Unraveling the Role of Bacteria in Nitrogen Cycling: Insights from Leaf Litter Decomposition in the Knyszyn Forest
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area, Experiment Design, and Litter Decomposition
2.2. DNA Extraction, Sequencing, and Bioinformatics
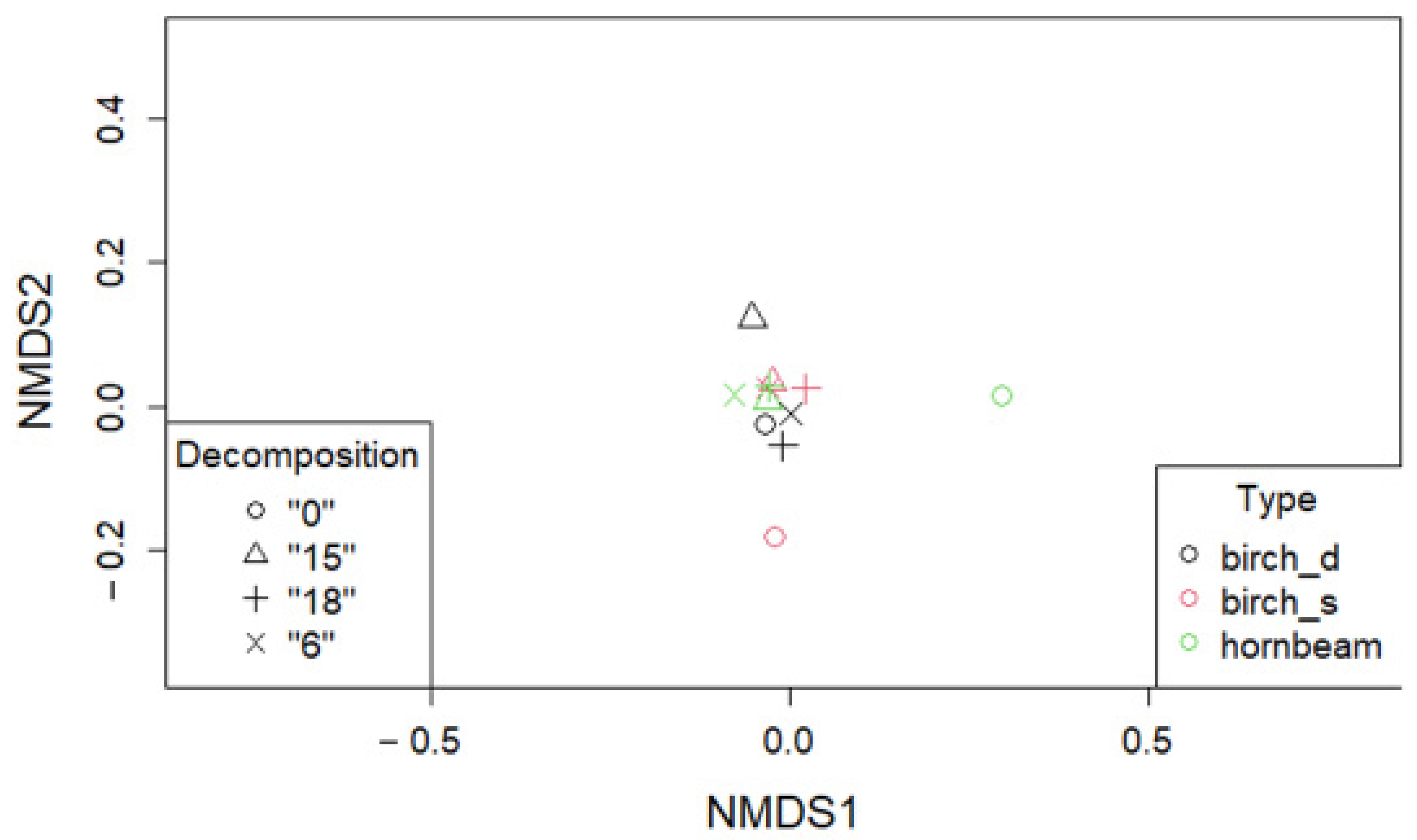
3. Results
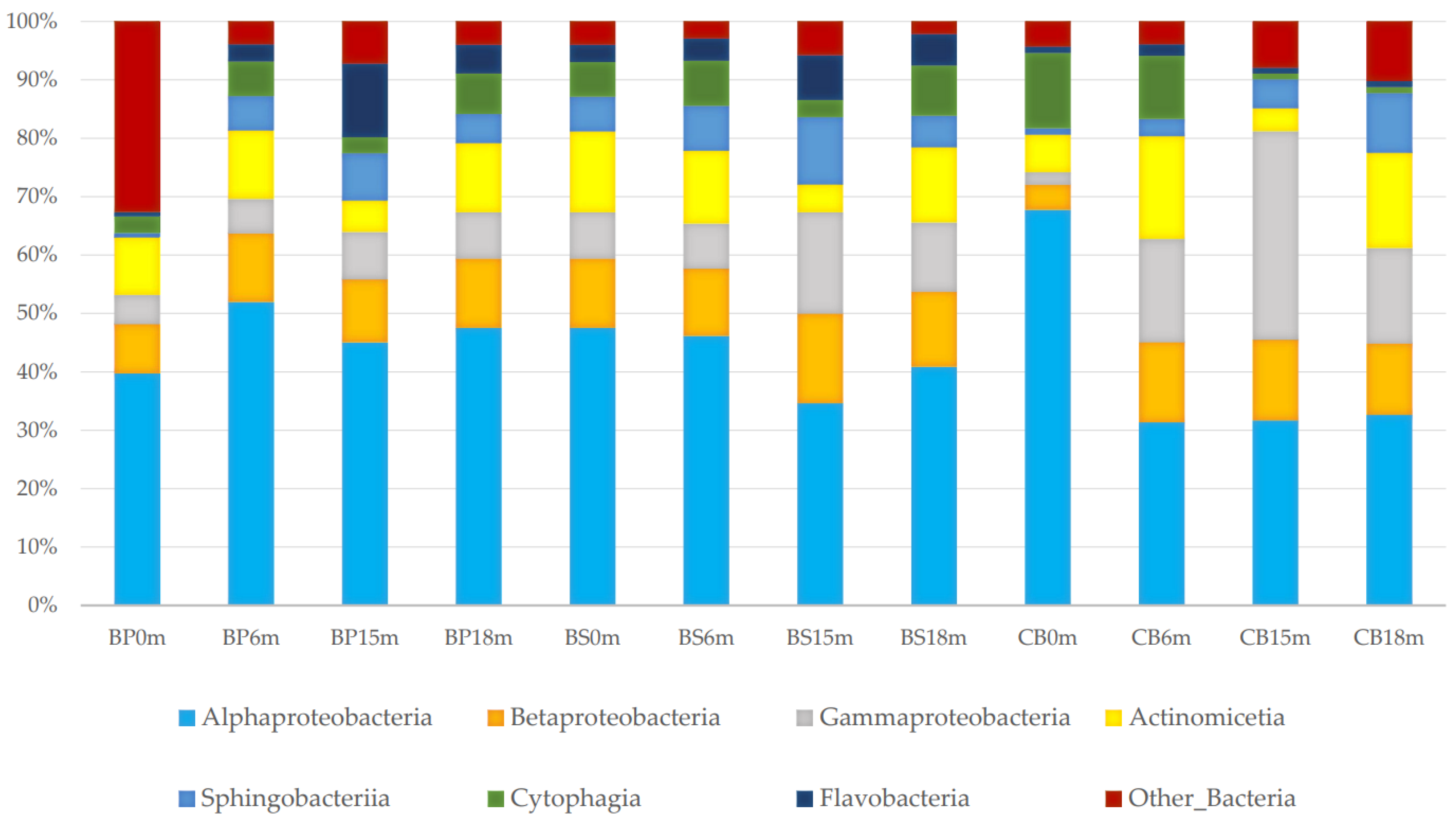
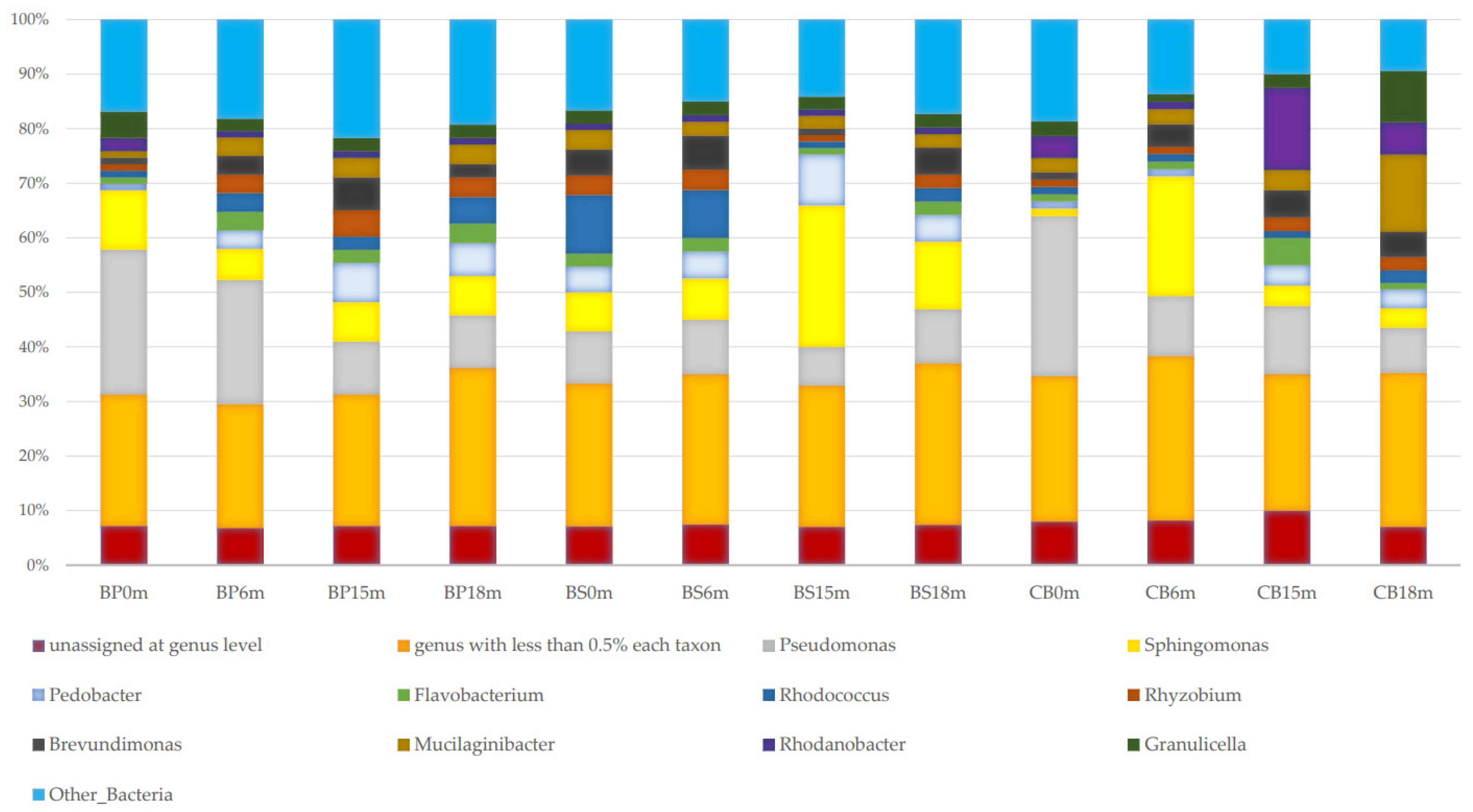
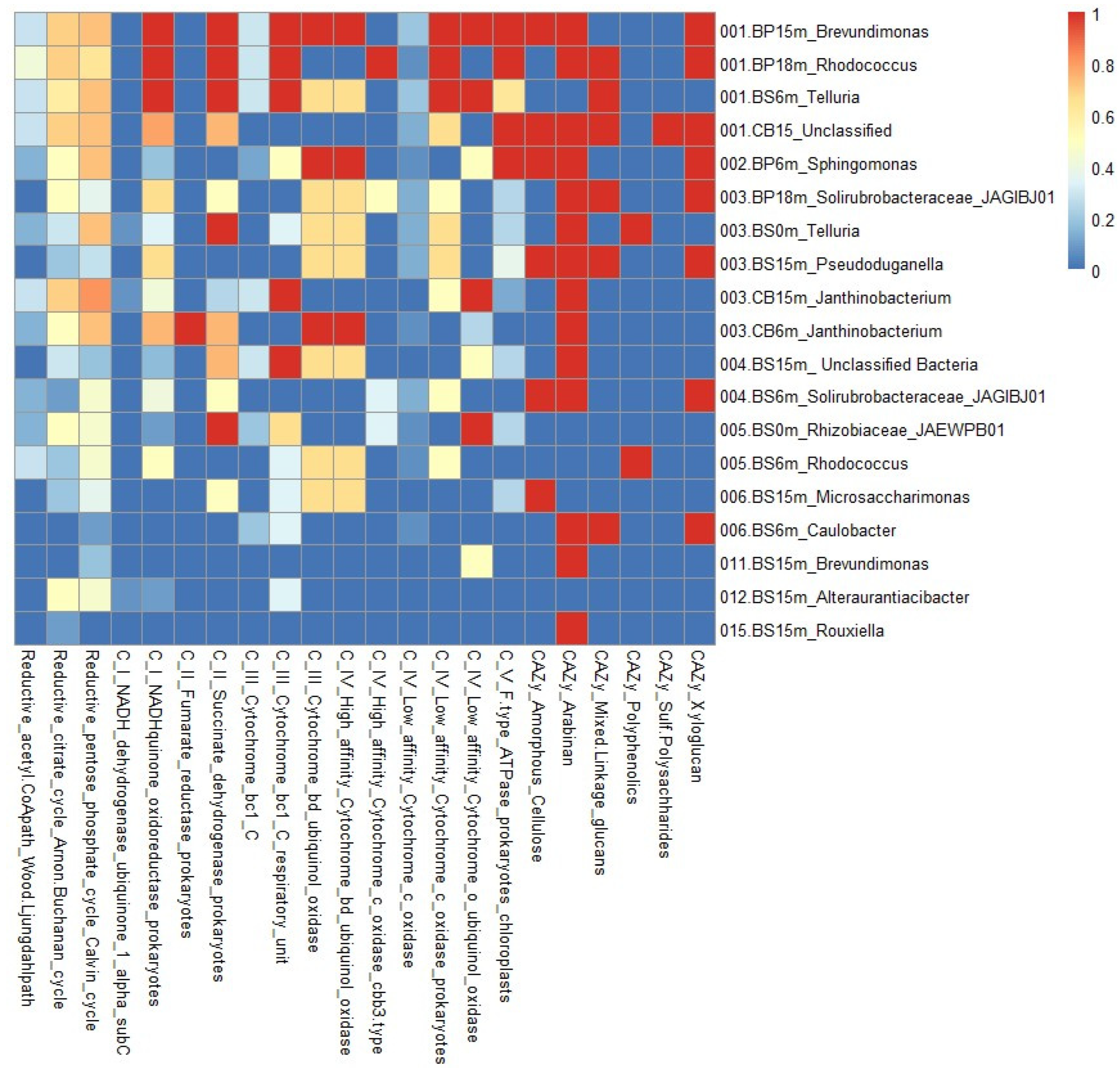
3.1. Taxonomic Composition and Structure of Bacterial Communities Using Kaiju
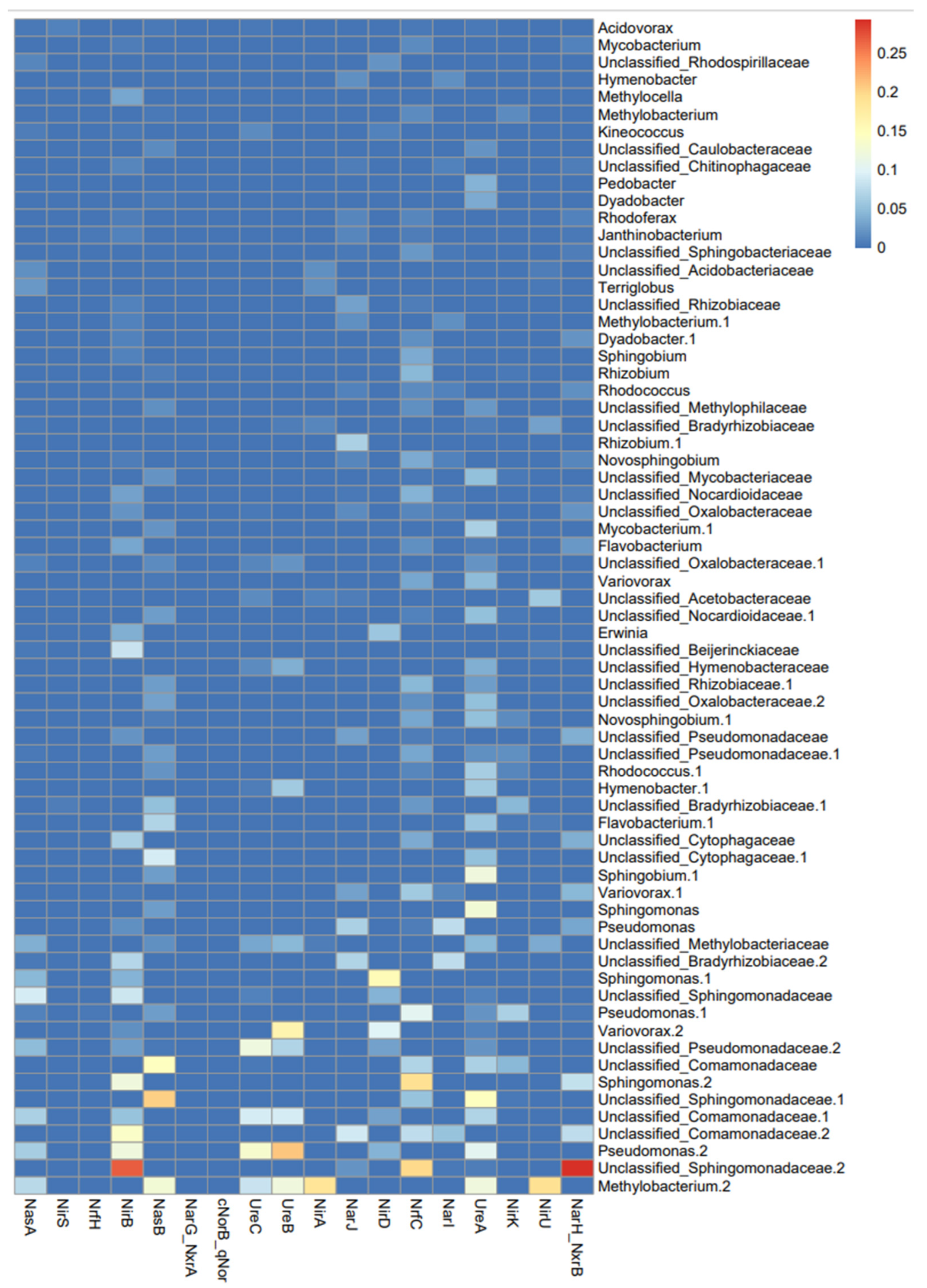
3.2. Functional Groups Analysis
3.3. Analysis of Bacterial MAGs (Metagenome-Assembled Genomes)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Otsing, E.; Barantal, S.; Anslan, S.; Koricheva, J.; Tedersoo, L. Litter Species Richness and Composition Effects on Fungal Richness and Community Structure in Decomposing Foliar and Root Litter. Soil Biol. Biochem. 2018, 125, 328–339. [Google Scholar] [CrossRef]
- Hobbie, S.E. Effects of Plant Species on Nutrient Cycling. Trends Ecol. Evol. 1992, 7, 336–339. [Google Scholar] [CrossRef] [PubMed]
- Hobbie, S.; Eddy, W.C.; Buyarsku, C.R.; Adair, C.; Ogdahl, M.L.; Weisenhorn, P. Responce of decomposing litter and its microbial community to multiple forms of nitrogen enrichment. Ecol. Monogr. 2012, 82, 389–405. [Google Scholar] [CrossRef]
- Schroeter, S.A.; Eveillard, D.; Chaffron, S.; Zoppi, J.; Bernd Kampe, B.; Lohmann, P.; Jehmlich, N.; von Bergen, M.; Sanchez-Arcos, C.; Pohnert, G.; et al. Microbial community functioning during plant litter decomposition. Sci. Rep. 2022, 12, 7451. [Google Scholar] [CrossRef] [PubMed]
- Wardle, D.A.; Yeates, G.W.; Barker, G.M.; Bonner, K.I. The Influence of Plant Litter Diversity on Decomposer Abundance and Diversity. Soil Biol. Biochem. 2006, 38, 1052–1062. [Google Scholar] [CrossRef]
- Yang, X.; Wang, X.; Xiao, S.; Liu, Z.; Zhou, X.; Du, G.; Liu, K.; Wang, Y.; Chen, S.; Nielsen, U.N. Dominant Plants Affect Litter Decomposition Mainly through Modifications of the Soil Microbial Community. Soil Biol. Biochem. 2021, 161, 108399. [Google Scholar] [CrossRef]
- Aslani, F.; Tedersoo, L.; Põlme, S.; Knox, O.; Bahram, M. Global Patterns and Determinants of Bacterial Communities Associated with Ectomycorrhizal Root Tips of Alnus Species. Soil Biol. Biochem. 2020, 148, 107923. [Google Scholar] [CrossRef]
- Hobara, S.; Osono, T.; Hirose, D.; Noro, K.; Hirota, M.; Benner, R. The roles of microorganisms in litter decomposition and soil formation. Biogeochemistry 2014, 118, 471–486. [Google Scholar] [CrossRef]
- Gupta, R.; Gupta, N. Nitrogen Assimilation and Dissimilation. In Fundamentals of Bacterial Physiology and Metabolism; Springer: Singapore, 2021. [Google Scholar] [CrossRef]
- Drzymulska, D. Palaeoenvironmental Aspects of the Genesis and Early Development of the Taboły Mire, North-Eastern Poland. Geologija 2011, 53, 121–129. Available online: https://www.researchgate.net/publication/273454337_Palaeoenvironmental_aspects_of_the_genesis_and_early_development_of_the_Taboly_mire_North-Eastern_Poland (accessed on 28 March 2024). [CrossRef]
- Smith, J.K.; Brown, A.R.; Wilson, C.D. Microbial Contributions to Nitrogen Cycling in Forest Soils: A Review. Forests 2018, 9, 123. [Google Scholar] [CrossRef]
- Jones, E.F.; White, B.T.; Johnson, L.M. Role of Bacterial Genera in Leaf Litter Decomposition: Insights from Metagenomic Studies. Microorganisms 2020, 8, 456. [Google Scholar] [CrossRef] [PubMed]
- IUSS Working Group WRB. World Reference Base for Soil Resources 2014, update 2015. In International Soil Classification System for Naming Soils and Creating Legends for Soil Maps; Food and Agriculture Organization of the United Nations: Rome, Italy, 2015. [Google Scholar]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1 km Spatial Resolution Climate Surfaces for Global Land Areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Isidorov, V.; Maslowiecka, J.; Sarapultseva, P. Bidirectional Emission of Organic Compounds by Decaying Leaf Litter of a Number of Forest-Forming Tree Species in the Northern Hemisphere. Geoderma 2024, 443, 116812. [Google Scholar] [CrossRef]
- Khomutovska, N.; de los Ríos, A.; Jasser, I. Diversity and colonization strategies of endolithic cyanobacteria in the cold mountain desert of Pamir. Microorganisms 2021, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 12 June 2023).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Luo, R.; Liu, C.M.; Leung, C.M.; Ting, H.F.; Sadakane, K.; Yamashita, H.; Lam, T.W. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef]
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and Sensitive Taxonomic Classification for Metagenomics with Kaiju. Nat. Commun. 2016, 7, 11257. [Google Scholar] [CrossRef]
- Kang, D.D.; Li, F.; Kirton, E.; Thomas, A.; Egan, R.; An, H.; Wang, Z. MetaBAT 2: An Adaptive Binning Algorithm for Robust and Efficient Genome Reconstruction from Metagenome Assemblies. PeerJ 2019, 7, e7359. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2014, 25, 1043–1055. [Google Scholar] [CrossRef]
- Olm, M.R.; Brown, C.T.; Brooks, B.; Banfield, J.F. dRep: A tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017, 11, 2864–2868. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.; Madden, T.; Schaffer, A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Sieber, C.M.K.; Probst, A.J.; Sharrar, A.; Thomas, B.C.; Hess, M.; Tringe, S.G.; Banfield, J.F. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 2018, 3, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Chuvochina, M.; Rinke, C.; Mussig, A.J. GTDB: An ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2021, 50, D785–D794. [Google Scholar] [CrossRef] [PubMed]
- Chivian, D.; Jungbluth, S.P.; Dehal, P.S.; Wood-Charlson, E.M.; Canon, R.S.; Allen, B.H.; Clark, M.M.; Gu, T.; Land, M.L.; Price, G.A.; et al. Metagenome-assembled genome extraction and analysis from microbiomes using KBase. Nat. Protoc. 2023, 18, 208–238. [Google Scholar] [CrossRef] [PubMed]
- Kazakov, A.; Novichkov, P. Fama: A Computational Tool for Comparative Analysis of Shotgun Metagenomic Data. Great Lakes Bioinformatics Conference (Poster Presentation). 2019. Available online: https://iseq.lbl.gov/mydocs/fama_glbio2019_poster.pdf (accessed on 20 February 2024).
- Shaffer, M.; Borton, M.A.; McGivern, B.B.; Zayed, A.A.; La Rosa, S.L.; Solden, L.M.; Liu, P.; Narrowe, A.B.; Rodríguez-Ramos, J.; Bolduc, B.; et al. DRAM for Distilling Microbial Metabolism to Automate the Curation of Microbiomes. bioRxiv 2020, 48, 8883–8900. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/ (accessed on 16 January 2024).
- Tláskal, V.; Voříšková, J.; Baldrian, P. Bacterial succession on decomposing leaf litter exhibits a specific occurrence pattern of cellulolytic taxa and potential decomposers of fungal mycelia. FEMS Microbiol. Ecol. 2016, 1, fiw177. [Google Scholar] [CrossRef]
- Cornwell, W.K.; Cornelissen, J.H.C.; Amatangelo, K.; Dorrepaal, E.; Eviner, V.T.; Godoy, O.; Hobbie, S.E.; Hoorens, B.; Kurokawa, H.; Pérez-Harguindeguy, N.; et al. Plant species traits are the predominant control on litter decomposition rates within biomes worldwide. Ecol. Lett. 2008, 11, 1065–1071. [Google Scholar] [CrossRef]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From Diversity and Genomics to Functional Role in Environmental Remediation and Plant Growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, T.; Chowdhury, S.P.; Uhl, J.; Rothballer, M. Genome-Based Characterization of Plant-Associated Rhodococcus qingshengii RL1 Reveals Stress Tolerance and Plant–Microbe Interaction Traits. Front. Microbiol. 2021, 12, 708605. [Google Scholar] [CrossRef] [PubMed]
- Francis, I.M.; Vereecke, D. Plant-Associated Rhodococcus Species, for Better and for Worse. In Biology of Rhodococcus; Alvarez, H., Ed.; Springer: Cham, Switzerland, 2019; pp. 359–377. [Google Scholar] [CrossRef]
- Gamit, H.; Amaresan, N. Role of Methylotrophic Bacteria in Managing Abiotic Stresses for Enhancing Agricultural Production. Pedosphere 2023, 33, 49–60. [Google Scholar] [CrossRef]
- Swapnil, P.; Meena, M.; Harish; Marwal, A.; Vijayalakshmi, S.; Zehra, A. Their Different Strategies to Mitigate Biotic and Abiotic Stress Responses. In Plant-Microbe Interaction—Recent Advances in Molecular and Biochemical Approaches; Academic Press: Cambridge, MA, USA, 2023; pp. 389–410. ISBN 9780323918756. [Google Scholar] [CrossRef]
- López-Mondéjar, R.; Zühlke, D.; Becher, D.; Riedel, K.; Baldrian, P. Pedobacter. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015. [Google Scholar]
- López-Mondéjar, R.; Zühlke, D.; Becher, D.; Riedel, K.; Baldrian, P. Cellulose and Hemicellulose Decomposition by Forest Soil Bacteria Proceeds by the Action of Structurally Variable Enzymatic Systems. Sci. Rep. 2016, 6, 25279. [Google Scholar] [CrossRef] [PubMed]
- Coluccia, M.; Besaury, L. Acidobacteria members harbor an abundant and diverse carbohydrate-active enzymes (cazyme) and secreted protease repertoire, key factors for potential efficient biomass degradation. Mol. Genet. Genomics 2023, 298, 1135–1154. [Google Scholar] [CrossRef] [PubMed]
- Tindall, B.J. Brevundimonas gen. nov., a new genus to accommodate the aerobic endospore-forming bacteria Paenibacillus brevis. Int. J. Syst. Evol. Microbiol. 2004, 54, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Im, W.T. Solirubrobacter. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015. [Google Scholar]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecol. Lett. 2007, 10, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Aylward, F.O.; Ferrari, B.C. Draft genome sequences of Sphingomonas sp. strains GTIN11 and GTIN21, isolated from groundwater samples from the Oak Ridge Field Research Center. Stand Genomic Sci. 2012, 7, 29–41. [Google Scholar]
- Alvarez, H.M.; Silva, R.A.; Cesari, A.C.; Zamit, A.L.; Peressutti, S.R.; Reichelt, R.; Keller, U.; Malkus, U.; Rasch, C.; Maskow, T.; et al. Physiological and morphological responses of an estuarine bacterium to polymers of diesel oil. Microb. Ecol. 1996, 32, 155–166. [Google Scholar] [CrossRef]
- Takeuchi, M.; Hamana, K.; Hiraishi, A. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int. J. Syst. Evol. Microbiol. 2001, 51, 1405–1417. [Google Scholar] [CrossRef] [PubMed]
- Hyeon, J.W.; Jeon, C.O. Galbiataella. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015. [Google Scholar]
- Poindexter, J.S. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 1964, 28, 231–295. [Google Scholar] [CrossRef] [PubMed]
- Kuypers, M.; Marchant, H.; Kartal, B. The Microbial Nitrogen-Cycling Network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khomutovska, N.; Jasser, I.; Isidorov, V.A. Unraveling the Role of Bacteria in Nitrogen Cycling: Insights from Leaf Litter Decomposition in the Knyszyn Forest. Forests 2024, 15, 1065. https://doi.org/10.3390/f15061065
Khomutovska N, Jasser I, Isidorov VA. Unraveling the Role of Bacteria in Nitrogen Cycling: Insights from Leaf Litter Decomposition in the Knyszyn Forest. Forests. 2024; 15(6):1065. https://doi.org/10.3390/f15061065
Chicago/Turabian StyleKhomutovska, Nataliia, Iwona Jasser, and Valery A. Isidorov. 2024. "Unraveling the Role of Bacteria in Nitrogen Cycling: Insights from Leaf Litter Decomposition in the Knyszyn Forest" Forests 15, no. 6: 1065. https://doi.org/10.3390/f15061065
APA StyleKhomutovska, N., Jasser, I., & Isidorov, V. A. (2024). Unraveling the Role of Bacteria in Nitrogen Cycling: Insights from Leaf Litter Decomposition in the Knyszyn Forest. Forests, 15(6), 1065. https://doi.org/10.3390/f15061065






