta, M.R.; Silva, P.A.; Mendes, G.C.; Alves, J.R.; Caetano, H.D.N.; Machado, J.P.B.; Brustolini, O.J.B.; Carpinetti, P.A.; Melo, B.P.; Silva, J.C.F.; et al. The Stress-Induced Soybean NAC Transcription Factor GmNAC81 Plays a Positive Role in Developmentally Programmed Leaf Senescence. Plant Cell Physiol. Cell Physiol. 2016, 57, 1098–1114. [Google Scholar] [CrossRef]
- Qian, Y.; Xi, Y.; Xia, L.; Qiu, Z.; Liu, L.; Ma, H. Membrane-Bound Transcription Factor ZmNAC074 Positively Regulates Abiotic Stress Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2023, 24, 16157. [Google Scholar] [CrossRef]
- Li, F.; Sun, H.J.; Jiao, Y.; Wang, F.L.; Yang, J.G.; Shen, L. Viral infection-induced endoplasmic reticulum stress and a membrane-associated transcription factor NbNAC089 are involved in resistance to virus in Nicotiana benthamiana. Plant Pathol. 2018, 67, 233–243. [Google Scholar] [CrossRef]
- Sukiran, N.L.; Ma, J.C.; Ma, H.; Su, Z. ANAC019 is required for recovery of reproductive development under drought stress in Arabidopsis. Plant Mol. Biol. 2019, 99, 161–174. [Google Scholar] [CrossRef]
- Alshareef, N.O.; Otterbach, S.L.; Allu, A.D.; Woo, Y.H.; de Werk, T.; Kamranfar, I.; Mueller-Roeber, B.; Tester, M.; Balazadeh, S.; Schmöckel, S.M. NAC transcription factors ATAF1 and ANAC055 affect the heat stress response in Arabidopsis. Sci. Rep. 2022, 12, 11264. [Google Scholar] [CrossRef]
- Huang, J.; Piater, L.; Dubery, I. The NAC transcription factor gene ANAC072 is differentially expressed in Arabidopsis thaliana in response to microbe-associated molecular pattern (MAMP) molecules. Physiol. Mol. Plant Pathol. 2012, 80, 19–27. [Google Scholar] [CrossRef]
- Fan, C. Genetic mechanisms of salt stress responses in halophytes. Plant Signal Behav. 2020, 15, 1704528. [Google Scholar] [CrossRef]
- Rahman, M.M.; Mostofa, M.G.; Keya, S.S.; Siddiqui, M.N.; Ansary, M.M.U.; Das, A.K.; Rahman, M.A.; Tran, L.S. Adaptive Mechanisms of Halophytes and Their Potential in Improving Salinity Tolerance in Plants. Int. J. Mol. Sci. 2021, 22, 10733. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, C.; Wuriyanghan, H.; Lei, Z.; Tang, Y.; Zhang, H.; Zhao, X. Exogenous Selenium Endows Salt-Tolerant and Salt-Sensitive Soybeans with Salt Tolerance through Plant-Microbial Coactions. Agronomy 2023, 13, 2271. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. Acids Res. 2015, 44, D279–D285. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Yu, C.; Chen, Y.; Lu, C.; Hwang, J. Prediction of Protein Subcellular Localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Guo, A.; Zhu, Q.; Chen, X.; Luo, J. GSDS: A gene structure display server. Yi Chuan = Hered./Zhongguo Yi Chuan Xue Hui Bian Ji 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.; Marra, M. CIRCOS: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xie, T.; Chen, C.; Luan, A.; Long, J.; Li, C.; Ding, Y.; He, Y. Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple (Ananas comosus). BMC Genom. 2017, 18, 503. [Google Scholar] [CrossRef]
- Yang, Z.; He, F.; An, Y.; Zhang, N.; Fan, S.; Tang, M.; Li, K. Genome-Wide Identification and Expression Analysis of Salt Tolerance-Associated WRKY Family Genes in Cyclocarya paliurus. Forests 2023, 14, 1771. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, L.; Liu, Y.; Shang, X.; Fang, S. Identification and Expression Analysis of R2R3-MYB Family Genes Associated with Salt Tolerance in Cyclocarya paliurus. Int. J. Mol. Sci. 2022, 23, 3429. [Google Scholar] [CrossRef]
- Merkwitz, C.; Blaschuk, O.; Schulz, A.; Ricken, A.M. Comments on Methods to Suppress Endogenous β-Galactosidase Activity in Mouse Tissues Expressing the LacZ Reporter Gene. J. Histochem. Cytochem. 2016, 64, 579–586. [Google Scholar] [CrossRef]
- Liao, G.; Duan, Y.; Wang, C.; Zhuang, Z.; Wang, H. Genome-Wide Identification, Characterization, and Expression Analysis of the NAC Gene Family in Litchi chinensis. Genes 2023, 14, 1416. [Google Scholar] [CrossRef]
- Guo, H.; Cui, Z.; Zhang, Y.; Wang, C. Sequence characterization and expression analysis of NAC genes from Betula platyphylla. Trees 2017, 31, 1919–1931. [Google Scholar] [CrossRef]
- Song, H.; Wang, Q.; Zhang, Z.; Lin, K.; Pang, E. Identification of clade-wide putative cis-regulatory elements from conserved non-coding sequences in Cucurbitaceae genomes. Hortic. Res. 2023, 10, uhad038. [Google Scholar] [CrossRef]
- Zhang, J.; Yuan, T.; Duan, X.; Wei, X.; Shi, T.; Li, J.; Russell, S.D.; Gou, X. Cis-Regulatory Elements Determine Germline Specificity and Expression Level of an Isopentenyltransferase Gene in Sperm Cells of Arabidopsis. Plant Physiol. 2016, 170, 1524–1534. [Google Scholar] [CrossRef]
- Juneja, P.; Quinn, A.; Jiggins, F.M. Latitudinal clines in gene expression and cis-regulatory element variation in Drosophila melanogaster. BMC Genom. 2016, 17, 981. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, L.; Wu, D.; Zhang, N.; Hua, J. Polymorphisms in cis-elements confer SAUR26 gene expression difference for thermo-response natural variation in Arabidopsis. New Phytol. 2021, 229, 2751–2764. [Google Scholar] [CrossRef]
- Yan, J.; Tong, T.; Li, X.; Chen, Q.; Dai, M.; Niu, F.; Yang, M.; Deyholos, M.K.; Yang, B.; Jiang, Y. A Novel NAC-Type Transcription Factor, NAC87, from Oilseed Rape Modulates Reactive Oxygen Species Accumulation and Cell Death. Plant Cell Physiol. 2017, 59, 290–303. [Google Scholar] [CrossRef]
- Niu, F.; Wang, C.; Yan, J.; Guo, X.; Wu, F.; Yang, B.; Deyholos, M.K.; Jiang, Y. Functional characterization of NAC55 transcription factor from oilseed rape (Brassica napus L.) as a novel transcriptional activator modulating reactive oxygen species accumulation and cell death. Plant Mol. Biol. 2016, 92, 89–104. [Google Scholar] [CrossRef]
- Bi, Y.; Wang, H.; Yuan, X.; Yan, Y.; Li, D.; Song, F. The NAC transcription factor ONAC083 negatively regulates rice immunity against Magnaporthe oryzae by directly activating transcription of the RING-H2 gene OsRFPH2-6. J. Integr. Plant Biol. 2023, 65, 854–875. [Google Scholar] [CrossRef]
- Bai, X.; Zhou, H.; Luo, D.; Chen, D.; Fan, J.; Shao, X.; Zhou, J.; Liu, W. A Rational Combination of Cyclocarya paliurus Triterpene Acid Complex (TAC) and Se-Methylselenocysteine (MSC) Improves Glucose and Lipid Metabolism via the PI3K/Akt/GSK3β Pathway. Molecules 2023, 28, 5499. [Google Scholar] [CrossRef]
- He, F.; Li, Y.; Guo, Z.; Chen, J. α-Glucosidase inhibitors screening from Cyclocarya paliurus based on spectrum–effect relationship and UPLC–MS/MS. Biomed. Chromatogr. 2022, 36, e5313. [Google Scholar] [CrossRef]
- Xie, L.; Huang, Z.; Qin, L.; Yu, Q.; Chen, Y.; Zhu, H.; Xie, J. Effects of sulfation and carboxymethylation on Cyclocarya paliurus polysaccharides: Physicochemical properties, antitumor activities and protection against cellular oxidative stress. Int. J. Biol. Macromol. 2022, 204, 103–115. [Google Scholar] [CrossRef]
- Burssens, S.; Himanen, K.; van de Cotte, B.; Beeckman, T.; Van Montagu, M.; Inzé, D.; Verbruggen, N. Expression of cell cycle regulatory genes and morphological alterations in response to salt stress in Arabidopsis thaliana. Planta 2000, 211, 632–640. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, C.; Yao, J.; Zhang, Y.; Zhang, Y.; Deng, S.; Zhao, N.; Sa, G.; Zhou, X.; Lu, C.; et al. Populus euphratica JRL Mediates ABA Response, Ionic and ROS Homeostasis in Arabidopsis under Salt Stress. Int. J. Mol. Sci. 2019, 20, 815. [Google Scholar] [CrossRef]
- Han, K.; Zhao, Y.; Sun, Y.; Li, Y. NACs, generalist in plant life. Plant Biotechnol. J. 2023, 21, 2433–2457. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, C.; Li, Z.; Sun, J.; Deng, Z.; Wen, L.; Li, X.; Guo, Y. Potato NAC Transcription Factor StNAC053 Enhances Salt and Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2021, 22, 2568. [Google Scholar] [CrossRef]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive Analysis of NAC Family Genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Zhang, W.; Ruan, J.; Ho, T.D.; You, Y.; Yu, T.; Quatrano, R.S. Cis-regulatory element based targeted gene finding: Genome-wide identification of abscisic acid- and abiotic stress-responsive genes in Arabidopsis thaliana. Bioinformatics 2005, 21, 3074–3081. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).