Environmental Response of Tree Species Distribution in Northeast China with the Joint Species Distribution Model
Abstract
:1. Introduction
2. Materials and Methods
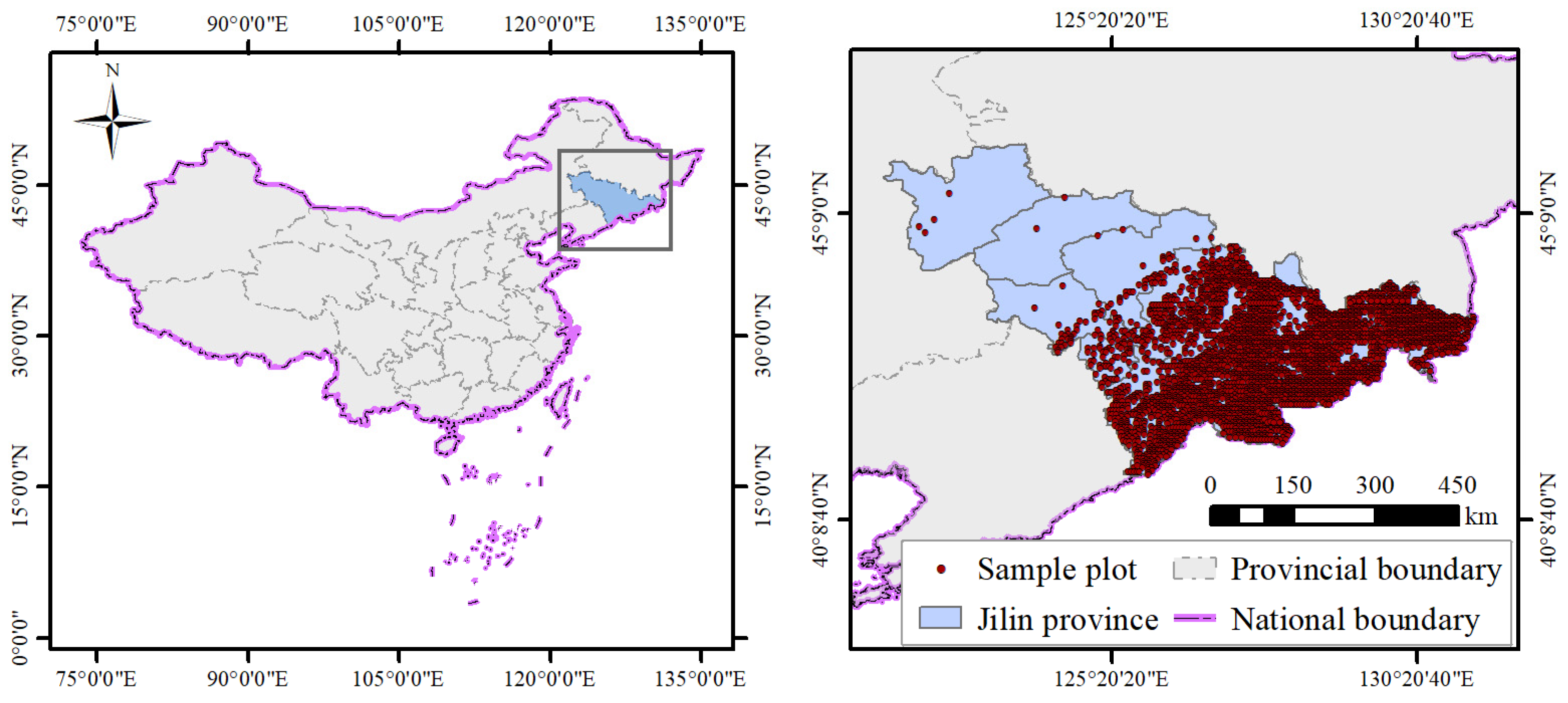
2.1. Study Area
2.2. Data Collection
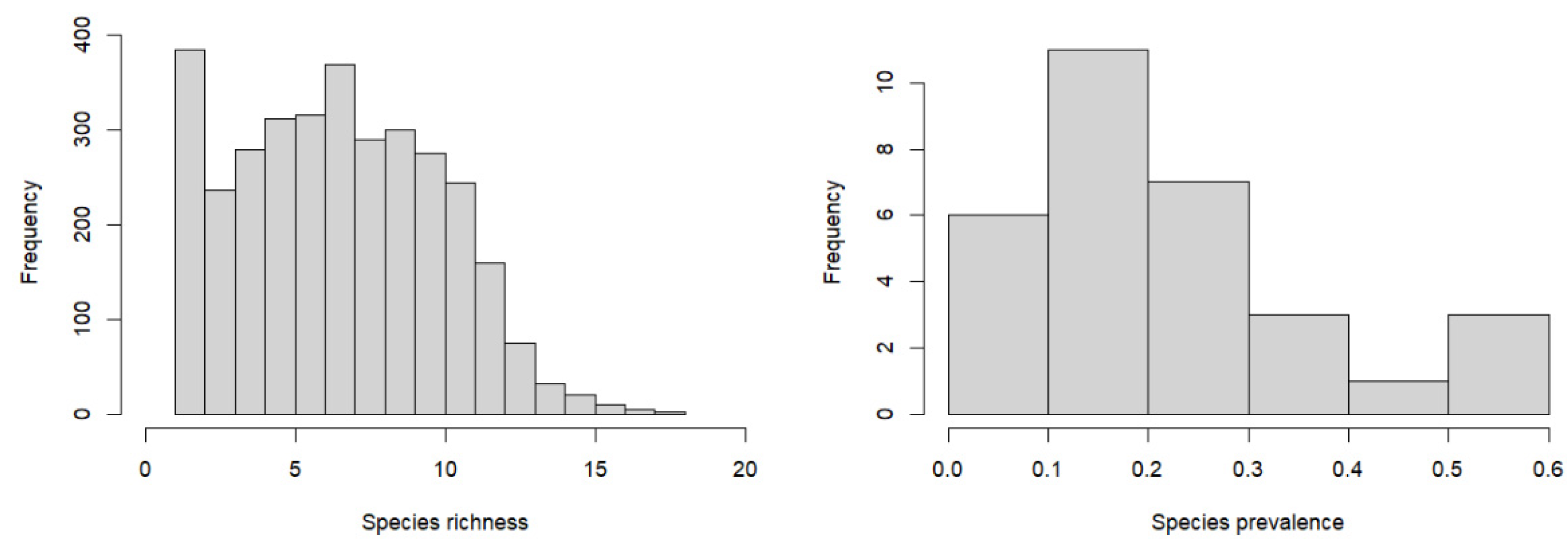
2.2.1. Plot Data
2.2.2. Climate Data
2.2.3. Soil Data
2.2.4. Tree Species Trait Factors Data
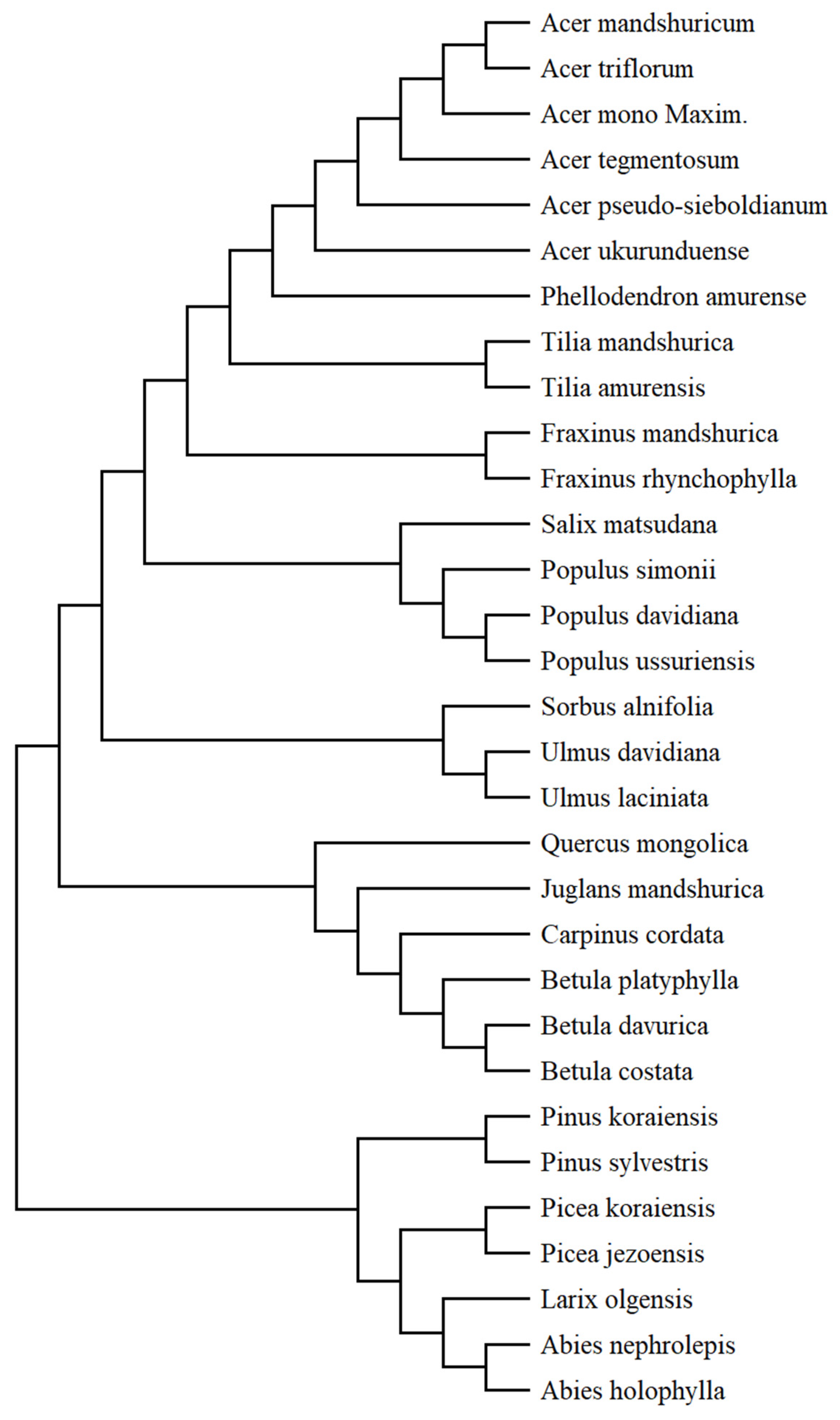
2.2.5. Tree Species Phylogenetic Data
2.3. Methods
2.3.1. Model Structure Setup and Fitting
2.3.2. Variable Selection
2.3.3. Model Evaluation Metrics
2.3.4. Data Analysis Tools
3. Results
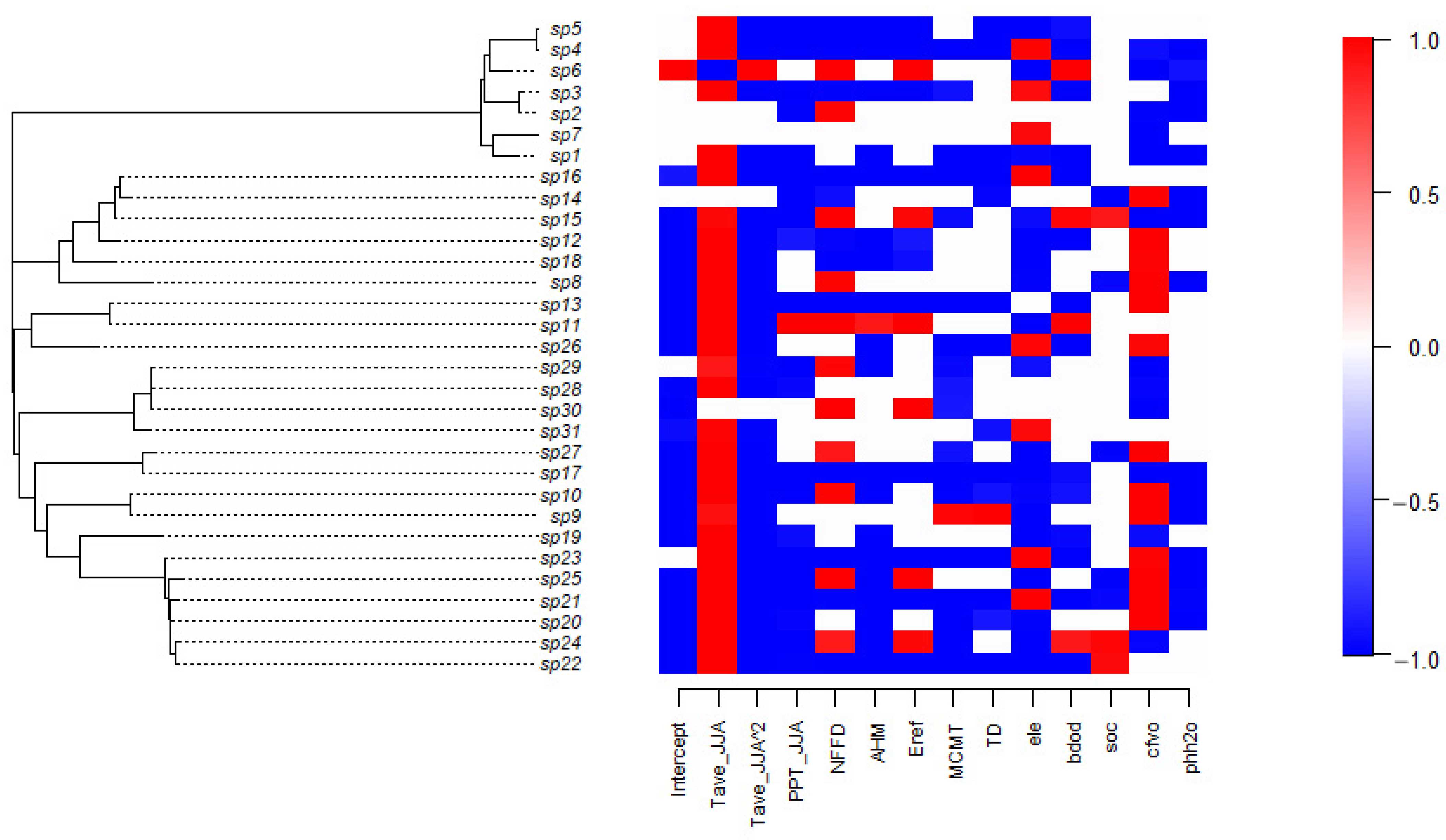
3.1. Tree Species Distribution Patterns
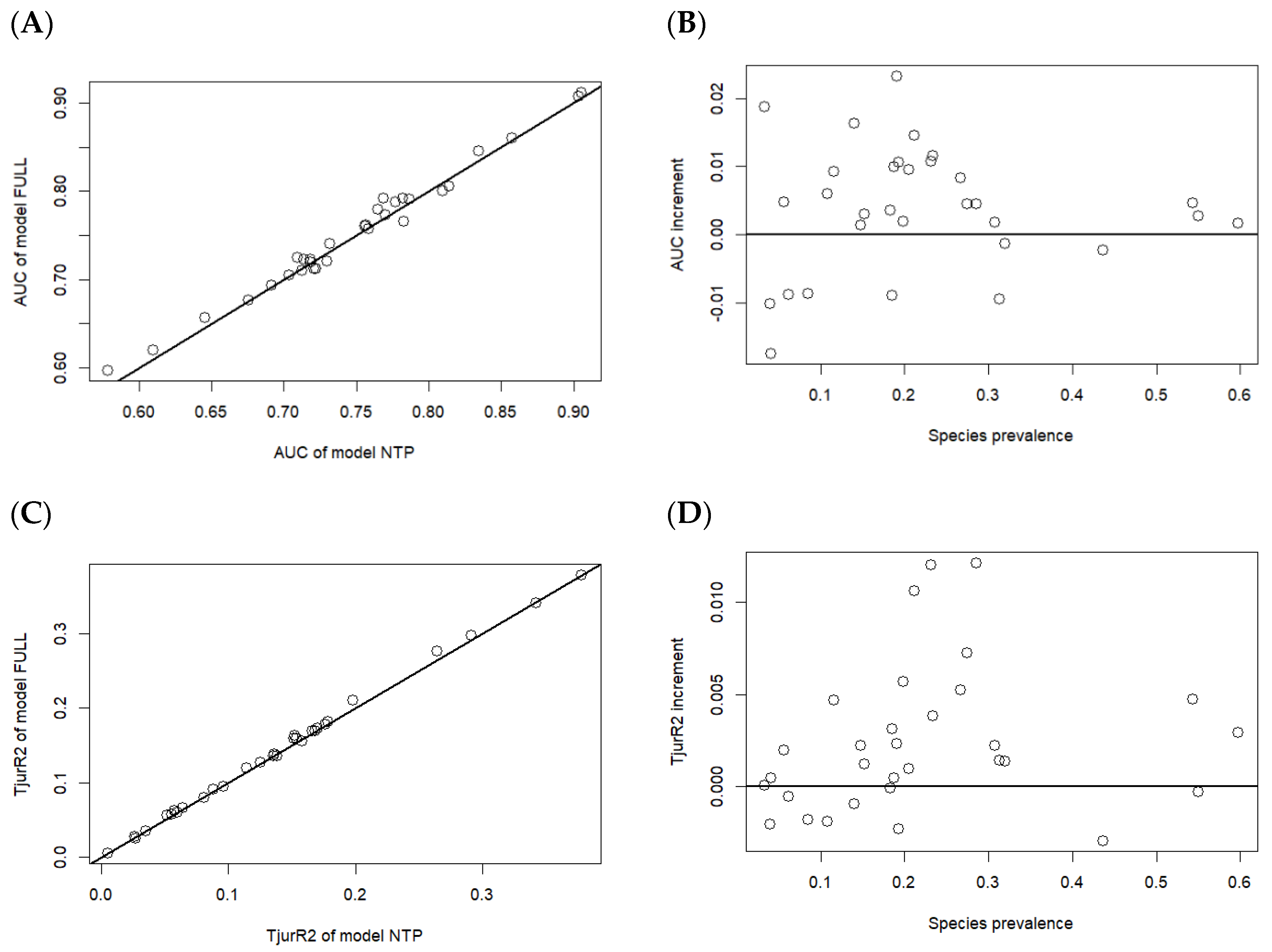
3.2. Model Interpretability and Predictive Power
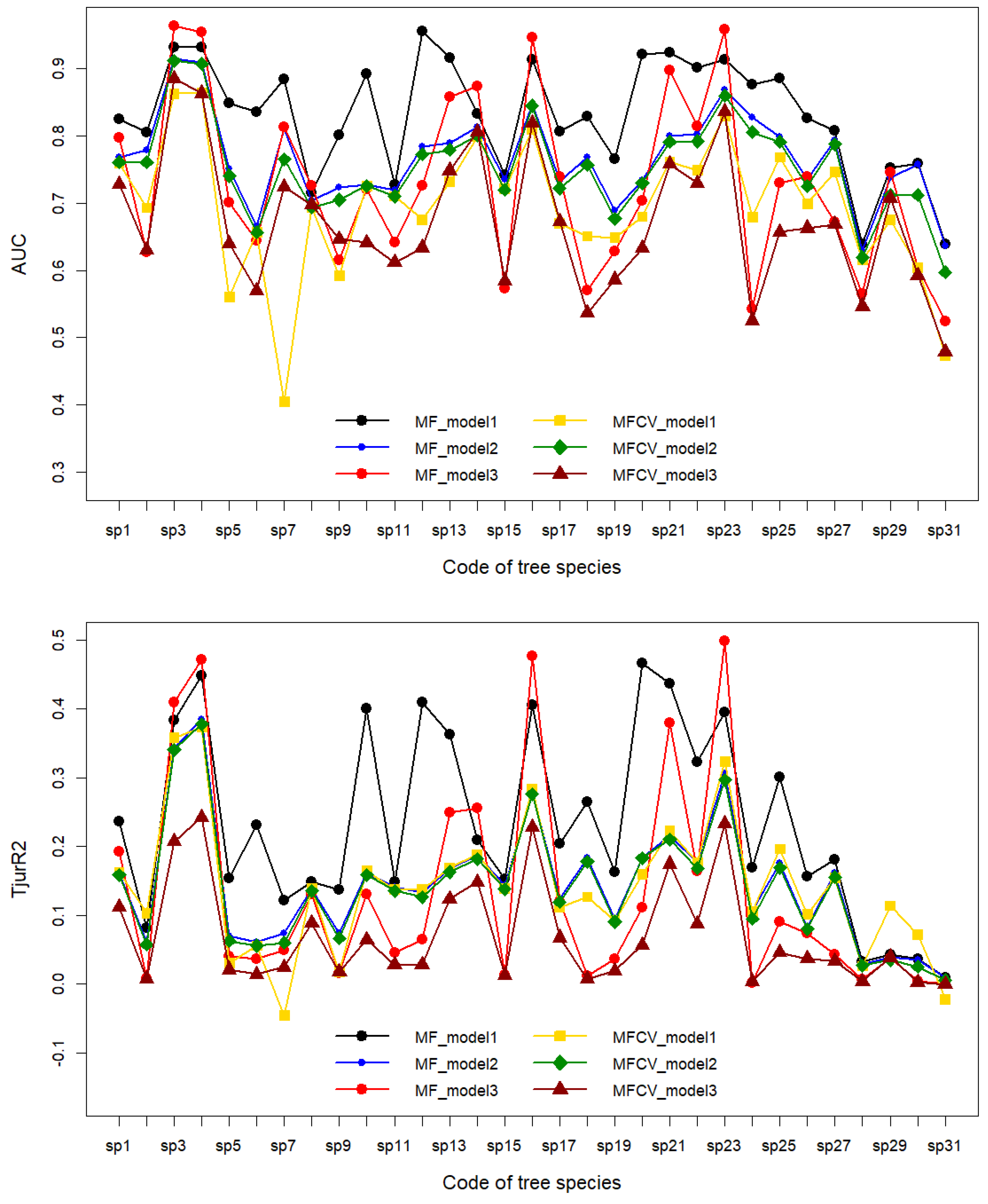
3.2.1. Overall Evaluation of Interpretability and Predictive Power of the Three Models
3.2.2. Evaluation of Interpretability and Predictive Power by Tree Species
3.3. Variance Contribution of Environmental Covariates
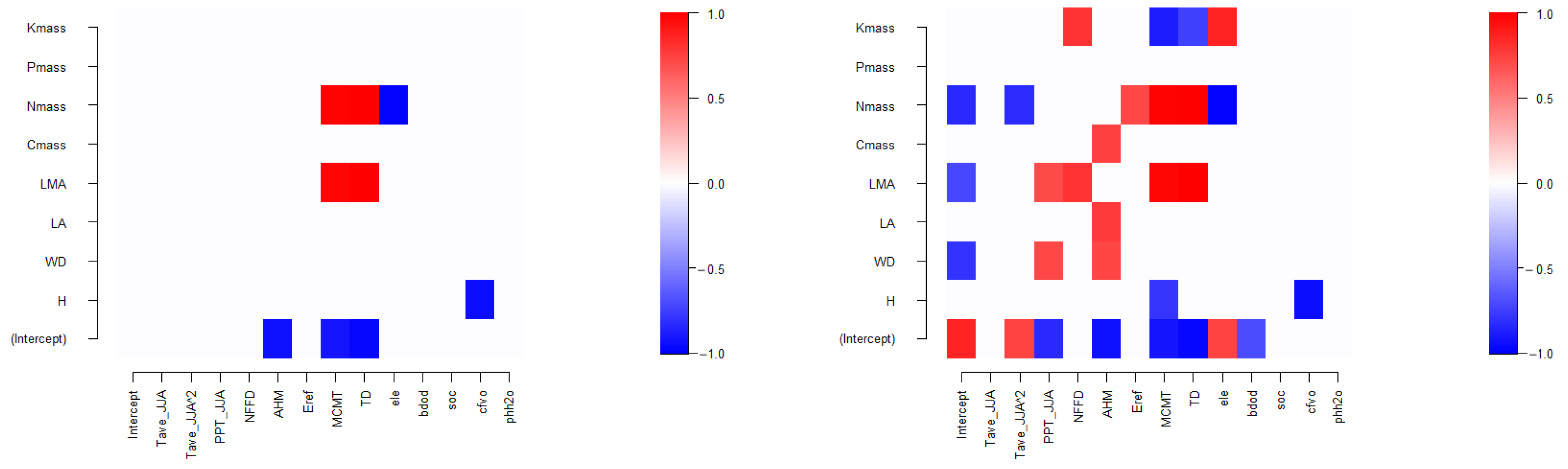
3.4. Tree Species Niches
4. Discussion
4.1. Relationship between Tree Species Niches and Traits—Phylogeny
4.2. Impact of Introducing Tree Species Traits and Phylogenetic Trees on Prediction
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Duan, G.; Lei, X.; Zhang, X.; Liu, X. Site index modeling of larch using a mixed-effects model across regional site types in northern China. Forests 2022, 13, 815. [Google Scholar] [CrossRef]
- Hu, X.F.; Duan, G.S.; Zhang, H.R. Modelling individual tree diameter growth of Quercus mongolica secondary forest in the northeast of China. Sustainability 2021, 13, 4533. [Google Scholar] [CrossRef]
- National Forestry and Grassland Administration. China Forest Resources Report 2014–2018; China Forestry Publishing House: Beijing, China, 2019. (In Chinese)
- Liu, D. Research on Quantitative Place Suitability and Tree Suitability Based on Distribution Suitability and Potential Productivity; Chinese Academy of Forestry: Beijing, China, 2018. (In Chinese) [Google Scholar]
- Lan, J.; Lei, X.; He, X.; Gao, W.; Guo, H. Stand density, climate and biodiversity jointly regulate the multifunctionality of natural forest ecosystems in northeast China. Eur. J. For. Res. 2023, 142, 493–507. [Google Scholar] [CrossRef]
- Leng, W.; He, H.S.; Bu, R.; Dai, L.; Hu, Y.; Wang, X. Predicting the distributions of suitable habitat for three larch species under climate warming in northeastern China. For. Ecol. Manag. 2008, 254, 420–428. [Google Scholar] [CrossRef]
- Wang, X.; Fang, J.; Sanders, N.J.; White, P.S.; Tang, Z. Relative importance of climate vs local factors in shaping the regional patterns of forest plant richness across northeast China. Ecography 2009, 32, 133–142. [Google Scholar] [CrossRef]
- Yin, X.J.; Zhou, G.S.; Sui, X.H.; He, Q.J.; Li, R.P. Dominant climatic factors and their thresholds in the geographical distribution of Quercus mongolica. Acta Ecol. Sin. 2013, 33, 103–109. (In Chinese) [Google Scholar]
- Liu, D.; Li, Y.T.; Hong, L.X.; Guo, H.; Xie, Y.S.; Zhang, Z.L.; Lei, X.D.; Tang, S.Z. Potential distribution suitability of major natural forests in Jilin Province based on maximum entropy model. Sci. Silvernica Sin. 2018, 54, 1–15. (In Chinese) [Google Scholar]
- Pugnaire, F.I. Positive Plant Interactions and Community Dynamics; CRC Press: Boca Raton, FL, USA, 2010. [Google Scholar]
- Wisz, M.S.; Pottier, J.; Kissling, W.D.; Pellissier, L.; Lenoir, J.; Damgaard, C.F.; Dormann, C.F.; Forchhammer, M.C.; Grytnes, J.-A.; Guisan, A.; et al. The role of biotic interactions in shaping distributions and realised assemblages of species: Implications for species distribution modelling. Biol. Rev. 2013, 88, 15–30. [Google Scholar] [CrossRef]
- Barrero, A.; Ovaskainen, O.; Traba, J.; Gómez-Catasús, J. Co-occurrence patterns in a steppe bird community: Insights into the role of dominance and competition. Oikos 2023, 2023, e09780. [Google Scholar] [CrossRef]
- Ovaskainen, O.; Soininen, J. Making more out of sparse data: Hierarchical modeling of species communities. Ecology 2011, 92, 289–295. [Google Scholar] [CrossRef]
- Randin, C.F.; Ashcroft, M.B.; Bolliger, J.; Cavender-Bares, J.; Coops, N.C.; Dullinger, S.; Dirnböck, T.; Eckert, S.; Ellis, E.; Fernández, N.; et al. Monitoring biodiversity in the Anthropocene using remote sensing in species distribution models. Remote Sens. Environ. 2020, 239, 111626. [Google Scholar] [CrossRef]
- Chardon, N.I.; Pironon, S.; Peterson, M.L.; Doak, D.F. Incorporating intraspecific variation into species distribution models improves distribution predictions, but cannot predict species traits for a wide-spread plant species. Ecography 2020, 43, 60–74. [Google Scholar] [CrossRef]
- Ovaskainen, O.; Abrego, N. Joint Species Distribution Modelling with Applications in R; Cambridge University Press: Cambridge, UK, 2020. [Google Scholar]
- Clark, J.S.; Gelfand, A.E.; Woodall, C.W.; Zhu, K. More than the sum of the parts: Forest climate response from joint species distribution models. Ecol. Appl. 2014, 24, 990–999. [Google Scholar] [CrossRef] [PubMed]
- Warton, D.I.; Blanchet, F.G.; O’hara, R.B.; Ovaskainen, O.; Taskinen, S.; Walker, S.C.; Hui, F.K. So many variables: Joint modeling in community ecology. Trends Ecol. Evol. 2015, 30, 766–779. [Google Scholar] [CrossRef] [PubMed]
- Harris, D.J. Generating realistic assemblages with a joint species distribution model. Methods Ecol. Evol. 2015, 6, 465–473. [Google Scholar] [CrossRef]
- Hui, F.K.C. Boral-Bayesian ordination and regression analysis of multivariate abundance data in r. Methods Ecol. Evol. 2016, 7, 744–750. [Google Scholar] [CrossRef]
- Ovaskainen, O.; Tikhonov, G.; Norberg, A.; Blanchet, F.G.; Duan, L.; Dunson, D.; Roslin, T.; Abrego, N. How to make more out of community data? A conceptual framework and its implementation as models and software. Ecol. Lett. 2017, 20, 561–576. [Google Scholar] [CrossRef] [PubMed]
- Zurell, D.; Pollock, L.J.; Thuiller, W. Do joint species distribution models reliably detect interspecific interactions from co-occurrence data in homogenous environments. Ecography 2018, 41, 1812–1819. [Google Scholar] [CrossRef]
- Wang, T.; Wang, G.; Innes, J.L.; Seely, B.; Chen, B. ClimateAP: An application for dynamic local downscaling of historical and future climate data in Asia Pacific. Front. Agric. Sci. Eng. 2017, 4, 448–458. [Google Scholar] [CrossRef]
- Poggio, L.; de Sousa, L.M.; Batjes, N.H.; Heuvelink, G.B.M.; Kempen, B.; Ribeiro, E.; Rossiter, D. SoilGrids 2.0: Producing soil information for the globe with quantified spatial uncertainty. Soil 2021, 7, 217–240. [Google Scholar] [CrossRef]
- Tikhonov, G.; Duan, L.; Abrego, N.; Newell, G.; White, M.; Dunson, D.; Ovaskainen, O. Computationally efficient joint species distribution modeling of big spatial data. Ecology 2019, 101, e02929. [Google Scholar] [CrossRef] [PubMed]
- Norberg, A.; Abrego, N.; Blanchet, F.G.; Adler, F.R.; Anderson, B.J.; Anttila, J.; Araújo, M.B.; Dallas, T.; Dunson, D.; Elith, J.; et al. A comprehensive evaluation of predictive performance of 33 species distribution models at species and community levels. Ecol. Monogr. 2019, 89, e01370. [Google Scholar] [CrossRef]
- Tjur, T. Coefficients of determination in logistic regression models a new proposal: The coefficient of discrimination. Am. Stat. 2009, 63, 366–372. [Google Scholar] [CrossRef]
- Valavi, R.; Guillera-Arroita, G.; Lahoz-Monfort, J.J.; Elith, J. Predictive performance of presence-only species distribution models: A benchmark study with reproducible code. Ecol. Monogr. 2022, 92, e01486. [Google Scholar] [CrossRef]
- Tikhonov, G.; Opedal, H.; Abrego, N.; Lehikoinen, A.; de Jonge, M.M.J.; Oksanen, J.; Ovaskainen, O. Joint species distribution modelling with the R-package Hmsc. Methods Ecol. Evol. 2020, 11, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Abrego, N.; Ovaskainen, O. Evaluating the predictive performance of presence–absence models: Why can the same model appear excellent or poor? Ecol. Evol. 2023, 13, e10784. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; François, R.; Henry, L.; Müller, K. dplyr: A Grammar of Data Manipulation. R Package Version 1.1.2. 2023. Available online: https://CRAN.R-project.org/package=dplyr (accessed on 15 October 2023).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Tikhonov, G.; Ovaskainen, O.; Oksanen, J.; De Jonge, M.; Opedal, O.; Dallas, T. Hmsc: Hierarchical Model of Species Communities. R Package Version 3.0-13. 2022. Available online: https://CRAN.R-project.org/package=Hmsc (accessed on 15 October 2023).
- Zhang, C.; Chen, Y.; Xu, B.; Xue, Y.; Ren, Y. Improving prediction of rare species’ distribution from community data. Sci. Rep. 2020, 10, 12230. [Google Scholar] [CrossRef]
- Blanco-Cano, L.; Navarro-Cerrillo, R.M.; González-Moreno, P. Biotic and abiotic effects determining the resilience of conifer mountain forests: The case study of the endangered Spanish fir. For. Ecol. Manag. 2022, 520, 120356. [Google Scholar] [CrossRef]
| Variable | Unit | Mean | Standard Deviation | Min | Max |
|---|---|---|---|---|---|
| Tave_JJA | °C | 19.01 | 1.38 | 12.60 | 22.80 |
| PPT_JJA | mm | 448.73 | 66.99 | 277.00 | 690.00 |
| NFFD | / | 172.10 | 10.37 | 129.00 | 202.00 |
| AHM | / | 19.18 | 3.76 | 7.90 | 44.00 |
| Eref | / | 688.20 | 32.97 | 401.00 | 839.00 |
| MCMT | °C | −15.89 | 0.94 | −19.10 | −10.50 |
| TD | °C | 36.22 | 1.29 | 32.00 | 40.50 |
| ele | m | 666.38 | 261.69 | 90.00 | 1860.00 |
| bdod | kg/dm3 | 132.57 | 3.43 | 118.84 | 144.05 |
| soc | g/kg | 269.52 | 56.43 | 65.96 | 516.38 |
| cfvo | cm3/100 cm3 | 235.78 | 37.38 | 81.78 | 407.60 |
| phh2o | pH | 60.25 | 2.22 | 52.23 | 83.14 |
| H | m | 24.68 | 9.56 | 8.00 | 50.00 |
| WD | g/cm3 | 0.50 | 0.11 | 0.32 | 0.71 |
| LA | m2 | 0.00 | 0.01 | 0.00 | 0.04 |
| LMA | kg/m2 | 0.08 | 0.08 | 0.02 | 0.38 |
| Cmass | g/kg | 407.84 | 82.73 | 240.20 | 512.77 |
| Nmass | g/kg | 17.67 | 7.01 | 2.20 | 30.86 |
| Pmass | g/kg | 1.65 | 0.65 | 0.67 | 3.86 |
| Kmass | g/kg | 12.42 | 6.31 | 5.02 | 30.25 |
| Models | Fitting | Cross Validation | ||
|---|---|---|---|---|
| AUC | Tjur R2 | AUC | Tjur R2 | |
| Model FULL | 0.8325 | 0.2326 | 0.6940 | 0.1412 |
| Model ENV | 0.7664 | 0.1454 | 0.7528 | 0.1399 |
| Model SPACE | 0.7297 | 0.1346 | 0.6719 | 0.0705 |
| Models | Climate | Site | Soil | Random |
|---|---|---|---|---|
| Model FULL | 0.5742 | 0.1275 | 0.0914 | 0.2069 |
| Model ENV | 0.7322 | 0.1617 | 0.1061 | 0.0000 |
| Model SPACE | 0.0000 | 0.0000 | 0.0000 | 1.0000 |
| Models | Model FULL | Model ENV | |
|---|---|---|---|
| Environmental covariates | Intercept | 0.3291 | 0.2521 |
| Tave_JJA | 0.0881 | 0.0659 | |
| Tave_JJA2 | 0.1521 | 0.0935 | |
| PPT_JJA | 0.1458 | 0.1254 | |
| NFFD | 0.0684 | 0.0839 | |
| AHM | 0.0541 | 0.0471 | |
| Eref | 0.0405 | 0.0494 | |
| MCMT | 0.4355 | 0.4569 | |
| TD | 0.4393 | 0.4522 | |
| ele | 0.3504 | 0.3229 | |
| bdod | 0.0418 | 0.0444 | |
| soc | 0.0394 | 0.0406 | |
| cfvo | 0.2119 | 0.1812 | |
| phh2o | 0.0677 | 0.0515 | |
| Species occurrence | 0.1832 | 0.1797 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yong, J.; Duan, G.; Chen, S.; Lei, X. Environmental Response of Tree Species Distribution in Northeast China with the Joint Species Distribution Model. Forests 2024, 15, 1026. https://doi.org/10.3390/f15061026
Yong J, Duan G, Chen S, Lei X. Environmental Response of Tree Species Distribution in Northeast China with the Joint Species Distribution Model. Forests. 2024; 15(6):1026. https://doi.org/10.3390/f15061026
Chicago/Turabian StyleYong, Juan, Guangshuang Duan, Shaozhi Chen, and Xiangdong Lei. 2024. "Environmental Response of Tree Species Distribution in Northeast China with the Joint Species Distribution Model" Forests 15, no. 6: 1026. https://doi.org/10.3390/f15061026







