Chemical and Microbial Differences of Root and Rhizosphere Soil among Different Provenances of Fokienia hodginsii
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area and Sampling
2.2. Quantification of Nutrients
2.3. Microbial Amplicon Sequencing
2.3.1. DNA Extraction and Quality Checking
2.3.2. PCR Amplification
2.3.3. High-Throughput Sequencing
2.4. Statistical Analysis
3. Results
3.1. Differences in Root Rhizosphere Soil (RS) Nutrient Contents Associated with Different Provenances
3.2. Diversity and Construction of Root Rhizosphere Microbes (RMs) Associated with Different Provenances
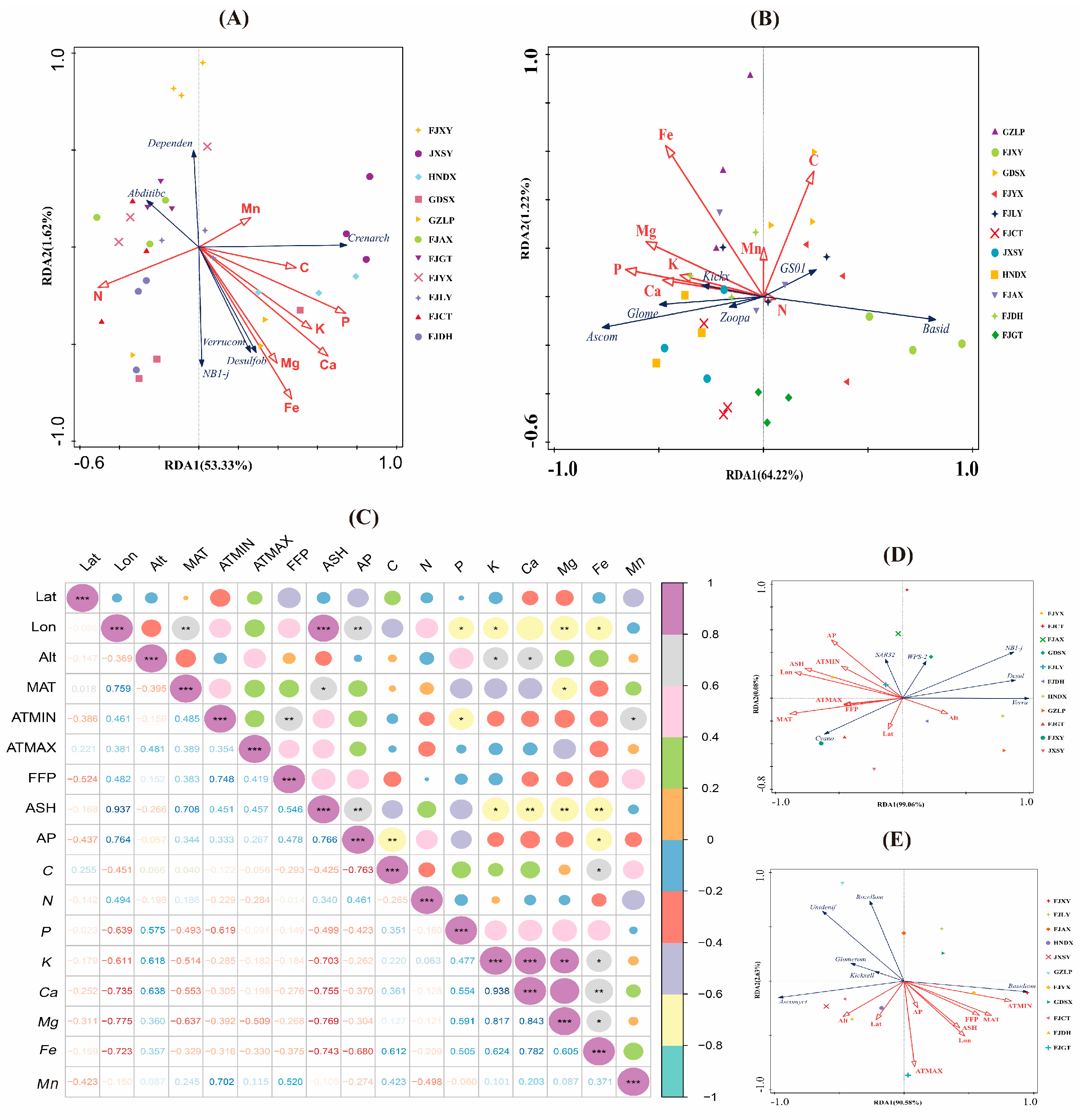
3.3. Connections between Root Rhizosphere Soil Nutrients and Microbes in Regard to Different Provenances
3.4. Diversity and Construction of Associated Microbiota (AM) in Different Provenances of F. hodginsii
3.5. Links between Root-Associated Microbiota and Nutrient Element Contents in Different Provenances of F. hodginsii
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cai, N.H.; Tang, J.R.; Li, Y.Q.; Chen, S.; Chen, L.; Xu, Y.L.; Li, G.Q. Effect of shading on growth and biomass of Fokienia hodginsi seedling. J. Fujian Agric. For. Univ. (Nat. Sci. Ed.) 2022, 44, 1305–1313. [Google Scholar]
- Chen, Z.S. Study on Physical Properties and Mechanical of Fokienia hodginsi. J. For. Environ. 1999, 3, 32–35. [Google Scholar]
- Lin, C.F.; Li, Z.; Niu, Z.P.; Zhang, Y.L.; Han, Y.G.; Chen, G.S. Litterfall nutrient dynamics in Fokienia hodginsi plantation. J. Fujian Agric. For. Univ. 2005, 1, 63–66. [Google Scholar]
- You, J.P. Water holding characteristics and ecological stoichiometric characteristics of carbon, nitrogen and phosphorus in litter of Fokienia hodginsii mixed forest. For. Sci. Technol. 2022, 47, 24–27. [Google Scholar]
- Yang, G.C. Study on the growth effect of different mixed ratio of Fokienia hodginsii and Pinus massoniana. Green Technol. 2022, 24, 99–102. [Google Scholar]
- Li, B.J.; Chen, Q.; Wang, X.X.; Rong, J.D.; Chen, L.G.; Zheng, Y.S. Differences in growth and nutrients between pure and mixed forests of Fokienia hodginsii at different ages. Northwest Bot. J. 2022, 42, 694–704. [Google Scholar]
- Tóth, Á. Bursaphelenchus xylophilus, the pinewood nematode: Its significance and a historical review. Acta Biol. Szeged. 2011, 55, 213–217. [Google Scholar]
- Liu, Q.; Wei, Y.; Xu, L.; Hao, Y.; Chen, X.; Zhou, Z. Transcriptomic Profiling Reveals Differentially Expressed Genes Associated with Pine Wood Nematode Resistance in Masson Pine (Pinus massoniana Lamb.). Sci. Rep. 2017, 7, 4693. [Google Scholar]
- Ponpandian, L.N.; Rim, S.O.; Shanmugam, G. Phylogenetic characterization of bacterial endophytes from four Pinus species and their nematicidal activity against the pine wood nematode. Sci. Rep. 2019, 9, 12457. [Google Scholar] [CrossRef]
- Liu, F.; Su, H.; Ding, T.; Huang, J.; Liu, T.; Ding, N.; Fang, G. Refined Assessment of Economic Loss from Pine Wilt Disease at the Subcompartment Scale. Forests 2023, 14, 139. [Google Scholar] [CrossRef]
- Yin, Q.; Chen, S.; Guo, W.; Huang, Y.; Huang, Y.; Zhou, R.; Fan, Q.; Liao, W. Pronounced genetic differentiation in Fokienia hodginsii revealed by simple sequence repeat markers. Ecol. Evol. 2018, 8, 10938–10951. [Google Scholar] [CrossRef]
- Zheng, W.; Gou, X.; Su, J.; Fan, H.; Yu, A.; Liu, W.; Deng, Y.; Manzanedo, R.D.; Fonti, P. Physiological and Growth Responses to Increasing Drought of an Endangered Tree Species in Southwest China. Forests 2019, 10, 514. [Google Scholar] [CrossRef]
- Chen, F.; Mu, J.; Huang, L.; Cui, X.Y.; Jin, Y.; Zhang, L. Features and succession analysis of Fokienia hodginsii communities in Sanchahe of Xishui National Nature Reserve, Guizhou Province. South. For. Sci. 2018, 46, 8–12. [Google Scholar]
- Li, Y.; Li, X.; Zhao, M.H.; Pang, Z.Y.; Wei, J.T.; Tigabu, M.; Chiang, V.L.; Sederoff, H.; Sederoff, R.; Zhao, X.Y. An Overview of the Practices and Management Methods for Enhancing Seed Production in Conifer Plantations for Commercial Use. Horticulturae 2021, 7, 252. [Google Scholar] [CrossRef]
- Huang, S.J.; Rong, J.D.; Zhang, L.H.; Yang, Y.; Jiang, J.J.; Zheng, Y.S. Research summarization of Fokienia hodginsii. Fujian For. Sci. Technol. 2013, 40, 236–242. [Google Scholar]
- Chen, Q.; Huang, X.; Jiang, D.H.; Ren, K.; Rong, J.D.; Chen, L.G.; Zheng, Y.S. Effects of shading on growth and biomass of Fokienia hodginsii seedlings. J. Fujian Agric. For. Univ. 2020, 49, 796–802. [Google Scholar]
- Zheng, R.H.; Yang, Z.W.; Liang, H.Y.; Xiao, X.X.; Li, W.Y. A Study on the provenance test of Fokienia hodginsi in seedling stage. J. Fujian For. Univ. 2001, 1, 40–44. [Google Scholar]
- Yang, Z.W.; Zheng, R.H.; Xiao, X.X.; Hou, B.X.; Cheng, Z.H.; Zeng, Z.G.; Xiao, F.M.; Li, X.M. A study on the genetic variation of seedling growth and root traits among provenance of Fokienia hodginsii. J. Nanjing For. Univ. 2001, 3, 26–30. [Google Scholar]
- Kuppler, J.; Albert, C.; Ames, G.; Armbruster, W.; Bönisch, G.; Boucher, F. Global gradients in intraspecific variation in vegetative and floral traits are partially associated with climate and species richness. Glob. Ecol. Biogeogr. 2020, 29, 10. [Google Scholar] [CrossRef]
- Cheng, Z.; Lei, S.; Li, Y.; Huang, W.; Ma, R.; Xiong, J.; Zhang, T.; Jin, L.; Haq, H.U.; Xu, X.; et al. Revealing the Variation and Stability of Bacterial Communities in Tomato Rhizosphere Microbiota. Microorganisms 2020, 8, 170. [Google Scholar] [CrossRef]
- Liu, S.; He, F.; Kuzyakov, Y.; Xiao, H.; Hoang, D.T.T.; Pu, S.; Razavi, B.S. Nutrients in the rhizosphere: A meta-analysis of content, availability, and influencing factors. Sci. Total Environ. 2022, 826, 153908. [Google Scholar] [CrossRef]
- White, J.F.; Kingsley, K.L.; Zhang, Q.; Verma, R.; Obi, N.; Dvinskikh, S.; Elmore, M.T.; Verma, S.K.; Gond, S.K.; Kowalski, K.P. Review: Endophytic microbes and their potential applications in crop management. Pest Manage. Sci. 2019, 75, 2558–2565. [Google Scholar] [CrossRef]
- Fadiji, A.E.; Ayangbenro, A.S.; Babalola, O.O. Organic Farming Enhances the Diversity and Community Structure of Endophytic Archaea and Fungi in Maize Plant: A Shotgun Approach. J. Soil Sci. Plant Nutr. 2020, 20, 2587–2599. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef]
- Hacquard, S.; Garrido-Oter, R.; González, A.; Spaepen, S.; Ackermann, G.; Lebeis, S.; McHardy, A.C.; Dangl, J.L.; Knight, R.; Ley, R.; et al. Microbiota and Host Nutrition across Plant and Animal Kingdoms. Cell Host Microbe 2015, 17, 603–616. [Google Scholar] [CrossRef]
- Verma, P.; Yadav, A.; Kumar, V.; Singh, D.P.; Saxena, A.K. Beneficial Plant-Microbes Interactions: Biodiversity of Microbes from Diverse Extreme Environments and Its Impact for Crop Improvement Plant-Microbe Interactions in Agro-Ecological Perspective; Springer: Cham, Switzerland, 2017; pp. 543–580. [Google Scholar]
- Liu, F.; Hewezi, T.; Lebeis, S.L.; Pantalone, V.; Grewal, P.S.; Staton, M.E. Soil indigenous microbiome and plant provenances cooperatively modify soybean rhizosphere microbiome assembly. BMC Microbiol. 2019, 19, 201. [Google Scholar] [CrossRef]
- Chang, J.; Sun, Y.; Tian, L.; Ji, L.; Luo, S.; Nasir, F.; Kuramae, E.E.; Tian, C. The Structure of Rhizosphere Fungal Communities of Wild and Domesticated Rice: Changes in Diversity and Co-occurrence Patterns. Front. Microbiol. 2021, 12, 610–823. [Google Scholar] [CrossRef]
- Chen, F.S.; Niklas, K.J.; Liu, Y.; Fang, X.M.; Wan, S.Z.; Wang, H. Nitrogen and phosphorus additions alter nutrient dynamics but not resorption efficiencies of Chinese fir leaves and twigs differing in age. Tree Physiol. 2015, 35, 1106–1117. [Google Scholar] [CrossRef]
- Khan, A.; Wang, Z.; Chen, Z.; Bu, J.; Adnan, M.Y.; Zhang, M. Investigation of soil nutrients and associated rhizobacterial communities in different sugarcane provenances in relation to sugar content. Chem. Biol. Technol. Agric. 2021, 8, 59. [Google Scholar] [CrossRef]
- Jiang, O.Y.; Li, L.; Duan, G.L.; Gustave, W.; Zhai, W.W.; Zou, L.N.; An, X.; Tang, X.J.; Xu, J.M. Root exudates increased arsenic mobility and altered microbial community in paddy soils. J. Environ. Sci. 2023, 127, 410–420. [Google Scholar] [CrossRef]
- Walker, J.J.; Pace, N.R. Phylogenetic composition of rocky mountain endolithic microbial ecosystems. Appl. Environ. Microbiol. 2007, 73, 3497–3504. [Google Scholar] [CrossRef]
- Dos Santos, M.; Kavamura, V.; Reynaldo, E.; Souza, D.; Da Silva, E.; May, A. Bacterial structure of agricultural soils with high and low yields. J. Plant Pathol. Microbiol. 2017, 8, 405. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Beckers, B.; Op De Beeck, M.; Thijs, S.; Truyens, S.; Weyens, N.; Boerjan, W. Performance of 16s rDNA primer pairs in the study of rhizosphere and endosphere bacterial microbiomes in metabarcoding studies. Front. Microbiol. 2016, 7, 650. [Google Scholar] [CrossRef]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR—Protocols and Applications—A Laboratory Manual; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End read merger. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Ballentine, B.; Greenberg, R. Common garden experiment reveals genetic control of phenotypic divergence between swamp sparrow subspecies that lack divergence in neutral provenances. PLoS ONE 2010, 5, e10229. [Google Scholar] [CrossRef]
- Yuan, S.; Guo, C.; Ma, L.; Wang, R.Z. Environmental conditions and genetic differentiation: What drives the divergence of coexisting Leymus chinensis ecotypes in a large-scale longitudinal gradient. J. Plant Ecol. 2016, 9, 616–628. [Google Scholar] [CrossRef]
- Wendling, M.; Büchi, L.; Amossé, C.; Sinaj, S.; Walter, A.; Charles, R. Influence of root and leaf traits on the uptake of nutrients in cover crops. Plant Soil 2016, 409, 419–434. [Google Scholar] [CrossRef]
- Gloy, J.; Herzschuh, U.; Kruse, S. Evolutionary adaptation of trees and modelled future larch forest extent in Siberia. Ecol. Modell. 2023, 478, 110278. [Google Scholar] [CrossRef]
- Guevara, M.Á.; Sánchez-Gómez, D.; Vélez, M.D.; de María, N.; Díaz, L.M.; Ramírez-Valiente, J.A.; Mancha, J.A.; Aranda, I.; Cervera, M.T. Epigenetic and Genetic Variability in Contrasting Latitudinal Fagus sylvatica L. provenances. Forests 2022, 13, 1971. [Google Scholar] [CrossRef]
- Wang, H.; Lin, S.; Dai, J.; Ge, Q. Modeling the effect of adaptation to future climate change on spring phenological trend of European beech (Fagus sylvatica L.). Sci. Total Environ. 2022, 846, 157540. [Google Scholar] [CrossRef]
- Rabarijaona, A.; Ponton, S.; Bert, D.; Ducousso, A.; Richard, B.; Levillain, J.; Brendel, O. Provenance Differences in Water-Use Efficiency Among Sessile Oak Populations Grown in a Mesic Common Garden. Front. For. Glob. Change 2022, 9, 199. [Google Scholar] [CrossRef]
- Kawecki, T.J.; Ebert, D. Conceptual issues in local adaptation. Ecol. Lett. 2004, 7, 1225–1241. [Google Scholar] [CrossRef]
- Zhou, Z.C.; Xie, Y.R.; Jin, G.Q.; Wu, J.F.; Chen, Y. Genetic response of Pinus massoniana provenances to phosphorus supply and nutrient characteristics of their rhizosphere soil. Sci. Silvae Sin. 2003, 6, 62–67. [Google Scholar]
- Ye, G.F.; Zhang, Y.; Luo, M.J.; Xu, j.S.; Yu, X.J. Effects of low phosphorus stress on nutritional status of Casuarina equisetifolia provenances at seedling stage. Straits Sci. 2008, 22, 51–52+59. [Google Scholar]
- Ling, Y.T. Analysis on the effect of release-thinning and pruning to mixed forest of Fokienia hodginsii and Pinus massoniana. Wuyi Sci. J. 2007, 23, 110–114. [Google Scholar]
- Su, Z.W. Effects of different densities on understory plants and soil fertility of Fokienia hodginsii. East China For. Manag. 2007, 4, 26–28. [Google Scholar]
- Ji, Z.J.; Quan, X.K.; Wang, C.K. Variations in leaf anatomy of Larix gmelinii reflect adaptation of its photosynthetic capacity to climate changes. Acta Ecol. Sin. 2013, 33, 6967–6974. [Google Scholar]
- Ahrens, C.W.; Andrew, M.E.; Mazanec, R.A.; Ruthrof, K.X.; Challis, A.; Hardy, G.; Byrne, M.; Tissue, D.T.; Rymer, P.D. Plant functional traits differ in adaptability and are predicted to be differentially affected by climate change. Ecol. Evol. 2019, 10, 232–248. [Google Scholar] [CrossRef]
- Gougherty, A.V.; Keller, S.R.; Fitzpatrick, M.C. Maladaptation, migration and extirpation fuel climate change risk in a forest tree species. Nat. Clim. Chang. 2021, 11, 166–171. [Google Scholar] [CrossRef]
- Gao, M.; Xiong, C.; Gao, C.; Tsui, C.K.M.; Wang, M.M.; Zhou, X.; Zhang, A.M.; Cai, L. Disease-induced changes in plant microbiome assembly and functional adaptation. Microbiome 2021, 9, 187. [Google Scholar] [CrossRef]
- Rolfe, S.A.; Griffiths, J.; Ton, J. Crying out for help with root exudates: Adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes. Curr. Opin. Microbiol. 2019, 49, 73–82. [Google Scholar] [CrossRef]
- Wen, T.; Zhao, M.; Liu, T.; Huang, Q.; Yuan, J.; Shen, Q. High abundance of Ralstonia solanacearum changed tomato rhizosphere microbiome and metabolome. BMC Plant Biol. 2020, 20, 166. [Google Scholar] [CrossRef]
- Dubey, A.; Malla, M.A.; Kumar, A. Taxonomical and functional bacterial community profiling in disease-resistant and disease-susceptible soybean provenances. Braz. J. Microbiol. 2022, 53, 1355–1370. [Google Scholar] [CrossRef]
- Singh, A.; Kumar, M.; Chakdar, H.; Pandiyan, K.; Kumar, S.C.; Zeyad, M.T.; Singh, B.N.; Ravikiran, K.T.; Mahto, A.; Srivastava, A.K.; et al. Influence of host genotype in establishing root associated microbiome of indica rice provenances for plant growth promotion. Front. Microbiol. 2022, 13, 1033158. [Google Scholar] [CrossRef]
- Li, Z.; Ma, L.; Zhang, Y.; Zhao, W.H.; Zhao, B.Z.; Zhang, J.B. Effect of wheat provenances with different resistance to Fusarium head blight on rhizosphere Fusarium graminearum abundance and microbial community composition. Plant Soil 2020, 448, 383–397. [Google Scholar] [CrossRef]
- Beschoren da Costa, P.; Benucci, G.M.N.; Chou, M.Y.; Van Wyk, J.; Chretien, M.; Bonito, G. Soil Origin and Plant Genotype Modulate Switchgrass Aboveground Productivity and Root Microbiome Assembly. mBio 2020, 13, e0007922. [Google Scholar] [CrossRef]
- Anderson, M.; Habiger, J. Characterization and identification of productivity-associated rhizobacteria in wheat. Appl. Environ. Microbiol. 2021, 78, 4434–4446. [Google Scholar] [CrossRef]
- Alahmad, A.; Edelman, L.; Castel, L.; Bernardon-Mery, A.; Laval, K.; Trinsoutrot-Gattin, I.; Thioye, B. Prebiotics: A Solution for Improving Plant Growth, Soil Health, and Carbon Sequestration. J. Soil Sci. Plant Nutr. 2023, 23, 6647–6669. [Google Scholar] [CrossRef]
- Wu, T.Y.; Wang, Y.H.; Wu, F.; Wu, X.Q. Dual inoculation with rhizosphere-promoting bacterium Bacillus cereus and beneficial fungus Peniophora cinerea improves salt stress tolerance and productivity in willow. Microbiol. Res. 2023, 268, 127280. [Google Scholar] [CrossRef]
- Guo, W.; Hao, H.; Zhang, W.H.; Hu, Z.H.; Leng, P.S. Ectomycorrhizal fungi enhance salt tolerance of Quercus mongolica by regulating ion balance. J. Ecol. 2022, 33, 3303–3311. [Google Scholar]
- Mautner, M.N.; Conner, A.J.; Killham, K.; Deamer, D.W. Biological potential of extraterrestrial materials Microbial and plant responses to nutrients in the Murchison carbonaceous meteorite. Icarus 1997, 129, 245–253. [Google Scholar] [CrossRef]
- Jinal, H.N.; Gopi, K.; Prittesh, P.; Kartik, V.P.; Amaresan, N. Phytoextraction of iron from contaminated soils by inoculation of iron-tolerant plant growth-promoting bacteria in Brassica juncea L. Czern. Environ. Sci. Pollut. Res. 2019, 26, 32815–32823. [Google Scholar] [CrossRef]
- Sharma, P.; Chaturvedi, P.; Chandra, R.; Kumar, S. Identification of heavy metals tolerant Brevundimonas sp. from rhizospheric zone of Saccharum munja L. and their efficacy in in-situ phytoremediation. Chemosphere 2022, 295, 133823. [Google Scholar] [CrossRef]
- Chelius, M.K.; Triplett, E.W. The Diversity of Archaea and Bacteria in Association with the Roots of Zea mays L. Microb. Ecol. 2001, 41, 252–263. [Google Scholar] [CrossRef]
- Simon, H.M.; Dodsworth, J.A.; Goodman, R.M. Crenarchaeota colonize terrestrial plant roots. Environ. Microbiol. 2000, 2, 495–505. [Google Scholar] [CrossRef]
- Orellana, L.H.; Francis, T.B.; Ferraro, M.; Hehemann, J.H.; Fuchs, B.M.; Amann, R.I. Verrucomicrobiota are specialist consumers of sulfated methyl pentoses during diatom blooms. ISME J. 2022, 16, 630–641. [Google Scholar] [CrossRef]
- Sichert, A.; Corzett, C.H.; Schechter, M.S.; Unfried, F.; Markert, S.; Becher, D.; Fernandez-Guerra, A.; Liebeke, M.; Schweder, T.; Polz, M.F.; et al. Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan. Nat. Microbiol. 2020, 5, 1026–1039. [Google Scholar] [CrossRef]
- He, S.; Stevens, S.L.R.; Chan, L.K.; Bertilsson, S.; Glavina Del Rio, T.; Tringe, S.G.; Malmstrom, R.R.; McMahon, K.D. Ecophysiology of Freshwater Verrucomicrobia Inferred from Metagenome-Assembled Genomes. mSphere 2017, 2, e00277-17. [Google Scholar] [CrossRef]
- Prathna, T.C. Significance of microbial volatiles in ecological health: Impact on wetland systems. In Volatiles and Metabolites of Microbes; Academic Press: Cambridge, MA, USA, 2021; pp. 163–176. [Google Scholar]
- Cao, M.M.; Wang, F.; Zhou, B.H.; Chen, H.L.; Yuan, R.F. Community Distribution of the Rhizospheric and Endophytic Bacteria of Phragmites australis and Their Limiting Factors in Iron Tailings. Huan Jing Ke Xue 2021, 42, 4998–5009. [Google Scholar]
- Li, Y.Y.; Pan, F.X.; Yao, H.Y. Response of symbiotic and asymbiotic nitrogen-fixing microorganisms to nitrogen fertilizer application. J. Soils Sediments 2019, 19, 1948–1958. [Google Scholar] [CrossRef]
- Fernandes, G.L.; Shenoy, B.D.; Damare, S.R. Diversity of bacterial community in the oxygen minimum zones of Arabian Sea and Bay of Bengal as deduced by Illumina sequencing. Front. Microbiol. 2020, 10, 3153. [Google Scholar] [CrossRef]
- Paingankar, M.S.; Ahire, K.; Mishra, P.; Rajpathak, S.; Deobagkar, D.D. Microbial diversity of the Arabian Sea in the oxygen minimum zones by metagenomics approach. BioRxiv 2019. BioRxiv:731828. [Google Scholar]
- Li, X.Y.; Duan, A.; Zhang, J.G. Site index for Chinese fir plantations varies with climatic and soil factors in southern China. J. For. Res. 2022, 33, 1765–1780. [Google Scholar] [CrossRef]
- Mao, L.; Zha, R.; Chen, S.; Zhang, J.; Jie, L.; Zha, X. Mixture Compound Fertilizer and Super Absorbent Polymer Application Significantly Promoted Growth and Increased Nutrient Levels in Pinus massoniana Seedlings and Soil in Seriously Eroded Degradation Region of Southern China. Front. Plant Sci. 2021, 12, 763175. [Google Scholar] [CrossRef]
- Tarin, M.W.K.; Fan, L.; Xie, D.; Tayyab, M.; Rong, J.; Chen, L.; Muneer, M.A.; Zheng, Y. Response of Soil Fungal Diversity and Community Composition to Varying Levels of Bamboo Biochar in Red Soils. Microorganisms 2021, 9, 1385. [Google Scholar] [CrossRef]
- Li, Y.Y.; Zhao, M.; Chen, W.W.; Du, H.Y.; Xie, X.D.; Wang, D.B.; Dai, Y.; Xia, Q.Y.; Wang, G.H. Comparative transcriptomic analysis reveals that multiple hormone signal transduction and carbohydrate metabolic pathways are affected by Bacillus cereus in Nicotiana tabacum. Genomics 2020, 112, 4254–4267. [Google Scholar] [CrossRef]
- Feng, J.; Rana, S.; Liu, Z.; Wang, Y.M.; Cai, Q.F.; Geng, X.D.; Zhou, H.N.; Zhang, T.; Wang, S.S.; Xue, X.Y.; et al. Diversity Analysis of Leaf Nutrient Endophytes and Metabolites in Dioecious Idesia polycarpa Maxim Leaves during Reproductive Stages. Life 2022, 12, 2041. [Google Scholar] [CrossRef]
- Li, F.; Zhang, X.M.; Gong, J.Y.; Liu, L.X.; Yi, Y. Specialized core bacteria associate with plants adapted to adverse environment with high calcium contents. PLoS ONE 2018, 13, e0194080. [Google Scholar] [CrossRef]
- Xiong, M.Y.; Jiang, W.; Zou, S.Z.; Kang, D.; Yan, X.C. Microbial carbohydrate-active enzymes influence soil carbon by regulating the of plant- and fungal-derived biomass decomposition in plateau peat wetlands under differing water conditions. Front. Microbiol. 2023, 14, 1266016. [Google Scholar] [CrossRef]
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, T.; Mao, H.; Jin, H.; Sun, Y.; Qi, Z. A Leymus chinensis histidine-rich Ca2+-binding protein binds Ca2+/Zn2+ and suppresses abscisic acid signaling in Arabidopsis. J. Plant Physiol. 2020, 252, 153–209. [Google Scholar] [CrossRef]
- Gu, S.; Yang, T.; Shao, Z.; Wang, T.; Cao, K.; Jousset, A.; Friman, V.P.; Mallon, C.; Mei, X.; Wei, Z.; et al. Siderophore-Mediated Interactions Determine the Disease Suppressiveness of Microbial Consortia. mSystems 2020, 5, e00811-19. [Google Scholar] [CrossRef]
- Kim, S.C.; Wang, X. Phosphatidic acid: An emerging versatile class of cellular mediators. Essays Biochem. 2020, 64, 533–546. [Google Scholar]
- Richard, C.T.; Elizabeth, A.H.; Pilon, S. Selenium transport and metabolism in plants: Phytoremediation and biofortification implications. J. Hazard. Mater. 2021, 4, 0304–3894. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.-L.; Zhu, T.; Wen, X.; Zhao, Q.; Chen, Y.; Wang, Y.-Z.; Li, J.; Su, S. Chemical and Microbial Differences of Root and Rhizosphere Soil among Different Provenances of Fokienia hodginsii. Forests 2024, 15, 1005. https://doi.org/10.3390/f15061005
Liu H-L, Zhu T, Wen X, Zhao Q, Chen Y, Wang Y-Z, Li J, Su S. Chemical and Microbial Differences of Root and Rhizosphere Soil among Different Provenances of Fokienia hodginsii. Forests. 2024; 15(6):1005. https://doi.org/10.3390/f15061005
Chicago/Turabian StyleLiu, Hao-Lan, Tengfei Zhu, Xinyi Wen, Qing Zhao, Yao Chen, Yun-Zi Wang, Jian Li, and Shunde Su. 2024. "Chemical and Microbial Differences of Root and Rhizosphere Soil among Different Provenances of Fokienia hodginsii" Forests 15, no. 6: 1005. https://doi.org/10.3390/f15061005
APA StyleLiu, H.-L., Zhu, T., Wen, X., Zhao, Q., Chen, Y., Wang, Y.-Z., Li, J., & Su, S. (2024). Chemical and Microbial Differences of Root and Rhizosphere Soil among Different Provenances of Fokienia hodginsii. Forests, 15(6), 1005. https://doi.org/10.3390/f15061005





