Short-Term Simulated Warming Changes the Beta Diversity of Bacteria in Taiga Forests’ Permafrost by Altering the Composition of Dominant Bacterial Phyla
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Sampling Procedures and Incubation Experiments
2.3. Analyses of Soil Physicochemical Properties
2.4. DNA Extraction, Amplification, and MiSeq Sequencing
2.5. Bioinformatic and Statistical Analyses
3. Results
3.1. Effect of Warming on Soil Physicochemical Properties
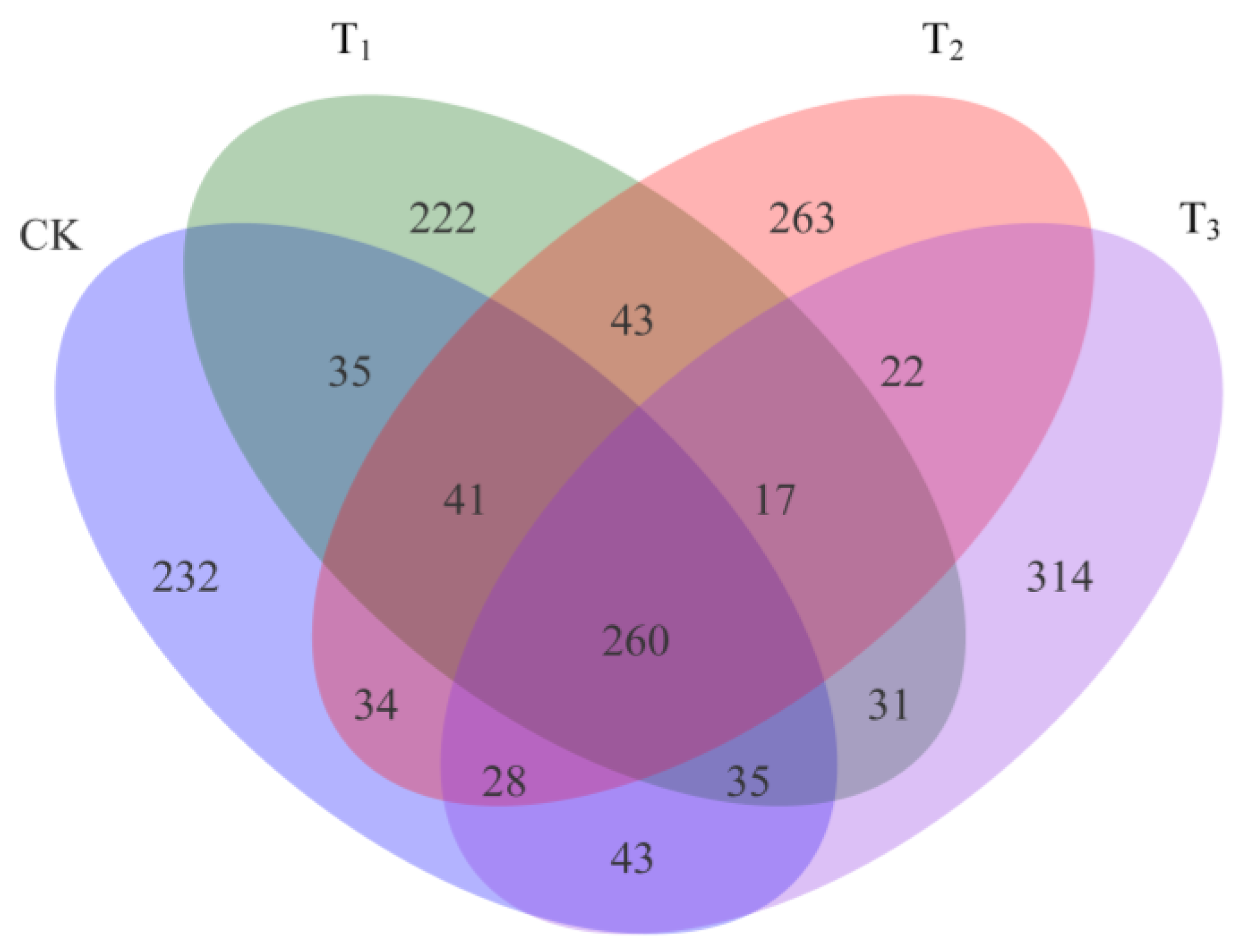
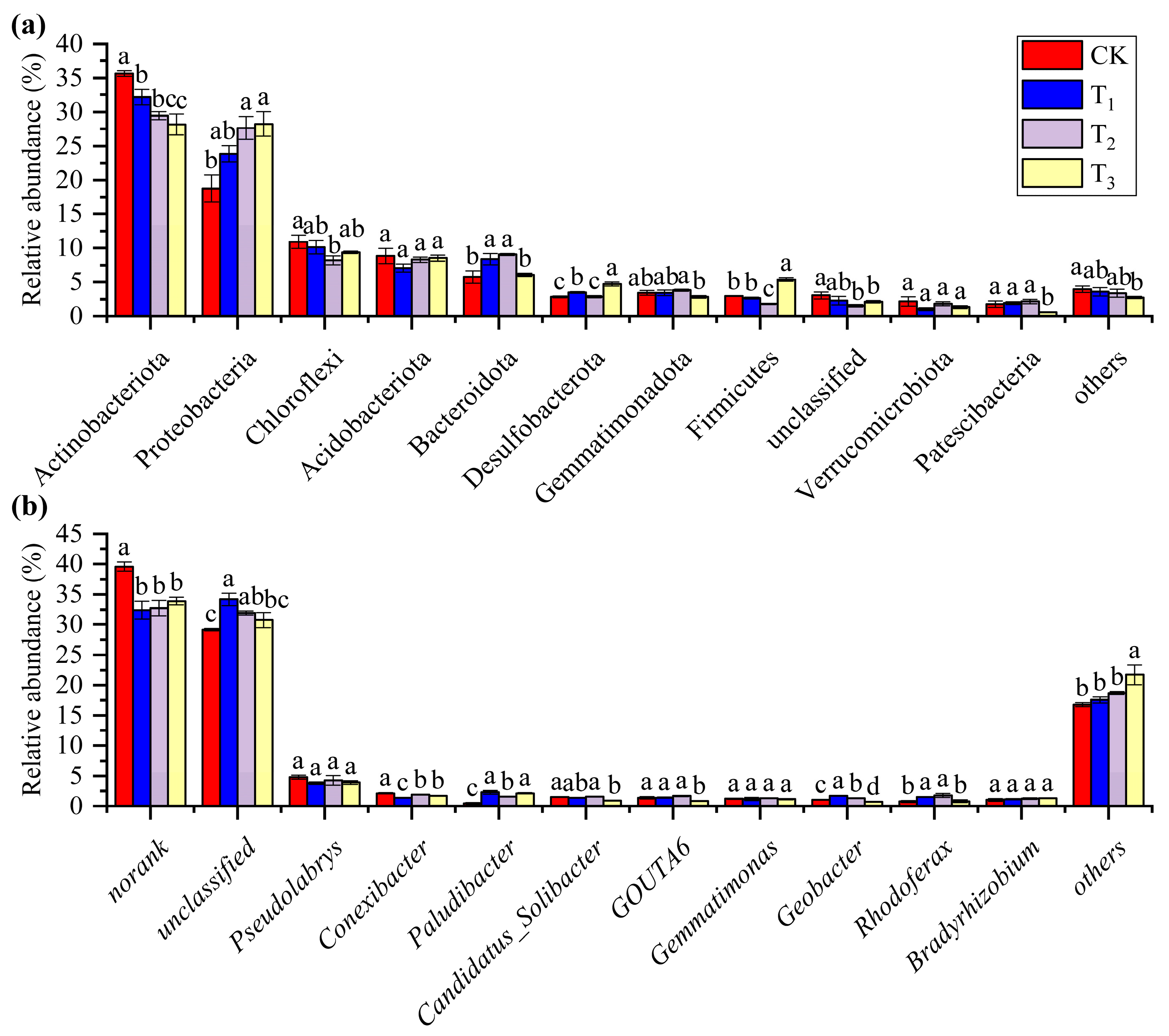
3.2. Heating Results in Differences in Bacterial Community Composition
3.3. Effects on Bacterial Community Diversity
3.3.1. Alpha Diversity
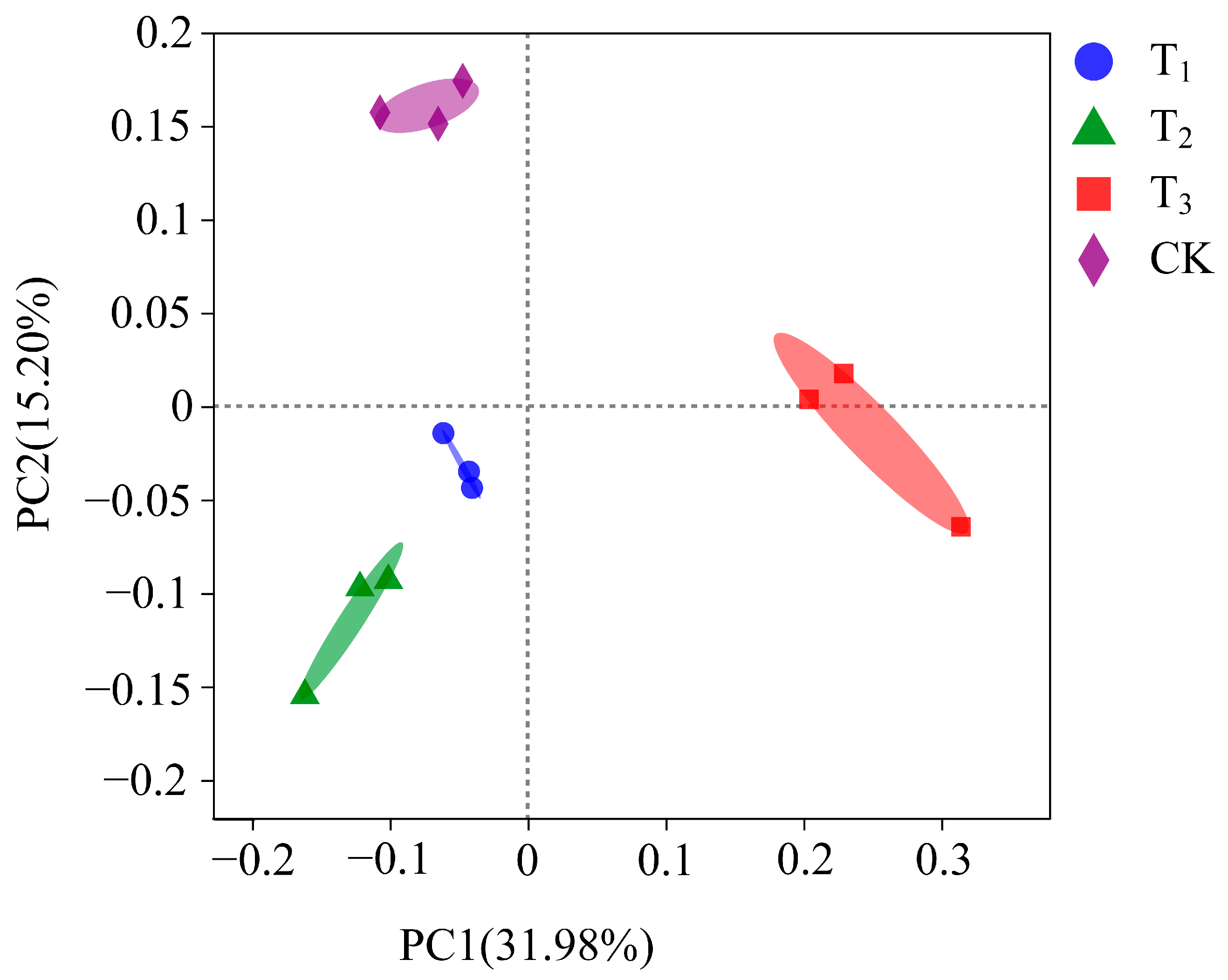
3.3.2. Beta Diversity
3.4. Correlation Analyses
3.4.1. Relationships between Community Composition and Alpha Diversity and Soil Properties
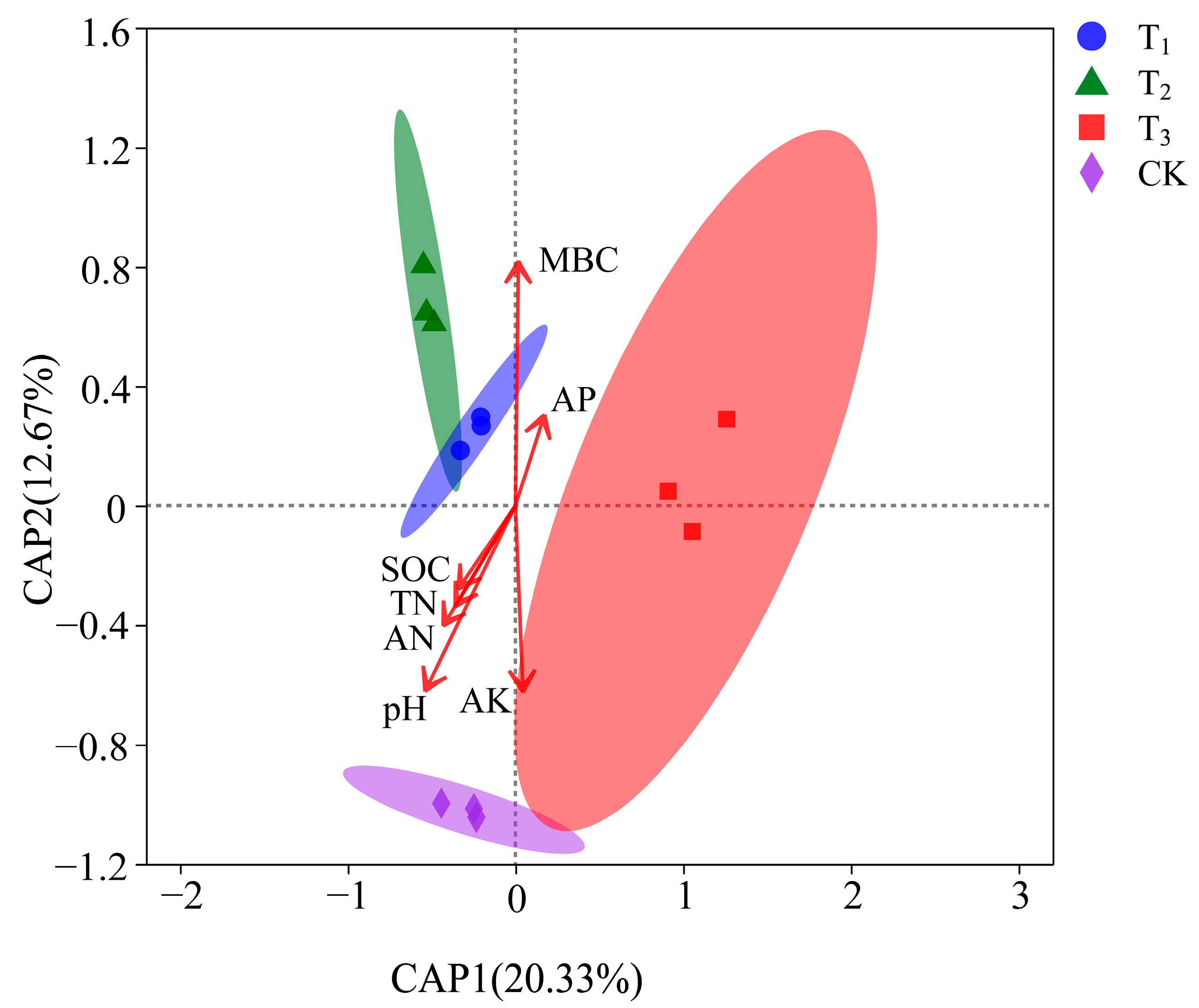
3.4.2. Relationship between Beta Diversity and Soil Physiochemistry
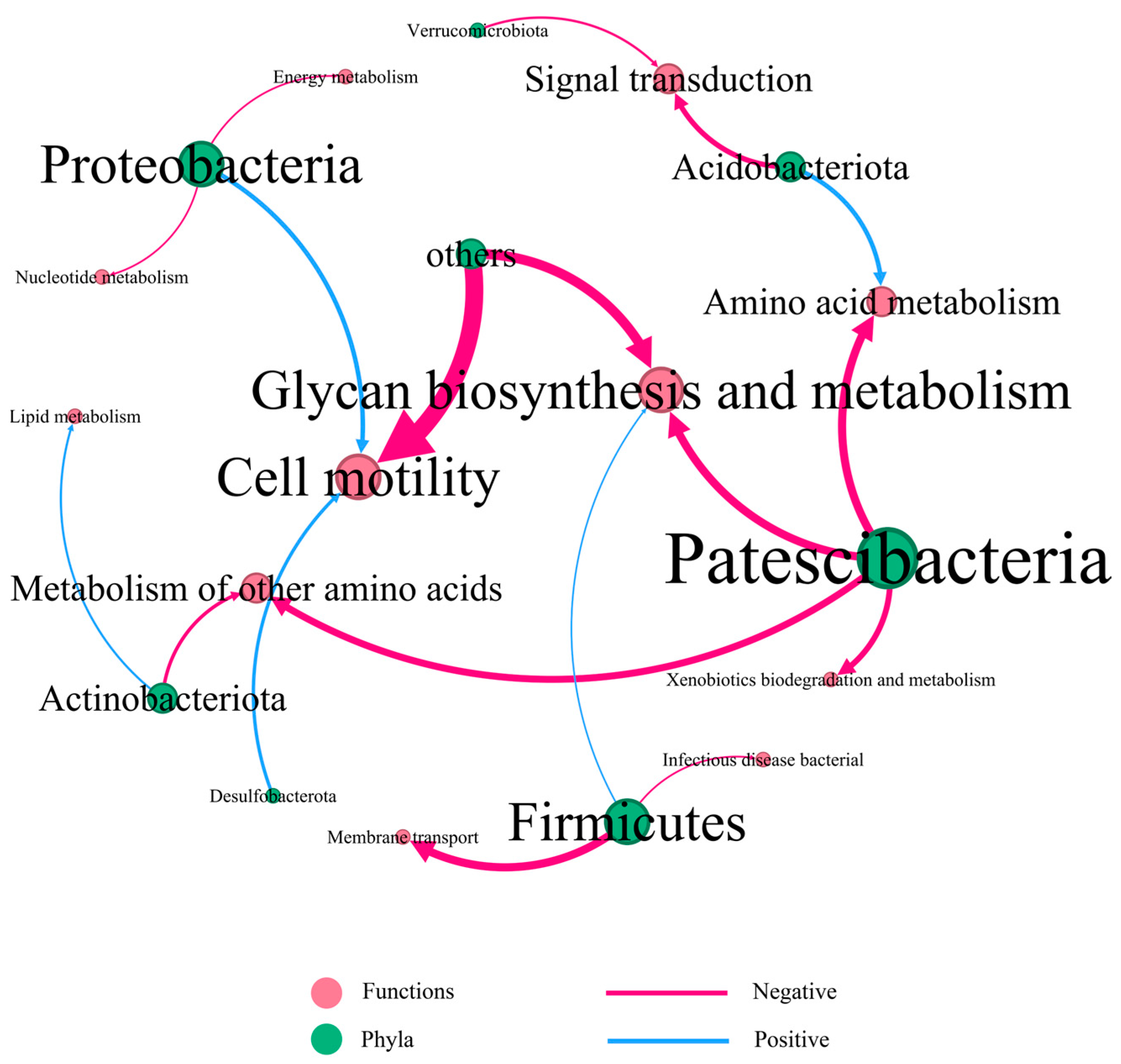
3.5. Bacterial Function Prediction
4. Discussion
4.1. Short-Term Warming Affects the Composition and Structure of the Bacterial Communities
4.2. Short-Term Warming Affects the Bacterial Community Diversity
4.3. Short-Term Warming Affects the Bacterial Community Functions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sunday, J.M. The pace of biodiversity change in a warming world. Nature 2020, 580, 460–461. [Google Scholar] [CrossRef] [PubMed]
- Masson-Delmotte, V.; Zhai, P.M.; Pirani, A.; Connors, S.L.; Péan, C.; Berger, S.; Caud, N.; Chen, Y.; Goldfarb, L.; Gomis, M.I.; et al. IPCC, 2021: Summary for Policymakers. In Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: New York, NY, USA, 2021; pp. 14–17. [Google Scholar] [CrossRef]
- Schuur, E.A.G.; McGuire, A.D.; Schaedel, C.; Grosse, G.; Harden, J.W.; Hayes, D.J.; Hugelius, G.; Koven, C.D.; Kuhry, P.; Lawrence, D.M.; et al. Climate change and the permafrost carbon feedback. Nature 2015, 520, 171–179. [Google Scholar] [CrossRef]
- Rantanen, M.; Karpechko, A.Y.; Lipponen, A.; Nordling, K.; Hyvärinen, O.; Ruosteenoja, K.; Vihma, T.; Laaksonen, A. The Arctic has warmed nearly four times faster than the globe since 1979. Commun. Earth Environ. 2022, 3, 168. [Google Scholar] [CrossRef]
- You, Q.L.; Cai, Z.Y.; Pepin, N.; Chen, D.L.; Ahrens, B.; Jiang, Z.H.; Wu, F.Y.; Kang, S.C.; Zhang, R.N.; Wu, T.H.; et al. Warming amplification over the Arctic Pole and Third Pole: Trends, mechanisms and consequences. Earth-Sci. Rev. 2021, 217, 103625. [Google Scholar] [CrossRef]
- Lyon, S.W.; Destouni, G.; Giesler, R.; Humborg, C.; Morth, M.; Seibert, J.; Karlsson, J.; Troch, P.A. Estimation of permafrost thawing rates in a sub-arctic catchment using recession flow analysis. Hydrol. Earth Syst. Sci. 2009, 13, 595–604. [Google Scholar] [CrossRef]
- Knoblauch, C.; Beer, C.; Schuett, A.; Sauerland, L.; Liebner, S.; Steinhof, A.; Rethemeyer, J.; Grigoriev, M.N.; Faguet, A.; Pfeiffer, E.M. Carbon dioxide and methane release following abrupt thaw of Pleistocene permafrost deposits in Arctic Siberia. J. Geophys. Res.-Biogeosci. 2021, 126, e2021JG006543. [Google Scholar] [CrossRef]
- Tanski, G.; Wagner, D.; Knoblauch, C.; Fritz, M.; Sachs, T.; Lantuit, H. Rapid CO2 release from eroding permafrost in seawater. Geophys. Res. Lett. 2019, 46, 11244–11252. [Google Scholar] [CrossRef]
- Wang, J.F.; Wu, Q.B. Annual soil CO2 efflux in a wet meadow during active layer freeze-thaw changes on the Qinghai-Tibet Plateau. Environ. Earth Sci. 2013, 69, 855–862. [Google Scholar] [CrossRef]
- Jansson, J.K.; Tas, N. The microbial ecology of permafrost. Nat. Rev. Microbiol. 2014, 12, 414–425. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, D.G.; Jiang, Z.Z.; Sun, P.; Xiao, H.L.; Wu, Y.; Chen, J.G. Changes in the soil microbial communities of alpine steppe at Qinghai-Tibetan Plateau under different degradation levels. Sci. Total Environ. 2018, 651, 2281–2291. [Google Scholar] [CrossRef]
- Zhang, B.G.; Wu, X.K.; Zhang, G.S.; Zhang, W.; Liu, G.X.; Chen, T.; Qin, Y.; Zhang, B.; Sun, L.K. Response of soil bacterial community structure to permafrost degradation in the upstream regions of the Shule River Basin, Qinghai-Tibet Plateau. Geomicrobiol. J. 2017, 34, 300–308. [Google Scholar] [CrossRef]
- Xue, K.; Yuan, M.M.; Shi, Z.J.; Qin, Y.J.; Deng, Y.; Cheng, L.; Wu, L.; He, Z.L.; Van Nostrand, J.D.; Bracho, R.; et al. Tundra soil carbon is vulnerable to rapid microbial decomposition under climate warming. Nat. Clim. Chang. 2016, 6, 595–600. [Google Scholar] [CrossRef]
- Perez-Mon, C.; Stierli, B.; Plötze, M.; Frey, B. Fast and persistent responses of alpine permafrost microbial communities to in situ warming. Sci. Total Environ. 2021, 807, 150720. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.F.; Liu, C.; Ma, D.L.; Wu, Y.F.; Man, H.R.; Wu, X.W.; Li, M.; Zang, S.Y. Organic carbon mineralization and bacterial community of active layer soils response to short-term warming in the Great Hing’an Mountains of Northeast China. Front. Microbiol. 2021, 12, 802213. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.T.; Kou, D.; Chen, Y.L.; Xue, K.; Ernakovich, J.G.; Chen, L.Y.; Yang, G.B.; Yang, Y. Altered microbial structure and function after thermokarst formation. Glob. Chang. Biol. 2021, 27, 823–835. [Google Scholar] [CrossRef] [PubMed]
- Mackelprang, R.; Waldrop, M.P.; DeAngelis, K.M.; David, M.M.; Chavarria, K.L.; Blazewicz, S.J.; Rubin, E.M.; Jansson, J.K. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 2011, 480, 368–371. [Google Scholar] [CrossRef]
- Jin, H.J.; Yu, Q.H.; Lü, L.Z.; Guo, D.X.; He, R.X.; Yu, S.P.; Sun, G.Y.; Li, Y.W. Degradation of permafrost in the Xing’anling Mountains, Northeastern China. Permafr. Periglac. Process. 2007, 18, 245–258. [Google Scholar] [CrossRef]
- Gao, W.F. Emission Characteristics and Influencing Factors of Greenhouse gas in the Continuous Permafrost Region of Daxing’an Mountains, Northeast China. Ph.D. Thesis, Northeast Forestry University, Harbin, China, 2019. [Google Scholar]
- Song, Y.Y.; Jiang, L.; Song, C.C.; Wang, X.W.; Ma, X.Y.; Zhang, H.; Tan, W.W.; Gao, J.L.; Hou, A.X. Microbial abundance and enzymatic activity from tussock and shrub soil in permafrost peatland after 6-year warming. Ecol. Indic. 2021, 126, 107589. [Google Scholar] [CrossRef]
- Feng, J.J.; Wang, C.; Lei, J.S.; Yang, Y.F.; Yan, Q.Y.; Zhou, X.S.; Tao, X.Y.; Ning, D.L.; Yuan, M.T.M.; Qin, Y.J.; et al. Warming-induced permafrost thaw exacerbates tundra soil carbon decomposition mediated by microbial community. Microbiome 2020, 8, 3. [Google Scholar] [CrossRef]
- Dong, X.F.; Liu, C.; Wu, X.D.; Man, H.R.; Wu, X.W.; Ma, D.L.; Li, M.; Zang, S.Y. Linking soil organic carbon mineralization with soil variables and bacterial communities in a permafrost-affected tussock wetland during laboratory incubation. Catena 2023, 221, 106783. [Google Scholar] [CrossRef]
- Dong, X.F.; Man, H.R.; Liu, C.; Wu, X.D.; Zhu, J.J.; Zheng, Z.C.; Ma, D.L.; Li, M.; Zang, S.Y. Changes in soil bacterial community along a gradient of permafrost degradation in Northeast China. Catena 2023, 222, 106870. [Google Scholar] [CrossRef]
- Jiang, L.; Song, Y.Y.; Sun, L.; Song, C.; Wang, X.; Ma, X.Y.; Liu, C.; Gao, J.L. Effects of warming on carbon emission and microbial abundances across different soil depths of a peatland in the permafrost region under anaerobic condition. Appl. Soil Ecol. 2020, 156, 103712. [Google Scholar] [CrossRef]
- Ren, J.S.; Song, C.C.; Hou, A.X.; Song, Y.Y.; Zhu, X.Y.; Cagle, G.A. Shifts in soil bacterial and archaeal communities during freeze-thaw cycles in a seasonal frozen marsh, Northeast China. Sci. Total Environ. 2018, 625, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.B.; Jiang, Y.B.; Zhou, T.; Cui, F.X.; Zhu, D.G.; Xu, F. Effects of litter fall on soil fungal diversity under snow cover in the Greater Xing’an Mountains. Res. Environ. Sci. 2022, 35, 1037–1044. [Google Scholar] [CrossRef]
- Jiang, Y.B.; Yang, L.B.; Wu, S.; Zhou, T. Warming changes the composition and diversity of fungal communities in permafrost. Ann. Microbiol. 2023, 73, 7. [Google Scholar] [CrossRef]
- Kim, H.M.; Jung, J.Y.; Yergeau, E.; Hwang, C.Y.; Hinzman, L.; Nam, S.; Hong, S.G.; Kim, O.S.; Chun, J.; Lee, Y.K. Bacterial community structure and soil properties of a subarctic tundra soil in Council, Alaska. FEMS Microbiol. Ecol. 2014, 89, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Shang, W.; Wu, X.D.; Zhao, L.; Yue, G.Y.; Zhao, Y.H.; Qiao, Y.P.; Li, Y.Q. Seasonal variations in labile soil organic matter fractions in permafrost soils with different vegetation types in the central Qinghai-Tibet Plateau. Catena 2016, 137, 670–678. [Google Scholar] [CrossRef]
- Hu, W.G.; Zhang, Q.; Li, D.Y.; Cheng, G.; Mu, J.; Wu, Q.B.; Niu, F.J.; An, L.Z.; Feng, H.Y. Diversity and community structure of fungi through a permafrost core profile from the Qinghai-Tibet Plateau of China. J. Basic Microbiol. 2015, 54, 1331–1341. [Google Scholar] [CrossRef]
- Wu, X.D.; Zhao, L.; Liu, G.M.; Xu, H.Y.; Zhang, X.L.; Ding, Y.J. Effects of permafrost thaw-subsidence on soil bacterial communities in the southern Qinghai-Tibetan Plateau. Appl. Soil Ecol. 2018, 128, 81–88. [Google Scholar] [CrossRef]
- Ade, L.J.; Hu, L.; Zi, H.B.; Wang, C.T.; Lerdau, M.; Dong, S.K. Effect of snowpack on the soil bacteria of alpine meadows in the Qinghai-Tibetan Plateau of China. Catena 2018, 164, 13–22. [Google Scholar] [CrossRef]
- Bates, S.T.; Berg-Lyons, D.; Caporaso, J.G.; Walters, W.A.; Knight, R.; Fierer, N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011, 5, 908–917. [Google Scholar] [CrossRef] [PubMed]
- Uroz, S.; Buée, M.; Murat, C.; Frey-Klett, P.; Martin, F. Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ. Microbiol. Rep. 2010, 2, 281–288. [Google Scholar] [CrossRef]
- Wang, X.S.; Guan, X.Y.; Zhang, X.J.; Xiang, S.Z.; Zhang, R.R.; Liu, M.Z. Microbial communities in petroleum-contaminated seasonally frozen soil and their response to temperature changes. Chemosphere 2020, 258, 127375. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Chen, K.L.; Wang, H.S.; Yang, Y. Effect of simulated warming on soil microorganism of bird island in Qinghai Lake. Microbiol. China 2021, 48, 722–732. [Google Scholar] [CrossRef]
- Gianoulis, T.A.; Raes, J.; Patel, P.V.; Bjornson, R.; Korbel, J.O.; Letunic, I.; Yamada, T.; Paccanaro, A.; Jensen, L.; Snyder, M.; et al. Quantifying environmental adaptation of metabolic pathways in metagenomics. Proc. Natl. Acad. Sci. USA 2009, 106, 1374–1379. [Google Scholar] [CrossRef] [PubMed]
- Steven, B.; Pollard, W.H.; Greer, C.W.; Whyte, L.G. Microbial diversity and activity through a permafrost/ground ice core profile from the Canadian high Arctic. Environ. Microbiol. 2008, 10, 3388–3403. [Google Scholar] [CrossRef] [PubMed]
- Yergeau, E.; Hogues, H.; Whyte, L.G.; Greer, C.W. The functional potential of high Arctic permafrost revealed by metagenomic sequencing, qPCR and microarray analyses. ISME J. 2010, 4, 1206–1214. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.S.; Hebsgaard, M.B.; Christensen, T.R.; Mastepanov, M.; Nielsen, R.; Munch, K.; Brand, T.; Thomas, M.; Gilbert, P.; Zuber, M.T.; et al. Ancient bacteria show evidence of DNA repair. Proc. Natl. Acad. Sci. USA 2007, 104, 14401–14405. [Google Scholar] [CrossRef] [PubMed]
- Spohn, M.; Klaus, K.; Wanek, W.; Richter, A. Microbial carbon use efficiency and biomass turnover times depending on soil depth—Implications for carbon cycling. Soil Biol. Biochem. 2016, 96, 74–81. [Google Scholar] [CrossRef]
- Axelrood, P.E.; Chow, M.L.; Radomski, C.C.; McDermott, J.M.; Davies, J. Molecular characterization of bacterial diversity from British Columbia forest soils subjected to disturbance. Can. J. Microbiol. 2002, 48, 655–674. [Google Scholar] [CrossRef]
- Maestre, F.T.; Delgado-Baquerizo, M.; Jeffries, T.C.; Eldridge, D.J.; Ochoa, V.; Gozalo, B.; Quero, J.L.; García-Gómez, M.; Gallardo, A.; Ulrich, W.; et al. Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc. Natl. Acad. Sci. USA 2015, 112, 15684–15689. [Google Scholar] [CrossRef]
- DeAngelis, K.M.; Allgaier, M.; Chavarria, Y.; Fortney, J.L.; Hugenholtz, P.; Simmons, B.; Sublette, K.; Silver, W.L.; Hazen, T.C. Characterization of trapped lignin-degrading microbes in tropical forest soil. PLoS ONE 2011, 6, e19306. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Wu, L.W.; Zhang, Y.; Guo, X.; Ning, D.L.; Zhou, X.S.; Feng, J.J.; Yuan, M.M.; Liu, S.; Guo, J.J.; Gao, Z.P.; et al. Reduction of microbial diversity in grassland soil is driven by long-term climate warming. Nat. Microbiol. 2022, 7, 1054–1062. [Google Scholar] [CrossRef]
- Sheik, C.S.; Beasley, W.H.; Elshahed, M.S.; Zhou, X.H.; Luo, Y.Q.; Krumholz, L.R. Effect of warming and drought on grassland microbial communities. ISME J. 2011, 5, 1692–1700. [Google Scholar] [CrossRef]
- Chen, Y.L.; Liu, F.T.; Kang, L.Y.; Zhang, D.Y.; Kou, D.; Mao, C.; Qin, S.Q.; Zhang, Q.W.; Yang, Y.H. Large-scale evidence for microbial response and associated carbon release after permafrost thaw. Glob. Chang. Biol. 2020, 27, 3218–3229. [Google Scholar] [CrossRef]
- Griffiths, R.I.; Thomson, B.C.; James, P.; Bell, T.; Bailey, M.; Whiteley, A.S. The bacterial biogeography of British soils. Environ. Microbiol. 2011, 13, 1642–1654. [Google Scholar] [CrossRef]
- Weedon, J.T.; Kowalchuk, G.A.; Aerts, R.; van Hal, J.; van Logtestijn, R.; Tas, N.; Röling, W.F.M.; van Bodegom, P.M. Summer warming accelerates sub-arctic peatland nitrogen cycling without changing enzyme pools or microbial community structure. Glob. Chang. Biol. 2012, 18, 138–150. [Google Scholar] [CrossRef]
- Rocca, J.D.; Simonin, M.; Blaszczak, J.R.; Ernakovich, J.G.; Gibbons, S.M.; Midani, F.S.; Washburne, A.D. The microbiome stress project: Toward a global meta-analysis of environmental stressors and their effects on microbial communities. Front. Microbiol. 2019, 9, 3272. [Google Scholar] [CrossRef]
- Jousset, A.; Bienhold, C.; Chatzinotas, A.; Gallien, L.; Gobet, A.; Kurm, V.; Küsel, K.; Rillig, M.C.; Rivett, D.W.; Salles, J.F.; et al. Where less may be more: How the rare biosphere pulls ecosystems strings. ISME J. 2017, 11, 853–862. [Google Scholar] [CrossRef]
- Rinnan, R.; Michelsen, A.; Bååth, E.; Jonasson, S. Fifteen years of climate change manipulations alter soil microbial communities in a subarctic heath ecosystem. Glob. Chang. Biol. 2007, 13, 28–39. [Google Scholar] [CrossRef]
- Siciliano, S.D.; Palmer, A.S.; Winsley, T.; Lamb, E.; Bissett, A.; Brown, M.V.; van Dorst, J.; Ji, M.K.; Ferrari, B.C.; Grogan, P.; et al. Soil fertility is associated with fungal and bacterial richness, whereas pH is associated with community composition in polar soil microbial communities. Soil Biol. Biochem. 2014, 78, 10–20. [Google Scholar] [CrossRef]
- Feng, Y.Z.; Grogan, P.; Caporaso, J.G.; Zhang, H.Y.; Lin, X.G.; Knight, R.; Chu, H.Y. pH is a good predictor of the distribution of anoxygenic purple phototrophic bacteria in Arctic soils. Soil Biol. Biochem. 2014, 74, 193–200. [Google Scholar] [CrossRef]
- Meyer, S.T.; Ebeling, A.; Eisenhauer, N.; Hertzog, L.; Hillebrand, H.; Milcu, A.; Pompe, S.; Abbas, M.; Bessler, H.; Buchmann, N.; et al. Effects of biodiversity strengthen over time as ecosystem functioning declines at low and increases at high biodiversity. Ecosphere 2016, 7, e1619. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Wang, L.; Qin, S.Q.; Kou, D.; Wang, S.Y.; Zheng, Z.H.; Peñuelas, J.; Yang, Y.H. Microbial nitrogen and phosphorus co-limitation across permafrost region. Glob. Chang. Biol. 2023, 29, 3910–3923. [Google Scholar] [CrossRef]
- Zhang, H.M.; Anders, P.; Samuel, F.; Elberling, B.; Jia, Z.J. Soil microbiomes modulate distinct patterns of soil respiration and methane oxidation in arctic active layer and permafrost. Acta Microbiol. Sin. 2017, 57, 839–855. [Google Scholar] [CrossRef]
- Wang, X.J.; Zhang, Z.C.; Yu, Z.Q.; Shen, G.F.; Cheng, H.F.; Tao, S. Composition and diversity of soil microbial communities in the alpine wetland and alpine forest ecosystems on the Tibetan Plateau. Sci. Total Environ. 2020, 747, 141358. [Google Scholar] [CrossRef]
- Pereira, L.B.; Vicentini, R.; Ottoboni, L.M.M. Changes in the bacterial community of soil from a neutral mine drainage channel. PLoS ONE 2014, 9, e96605. [Google Scholar] [CrossRef]
- Miransari, M. Soil microbes and the availability of soil nutrients. Acta Physiol. Plant. 2013, 35, 3075–3084. [Google Scholar] [CrossRef]
- Mori, T.; Wang, S.H.; Wang, Z.H.; Wang, C.; Mo, H.; Mo, J.M.; Lu, X.K. Testing potassium limitation on soil microbial activity in a sub-tropical forest. J. For. Res. 2019, 30, 2341–2347. [Google Scholar] [CrossRef]
- Moro, H.; Kunito, T.; Saito, T.; Yaguchi, N.; Sato, T. Soil microorganisms are less susceptible than crop plants to potassium deficiency. Arch. Agron. Soil Sci. 2014, 60, 1807–1813. [Google Scholar] [CrossRef]
- Li, H.; Yang, S.; Xu, Z.W.; Yan, Q.Y.; Li, X.B.; van Nostrand, J.D.; He, Z.L.; Yao, F.; Han, X.G.; Zhou, J.Z.; et al. Responses of soil microbial functional genes to global changes are indirectly influenced by aboveground plant biomass variation. Soil Biol. Biochem. 2017, 104, 18–29. [Google Scholar] [CrossRef]
- Pitta, D.W.; Pinchak, W.E.; Indugu, N.; Vecchiarelli, B.; Sinha, R.; Fulford, J.D. Metagenomic analysis of the rumen microbiome of steers with wheat-induced frothy bloat. Front. Microbiol. 2016, 7, 689. [Google Scholar] [CrossRef]
- Shanks, O.C.; Kelty, C.A.; Archibeque, S.; Jenkins, M.; Newton, R.J.; McLellan, S.L.; Huse, S.M.; Sogin, M.L. Community structures of fecal bacteria in cattle from different animal feeding operations. Appl. Environ. Microbiol. 2011, 77, 2992–3001. [Google Scholar] [CrossRef]
- Yadav, A.; Borrelli, J.C.; Elshahed, M.S.; Youssef, N.H. Genomic analysis of family uba6911 (group 18 Acidobacteria) expands the metabolic capacities of the phylum and highlights adaptations to terrestrial habitats. Appl. Environ. Microbiol. 2021, 87, e94721. [Google Scholar] [CrossRef]
- Li, Y.Y.; Xu, T.T.; Ai, Z.; Wei, L.L.; Ma, F. Diversity and predictive functional of Caragana jubata bacterial community in rhizosphere and non-rhizosphere soil at different altitudes. Environ. Sci. 2023, 44, 2304–2314. [Google Scholar] [CrossRef]
- Poncin, K.; Gillet, S.; De Bolle, X. Learning from the master: Targets and functions of the CtrA response regulator in Brucella abortus and other alpha-proteobacteria. FEMS Microbiol. Rev. 2018, 42, 500–513. [Google Scholar] [CrossRef]
- Peng, T.S. Effects of Phosphorus Addition on Soil Nutrient Elements and Microorganisms in Young Phoebe bournei Plantations. Master’s Thesis, Central South University of Forestry and Technology, Changsha, China, 2022. [Google Scholar]

| Treatment | Coverage | Sobs | ACE | Shannon | Simpson (×10−2) |
|---|---|---|---|---|---|
| CK | 1.00 ± 0.00 a | 392.67 ± 14.88 a | 392.78 ± 14.90 a | 5.22 ± 0.04 ab | 1.19 ± 0.09 a |
| T1 | 1.00 ± 0.00 a | 380.00 ± 16.86 a | 380.12 ± 16.97 a | 5.16 ± 0.06 b | 1.15 ± 0.13 ab |
| T2 | 1.00 ± 0.00 a | 382.33 ± 22.28 a | 382.39 ± 22.30 a | 5.27 ± 0.08 ab | 0.89 ± 0.07 ab |
| T3 | 0.99 ± 0.01 a | 438.33 ± 17.02 a | 438.91 ± 17.36 a | 5.36 ± 0.05 a | 0.86 ± 0.10 b |
| Method | Adonis | ANOSIM | MRPP | |||
|---|---|---|---|---|---|---|
| All groups | R2 | p | R | p | A | p |
| 0.56 | 0.001 *** | 0.91 | 0.001 *** | 0.27 | 0.001 *** | |
| Soil Factors | R2 | p |
|---|---|---|
| pH | 0.96 | 0.001 *** |
| TN | 0.33 | 0.161 |
| SOC | 0.25 | 0.272 |
| MBC | 0.96 | 0.001 *** |
| AN | 0.47 | 0.059 |
| AK | 0.56 | 0.032 *** |
| AP | 0.16 | 0.494 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Y.; Wu, S.; Yang, L.; Liu, Y.; Gao, M.; Ni, H. Short-Term Simulated Warming Changes the Beta Diversity of Bacteria in Taiga Forests’ Permafrost by Altering the Composition of Dominant Bacterial Phyla. Forests 2024, 15, 693. https://doi.org/10.3390/f15040693
Jiang Y, Wu S, Yang L, Liu Y, Gao M, Ni H. Short-Term Simulated Warming Changes the Beta Diversity of Bacteria in Taiga Forests’ Permafrost by Altering the Composition of Dominant Bacterial Phyla. Forests. 2024; 15(4):693. https://doi.org/10.3390/f15040693
Chicago/Turabian StyleJiang, Yunbing, Song Wu, Libin Yang, Yongzhi Liu, Mingliang Gao, and Hongwei Ni. 2024. "Short-Term Simulated Warming Changes the Beta Diversity of Bacteria in Taiga Forests’ Permafrost by Altering the Composition of Dominant Bacterial Phyla" Forests 15, no. 4: 693. https://doi.org/10.3390/f15040693
APA StyleJiang, Y., Wu, S., Yang, L., Liu, Y., Gao, M., & Ni, H. (2024). Short-Term Simulated Warming Changes the Beta Diversity of Bacteria in Taiga Forests’ Permafrost by Altering the Composition of Dominant Bacterial Phyla. Forests, 15(4), 693. https://doi.org/10.3390/f15040693






