Use of a Consumer-Grade UAV Laser Scanner to Identify Trees and Estimate Key Tree Attributes across a Point Density Range
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Field Measurements
2.3. LiDAR Data
2.3.1. Data Collection and Pre-Processing
2.3.2. Individual Tree Segmentation
2.3.3. Metric Extraction
2.3.4. Data Thinning
2.4. Tree Structural Attribute Prediction
2.4.1. Prediction of DBH
2.4.2. Prediction of Volume
2.5. Accuracy Assessment
3. Results
3.1. Accuracy of Individual Tree Segmentation
3.2. DTM and CHM Assessment
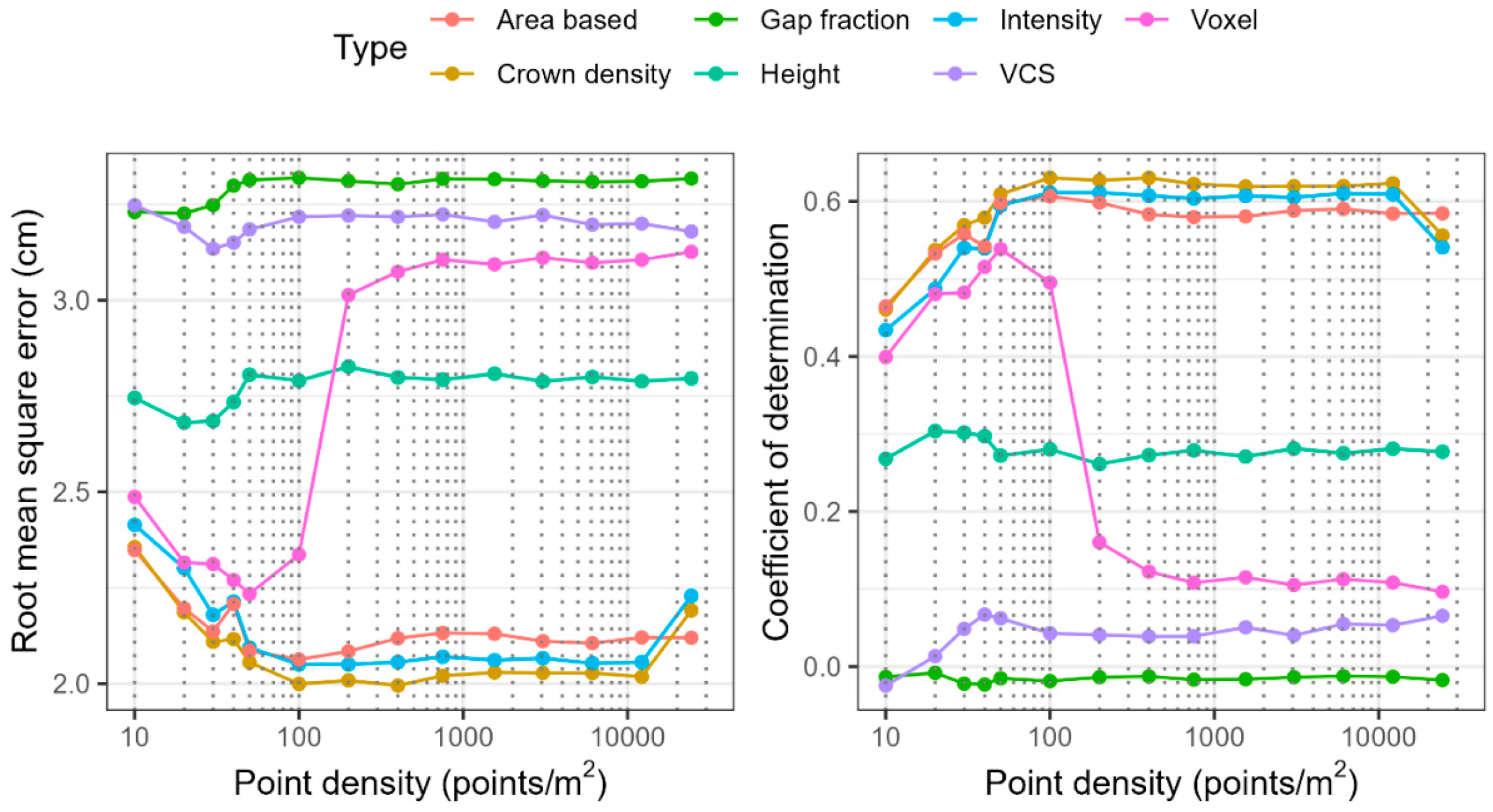
3.3. DBH Prediction
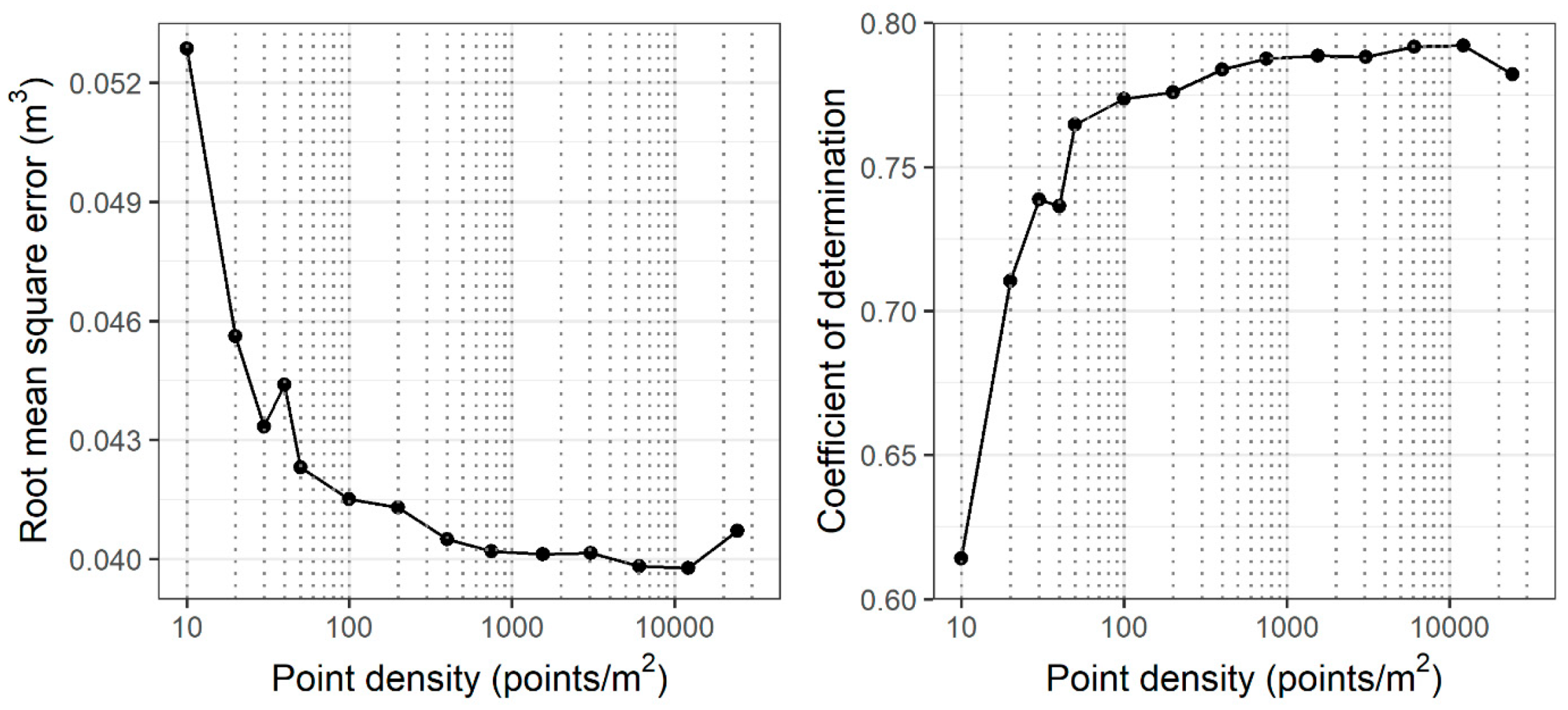
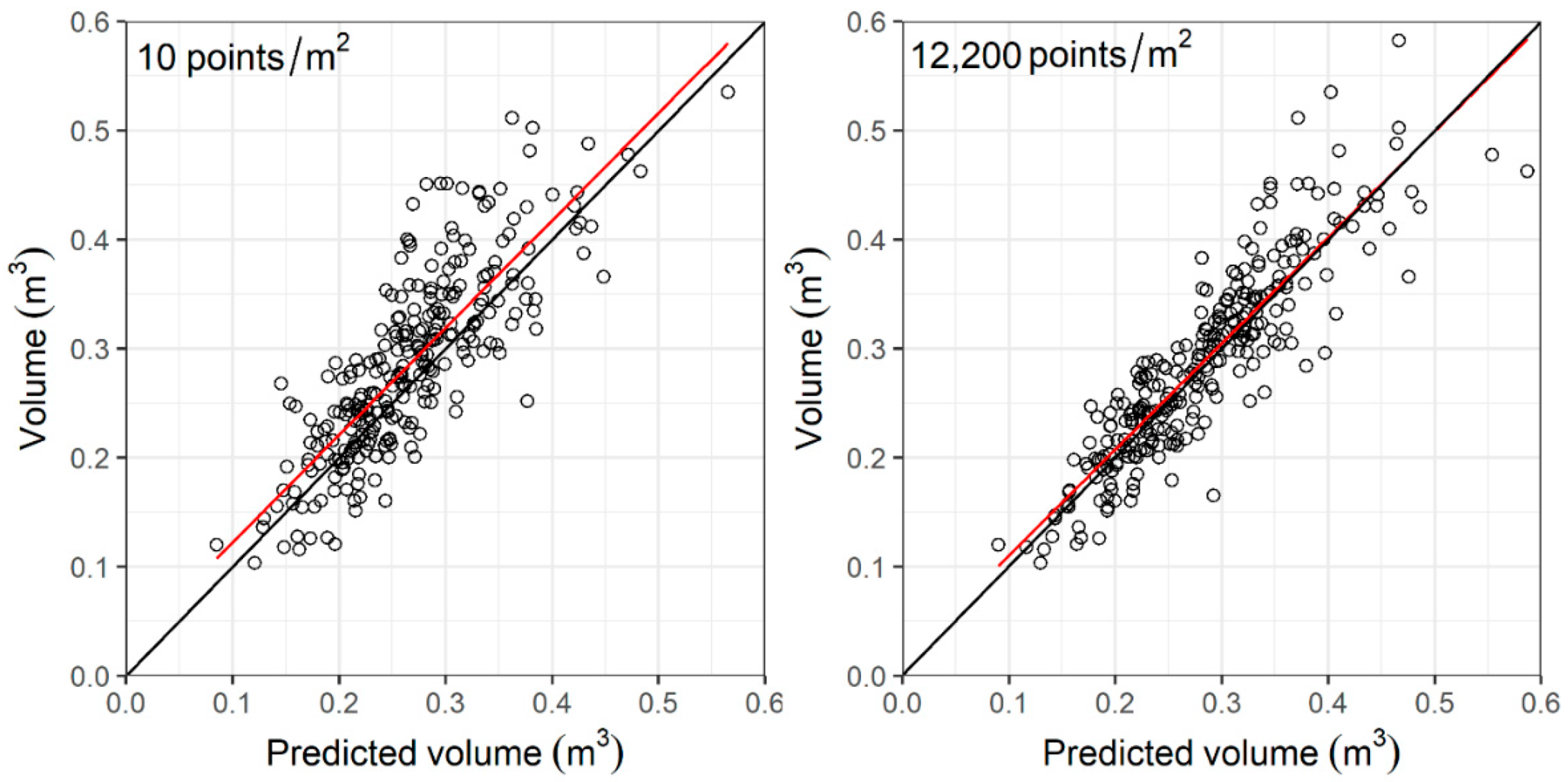
3.4. Volume Prediction
4. Discussion
4.1. Robustness of Individual Tree Segmentation, DTM and CHM across Varying Point Densities
4.2. Accuracy and Sensitivity of Forest Structural Attribute Estimation
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Parameter | Specification |
| Sensor | DJI-Zenmuse L1 LiDAR and Photogrammetry sensor |
| Capture date | 23 January 2023 |
| Flying speed (m/s) | 3 |
| Flying height above ground (m) | 55 |
| Distance between flight lines (m) | 10 |
| Number of scan layers | 2 (Gridded: Perpendicular to each other) |
| Scan mode | Repetitive scanning |
| LiDAR strike frequency (Hz) | 160 |
| Laser wavelength (nm) | 905 |
| Field of view (°) | 70.4 (horizontal) × 4.5 (vertical) |
| Beam divergence (°) | 0.03 (horizontal) × 0.28 (vertical) |
| Number of returns | 3 |
| Average (standard deviation) density of resulting point cloud (points/m2) | 15,966 (8500) |
| Class | Abbreviation | Description | R Package |
|---|---|---|---|
| Point-based metrics | |||
| 1. Standard height metrics | zmax | Maximum height above ground | lidR |
| zmean | Mean height above ground | ||
| zsd | Standard deviation of height distribution | ||
| zcv | Coefficient of variation of height distribution | ||
| zskew | Skewness of height distribution | ||
| zkurt | Kurtosis of height distribution | ||
| zentropy | Entropy of height distribution | ||
| zq(X), where X is a percentile, (e.g., zq95) | Percentile heights (5th, 10th, 15th, 20th, 25th, 30th, 35th, 40th, 45th, 50th, 55th, 60th, 65th, 70th, 75th, 80th, 85th, 90th, and 95th) | ||
| 2. Standard intensity metrics | itot | Sum of the intensity of returns | lidR |
| imax | Maximum intensity of returns | ||
| imean | Mean intensity of returns | ||
| isd | Standard deviation of intensity distribution | ||
| icv | Coefficient of variation of intensity distribution | ||
| iskew | Skewness of intensity distribution | ||
| ikurt | Kurtosis of intensity distribution | ||
| ipground | Intensity of ground returns | ||
| ipcumzq(X), e.g., ipcumzq90 | Percentage of intensity returned below the Xth height percentile | ||
| 3. Standard crown density metrics | zpcum(X), e.g., zpcum9, | Cumulative percentage of return in the Xth layer | lidR |
| pground | Percentage of returns classified as “ground” | ||
| pzabovezmean | Percentage of returns above the mean height of each tree | ||
| pzabove2 | Percentage of returns above 2 m height | ||
| p1st, p2nd, p3rd | Percentage of returns (first return–third return) | ||
| n | Total number of points | ||
| 4. Gap fraction and LAD metrics | gfp_m | Mean of gap fraction profile (layer thickness: 1 m) | lidR |
| gfp_sd | Standard deviation of gap fraction profile | ||
| gfp_IQR | Interquartile range of gap fraction profile | ||
| lad_m | Mean of leaf area density | ||
| lad_sd | Standard deviation of leaf area density | ||
| 5. Vertical canopy structural metrics | wpd_scale | Weibull probability distribution fitted to foliage profile: scale parameter α | fitdistrplus |
| wpd_shape | Weibull probability distribution fitted to foliage profile: shape parameter β | ||
| Area-based metrics | |||
| 6. 2D and 3D convex hull metrics | cvx2D_area | 2D area of individual crown convex hull | cxhull |
| cnx3D_vol | 3D volume of the individual tree convex hull | ||
| Voxel-based metrics (25 cm voxel res) | |||
| 7. Voxel-based vertical canopy structural metrics | filled_canopy | Filled canopy volume percentage: Proportion of total number of voxels containing points | NA |
| open_gap | Open gap volume percentage: Proportion of voxels containing no points above the canopy | ||
| closed_gap | Closed gap volume percentage: Proportion of voxels containing no points below the canopy | ||
| euphotic | Euphotic volume percentage: Proportion of voxels in the uppermost 65% of cells that contain points of a column | ||
| oligophotic | Oligophotic volume percentage: Proportion of voxels in the lower 35% of cells that contain points in a column | ||
| Maximum Density of Source Point Cloud (points/m2) | Point Cloud Ground Normalized Using the Highest Density DTM | Point Cloud Ground Normalized Using the Native DTM | ||||||
|---|---|---|---|---|---|---|---|---|
| Tree Peak Detection Accuracy | Crown Delineation Accuracy | Tree Peak Detection Accuracy | Crown Delineation Accuracy | |||||
| Precision | Recall | F1 | Proportion of Accurately Delineated Crowns | Precision | Recall | F1 | Proportion of Accurately Delineated Crowns | |
| 10 | 0.98 | 0.95 | 0.96 | 0.96 | 0.97 | 0.95 | 0.96 | 0.96 |
| 20 | 0.98 | 0.95 | 0.96 | 0.98 | 0.98 | 0.95 | 0.96 | 0.98 |
| 30 | 0.98 | 0.95 | 0.96 | 0.99 | 0.98 | 0.95 | 0.96 | 0.99 |
| 40 | 0.98 | 0.95 | 0.96 | 0.99 | 0.98 | 0.95 | 0.96 | 0.99 |
| 50 | 0.97 | 0.96 | 0.96 | 0.99 | 0.97 | 0.96 | 0.96 | 0.99 |
| 100 | 0.97 | 0.97 | 0.97 | 0.99 | 0.97 | 0.96 | 0.96 | 0.99 |
| 200 | 0.97 | 0.97 | 0.97 | 0.99 | 0.97 | 0.97 | 0.97 | 0.99 |
| 400 | 0.97 | 0.98 | 0.97 | 0.99 | 0.97 | 0.98 | 0.97 | 0.99 |
| 750 | 0.96 | 0.98 | 0.97 | 0.99 | 0.96 | 0.98 | 0.97 | 0.99 |
| 1550 | 0.96 | 0.98 | 0.97 | 0.99 | 0.96 | 0.98 | 0.97 | 0.99 |
| 3050 | 0.96 | 0.98 | 0.97 | 0.99 | 0.96 | 0.98 | 0.97 | 0.99 |
| 6100 | 0.96 | 0.98 | 0.97 | 0.99 | 0.96 | 0.98 | 0.97 | 0.99 |
| 12,200 | 0.96 | 0.98 | 0.97 | 0.99 | 0.96 | 0.98 | 0.97 | 0.99 |
| 24,450 | NA | NA | NA | NA | 0.96 | 0.98 | 0.97 | 0.99 |
References
- FAO. Global Forest Resources Assessment 2020—Key Findings; FAO: Rome, Italy, 2020; p. 5. [Google Scholar] [CrossRef]
- Bukoski, J.J.; Cook-Patton, S.C.; Melikov, C.; Ban, H.; Chen, J.L.; Goldman, E.D.; Harris, N.L.; Potts, M.D. Rates and drivers of aboveground carbon accumulation in global monoculture plantation forests. Nat. Commun. 2022, 13, 4206. [Google Scholar] [CrossRef]
- NZFOA. New Zealand Forestry Industry, Facts and Figures 2021/2022; New Zealand Plantation Forest Industry: Wellington, New Zealand, 2022. [Google Scholar]
- Koutika, L.; Matondo, R.; Mabiala-Ngoma, A.; Tchichelle, V.S.; Toto, M.; Madzoumbou, J.; Akana, J.A.; Gomat, H.Y.; Mankessi, F.; Mbou, A.T.; et al. Sustaining Forest Plantations for the United Nations’ 2030 Agenda for Sustainable Development. Sustainability 2022, 14, 4624. [Google Scholar] [CrossRef]
- Vidal, C.; Alberdi, I.; Redmond, J.; Vestman, M.; Lanz, A.; Schadauer, K. The role of European National Forest Inventories for international forestry reporting. Ann. For. Sci. 2016, 73, 793–806. [Google Scholar] [CrossRef]
- Roise, J.P.; Cubbage, F.W.; Abt, R.C.; Siry, J.P. Regulation of Timber Yield for Sustainable Management of Industrial Forest Plantations—Theory and Practice. In Sustainable Forest Management; Springer: Dordrecht, The Netherlands, 2000; pp. 217–255. [Google Scholar]
- Maltamo, M. Estimation of timber volume and stem density based on scanning laser altimetry and expected tree size distribution functions. Remote Sens. Environ. 2004, 90, 319–330. [Google Scholar] [CrossRef]
- Zhao, D.; Kane, M.; Borders, B.E. Growth responses to planting density and management intensity in loblolly pine plantations in the southeastern USA Lower Coastal Plain. Ann. For. Sci. 2011, 68, 625–635. [Google Scholar] [CrossRef]
- Kanninen, M.; Pérez, D.; Montero, M.; Víquez, E. Intensity and timing of the first thinning of Tectona grandis plantations in Costa Rica: Results of a thinning trial. For. Ecol. Manag. 2004, 203, 89–99. [Google Scholar] [CrossRef]
- Hébert, F.; Krause, C.; Plourde, P.-Y.; Achim, A.; Prégent, G.; Ménétrier, J. Effect of Tree Spacing on Tree Level Volume Growth, Morphology, and Wood Properties in a 25-Year-Old Pinus banksiana Plantation in the Boreal Forest of Quebec. Forests 2016, 7, 276. [Google Scholar] [CrossRef]
- Zhu, Z.; Kleinn, C.; Nölke, N. Assessing tree crown volume—A review. For. Int. J. For. Res. 2021, 94, 18–35. [Google Scholar] [CrossRef]
- Zhen, Z.; Quackenbush, L.; Zhang, L. Trends in Automatic Individual Tree Crown Detection and Delineation—Evolution of LiDAR Data. Remote Sens. 2016, 8, 333. [Google Scholar] [CrossRef]
- Brūmelis, G.; Dauškane, I.; Elferts, D.; Strode, L.; Krama, T.; Krams, I. Estimates of Tree Canopy Closure and Basal Area as Proxies for Tree Crown Volume at a Stand Scale. Forests 2020, 11, 1180. [Google Scholar] [CrossRef]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote Sensing Technologies for Enhancing Forest Inventories: A Review. Can. J. Remote Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef]
- Torresan, C.; Berton, A.; Carotenuto, F.; Di Gennaro, S.F.; Gioli, B.; Matese, A.; Miglietta, F.; Vagnoli, C.; Zaldei, A.; Wallace, L. Forestry applications of UAVs in Europe: A review. Int. J. Remote Sens. 2016, 38, 2427–2447. [Google Scholar] [CrossRef]
- Puliti, S.; Ene, L.T.; Gobakken, T.; Næsset, E. Use of partial-coverage UAV data in sampling for large scale forest inventories. Remote Sens. Environ. 2017, 194, 115–126. [Google Scholar] [CrossRef]
- Štroner, M.; Urban, R.; Línková, L. A New Method for UAV Lidar Precision Testing Used for the Evaluation of an Affordable DJI ZENMUSE L1 Scanner. Remote Sens. 2021, 13, 4811. [Google Scholar] [CrossRef]
- Watt, M.S.; Meredith, A.; Watt, P.; Gunn, A. The influence of LiDAR pulse density on the precision of inventory metrics in young unthinned Douglas-fir stands during initial and subsequent LiDAR acquisitions. N. Z. J. For. Sci. 2014, 44, 18. [Google Scholar] [CrossRef]
- Pearse, G.D.; Watt, M.S.; Dash, J.P.; Stone, C.; Caccamo, G. Comparison of models describing forest inventory attributes using standard and voxel-based lidar predictors across a range of pulse densities. Int. J. Appl. Earth Obs. Geoinf. 2019, 78, 341–351. [Google Scholar] [CrossRef]
- Treitz, P.; Lim, K.; Woods, M.; Pitt, D.; Nesbitt, D.; Etheridge, D. LiDAR Sampling Density for Forest Resource Inventories in Ontario, Canada. Remote Sens. 2012, 4, 830–848. [Google Scholar] [CrossRef]
- Hansen, E.; Gobakken, T.; Næsset, E. Effects of Pulse Density on Digital Terrain Models and Canopy Metrics Using Airborne Laser Scanning in a Tropical Rainforest. Remote Sens. 2015, 7, 8453–8468. [Google Scholar] [CrossRef]
- Manuri, S.; Andersen, H.-E.; McGaughey, R.J.; Brack, C. Assessing the influence of return density on estimation of lidar-based aboveground biomass in tropical peat swamp forests of Kalimantan, Indonesia. Int. J. Appl. Earth Obs. Geoinf. 2017, 56, 24–35. [Google Scholar] [CrossRef]
- Jakubowski, M.K.; Guo, Q.; Kelly, M. Tradeoffs between lidar pulse density and forest measurement accuracy. Remote Sens. Environ. 2013, 130, 245–253. [Google Scholar] [CrossRef]
- Liu, K.; Shen, X.; Cao, L.; Wang, G.; Cao, F. Estimating forest structural attributes using UAV-LiDAR data in Ginkgo plantations. ISPRS J. Photogramm. Remote Sens. 2018, 146, 465–482. [Google Scholar] [CrossRef]
- Wang, Y.; Lehtomäki, M.; Liang, X.; Pyörälä, J.; Kukko, A.; Jaakkola, A.; Liu, J.; Feng, Z.; Chen, R.; Hyyppä, J. Is field-measured tree height as reliable as believed—A comparison study of tree height estimates from field measurement, airborne laser scanning and terrestrial laser scanning in a boreal forest. ISPRS J. Photogramm. Remote Sens. 2019, 147, 132–145. [Google Scholar] [CrossRef]
- Hartley, R.J.L.; Leonardo, E.M.; Massam, P.; Watt, M.S.; Estarija, H.J.; Wright, L.; Melia, N.; Pearse, G.D. An Assessment of High-Density UAV Point Clouds for the Measurement of Young Forestry Trials. Remote Sens. 2020, 12, 4039. [Google Scholar] [CrossRef]
- Roussel, J.-R.; Auty, D.; Coops, N.C.; Tompalski, P.; Goodbody, T.R.H.; Meador, A.S.; Bourdon, J.-F.; de Boissieu, F.; Achim, A. lidR: An R package for analysis of Airborne Laser Scanning (ALS) data. Remote Sens. Environ. 2020, 251, 112061. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Isenburg, M. LAStools—Efficient LiDAR Processing Software, Version 2.0.2; Rapidlasso GmbH: Gilching, Germany, 2019. [Google Scholar]
- Khosravipour, A.; Skidmore, A.K.; Isenburg, M.; Wang, T.; Hussin, Y.A. Generating pit—Free canopy height models from airborne lidar. Photogramm. Eng. Remote Sens. 2014, 80, 863–872. [Google Scholar] [CrossRef]
- Panagiotidis, D.; Abdollahnejad, A.; Surový, P.; Chiteculo, V. Determining tree height and crown diameter from high-resolution UAV imagery. Int. J. Remote Sens. 2017, 38, 2392–2410. [Google Scholar] [CrossRef]
- Plowright, A.; Roussel, J. Tools for Analyzing Remote Sensing Forest Data. 2021. Available online: https://cran.r-project.org/package=ForestTools (accessed on 20 March 2024).
- du Toit, F.; Coops, N.C.; Ratcliffe, B.; El-Kassaby, Y.A.; Lucieer, A. Modelling internal tree attributes for breeding applications in Douglas-fir progeny trials using RPAS-ALS. Sci. Remote Sens. 2023, 7, 100072. [Google Scholar] [CrossRef]
- du Toit, F.; Coops, N.C.; Tompalski, P.; Goodbody, T.R.H.; El-Kassaby, Y.A.; Stoehr, M.; Turner, D.; Lucieer, A. Characterizing variations in growth characteristics between Douglas-fir with different genetic gain levels using airborne laser scanning. Trees 2020, 34, 649–664. [Google Scholar] [CrossRef]
- du Toit, F.; Coops, N.C.; Ratcliffe, B.; El-Kassaby, Y.A. Generating Douglas-fir Breeding Value Estimates Using Airborne Laser Scanning Derived Height and Crown Metrics. Front. Plant Sci. 2022, 13, 893017. [Google Scholar] [CrossRef] [PubMed]
- Coops, N.C.; Hilker, T.; Wulder, M.A.; St-Onge, B.; Newnham, G.; Siggins, A.; Trofymow, J.A. Estimating canopy structure of Douglas-fir forest stands from discrete-return LiDAR. Trees 2007, 21, 295–310. [Google Scholar] [CrossRef]
- Lefsky, M.A.; Cohen, W.B.; Acker, S.A.; Parker, G.G.; Spies, T.A.; Harding, D. Lidar remote sensing of the canopy structure and biophysical properties of Douglas-fir western hemlock forests. Remote Sens. Environ. 1999, 70, 339–361. [Google Scholar] [CrossRef]
- Bouvier, M.; Durrieu, S.; Fournier, R.A.; Renaud, J.-P. Generalizing predictive models of forest inventory attributes using an area-based approach with airborne LiDAR data. Remote Sens. Environ. 2015, 156, 322–334. [Google Scholar] [CrossRef]
- Delignette-Muller, M.; Dutang, C.; Pouillot, R.; Denis, J.; Siberchicot, A. Fitdistrplus: Help to Fit of a Parametric Distribution to Non-Censored or Censored Data. 2023. Available online: https://CRAN.R-project.org/package=fitdistrplus (accessed on 20 March 2024).
- Lecigne, B.; Delagrange, S.; Messier, C. Exploring trees in three dimensions: VoxR, a novel voxel-based R package dedicated to analysing the complex arrangement of tree crowns. Ann. Bot. 2018, 121, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Wold, H. Estimation of Principal Components and Related Models by Iterative Least Squares; Multivariate Analysis; Krishnaiah, P.R., Ed.; Academic Press: New York, NY, USA, 1966. [Google Scholar]
- Geladi, P.; Kowalski, B.R. Partial least-squares regression: A tutorial. Anal. Chim. Acta 1986, 185, 1–17. [Google Scholar] [CrossRef]
- Wold, S.; Trygg, J.; Berglund, A.; Antti, H. Some recent developments in PLS modeling. Chemom. Intell. Lab. Systems. 2001, 58, 131–150. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Wright, M.N.; Ziegler, A. Ranger: A fast implementation of random forests for high dimensional data in C++ and R. J. Stat. Softw. 2014, 77, 1–17. [Google Scholar] [CrossRef]
- Kimberley, M.O.; Beets, P.N. National volume function for estimating total stem volume of Pinus radiata stands in New Zealand. N. Z. J. For. Sci. 2007, 37, 355–371. [Google Scholar]
- Cao, L.; Gao, S.; Li, P.; Yun, T.; Shen, X.; Ruan, H. Aboveground Biomass Estimation of Individual Trees in a Coastal Planted Forest Using Full-Waveform Airborne Laser Scanning Data. Remote Sens. 2016, 8, 729. [Google Scholar] [CrossRef]
- Larsen, M.; Eriksson, M.; Descombes, X.; Perrin, G.; Brandtberg, T.; Gougeon, F.A. Comparison of six individual tree crown detection algorithms evaluated under varying forest conditions. Int. J. Remote Sens. 2011, 32, 5827–5852. [Google Scholar] [CrossRef]
- Wu, X.; Shen, X.; Cao, L.; Wang, G.; Cao, F. Assessment of Individual Tree Detection and Canopy Cover Estimation using Unmanned Aerial Vehicle based Light Detection and Ranging (UAV-LiDAR) Data in Planted Forests. Remote Sens. 2019, 11, 908. [Google Scholar] [CrossRef]
- Popescu, S.C.; Wynne, R.H.; Nelson, R.F. Estimating plot-level tree heights with lidar: Local filtering with a canopy-height based variable window size. Comput. Electron. Agric. 2002, 37, 71–95. [Google Scholar] [CrossRef]
- Erikson, M.; Olofsson, K. Comparison of three individual tree crown detection methods. Mach. Vis. Appl. 2005, 16, 258–265. [Google Scholar] [CrossRef]
- Yu, X.; Hyyppä, J.; Vastaranta, M.; Holopainen, M.; Viitala, R. Predicting individual tree attributes from airborne laser point clouds based on the random forests technique. ISPRS J. Photogramm. Remote Sens. 2011, 66, 28–37. [Google Scholar] [CrossRef]
- Cosenza, D.N.; Gomes Pereira, L.; Guerra-Hernández, J.; Pascual, A.; Soares, P.; Tomé, M. Impact of Calibrating Filtering Algorithms on the Quality of LiDAR-Derived DTM and on Forest Attribute Estimation through Area-Based Approach. Remote Sens. 2020, 12, 918. [Google Scholar] [CrossRef]
- Ene, L.; Næsset, E.; Gobakken, T. Single tree detection in heterogeneous boreal forests using airborne laser scanning and area-based stem number estimates. Int. J. Remote Sens. 2012, 33, 5171–5193. [Google Scholar] [CrossRef]
- Torresani, M.; Rocchini, D.; Sonnenschein, R.; Zebisch, M.; Hauffe, H.C.; Heym, M.; Pretzsch, H.; Tonon, G. Height variation hypothesis: A new approach for estimating forest species diversity with CHM LiDAR data. Ecol. Indic. 2020, 117, 106520. [Google Scholar] [CrossRef]
- Huang, H.; Gong, P.; Cheng, X.; Clinton, N.; Li, Z. Improving Measurement of Forest Structural Parameters by Co-Registering of High Resolution Aerial Imagery and Low Density LiDAR Data. Sensors 2009, 9, 1541–1558. [Google Scholar] [CrossRef] [PubMed]
- Dalla Corte, A.P.; Rex, F.E.; Almeida, D.R.A.d.; Sanquetta, C.R.; Silva, C.A.; Moura, M.M.; Wilkinson, B.; Zambrano, A.M.A.; Cunha Neto, E.M.d.; Veras, H.F.P.; et al. Measuring Individual Tree Diameter and Height Using GatorEye High-Density UAV-Lidar in an Integrated Crop-Livestock-Forest System. Remote Sens. 2020, 12, 863. [Google Scholar] [CrossRef]
- Hayashi, R.; Weiskittel, A.; Sader, S. Assessing the Feasibility of Low-Density LiDAR for Stand Inventory Attribute Predictions in Complex Managed Forests of Northern Maine, U.S.A. Forests 2014, 5, 363–383. [Google Scholar] [CrossRef]
- Cățeanu, M.; Ciubotaru, A. The Effect of LiDAR Sampling Density on DTM Accuracy for Areas with Heavy Forest Cover. Forests 2021, 12, 265. [Google Scholar] [CrossRef]
- Fradette, M.-S.; Leboeuf, A.; Riopel, M.; Bégin, J. Method to Reduce the Bias on Digital Terrain Model and Canopy Height Model from LiDAR Data. Remote Sens. 2019, 11, 863. [Google Scholar] [CrossRef]
- Vauhkonen, J.; Korpela, I.; Maltamo, M.; Tokola, T. Imputation of single-tree attributes using airborne laser scanning-based height, intensity, and alpha shape metrics. Remote Sens. Environ. 2010, 114, 1263–1276. [Google Scholar] [CrossRef]
- Yao, W.; Krzystek, P.; Heurich, M. Tree species classification and estimation of stem volume and DBH based on single tree extraction by exploiting airborne full-waveform LiDAR data. Remote Sens. Environ. 2012, 123, 368–380. [Google Scholar] [CrossRef]
- Qi, Y.; Coops, N.C.; Daniels, L.D.; Butson, C.R. Comparing tree attributes derived from quantitative structure models based on drone and mobile laser scanning point clouds across varying canopy cover conditions. ISPRS J. Photogramm. Remote Sens. 2022, 192, 49–65. [Google Scholar] [CrossRef]
- Panagiotidis, D.; Abdollahnejad, A.; Slavík, M. 3D point cloud fusion from UAV and TLS to assess temperate managed forest structures. Int. J. Appl. Earth Obs. Geoinf. 2022, 112, 102917. [Google Scholar] [CrossRef]
- Brede, B.; Calders, K.; Lau, A.; Raumonen, P.; Bartholomeus, H.M.; Herold, M.; Kooistra, L. Non-destructive tree volume estimation through quantitative structure modelling: Comparing UAV laser scanning with terrestrial LIDAR. Remote Sens. Environ. 2019, 233, 111355. [Google Scholar] [CrossRef]
- Hartley, R.J.; Jayathunga, S.; Massam, P.D.; De Silva, D.; Estarija, H.J.; Davidson, S.J.; Wuraola, A.; Pearse, G.D. Assessing the potential of backpack-mounted mobile laser scanning systems for tree phenotyping. Remote Sens. 2022, 14, 3344. [Google Scholar] [CrossRef]
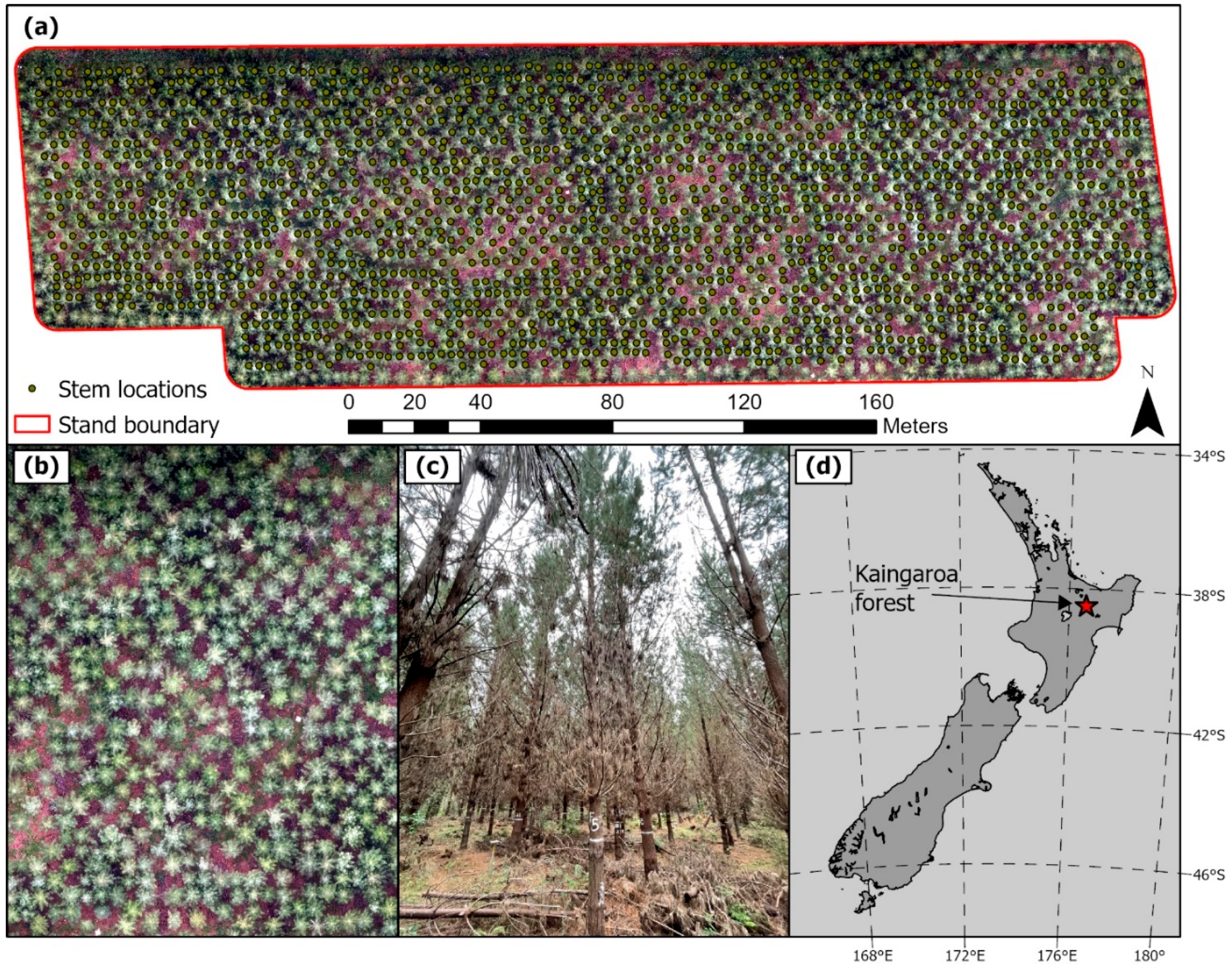


| Site Characteristics | |||
|---|---|---|---|
| Trial area | 3 ha | ||
| Establishment date | August 2014 | ||
| Planting spacing | 3.1 m | ||
| Total number of trees measured | 1744 | ||
| Number of trees excluding multileader, dead, and damaged trees | 1392 | ||
| Field measurements started on | 27 February 2023 | ||
| Field measurements completed on | 10 March 2023 | ||
| Attribute | Mean | Standard deviation | Range |
| Terrain attributes | |||
| Elevation (m) | 370 | 2.6 | 363–374 |
| Slope (°) | 7.8 | 6.5 | 0–23 |
| Tree structural attributes | |||
| DBH (cm) | 23.27 | 3.63 | 7.80–37.80 |
| Maximum height (m) | 17.66 | 1.38 | 11.36–21.52 |
| Total stem volume (m3) | 0.28 | 0.09 | 0.03–0.73 |
| Density of the Decimated Point Cloud (Points/m2) | DTM | CHM | ||
|---|---|---|---|---|
| RMSE (m) | MBE (m) | RMSE (m) | MBE (m) | |
| 10 | 0.11 | 0.06 | 3.60 | −3.12 |
| 20 | 0.09 | 0.05 | 2.64 | −2.29 |
| 30 | 0.08 | 0.05 | 2.44 | −2.13 |
| 40 | 0.08 | 0.04 | 2.33 | −2.03 |
| 50 | 0.07 | 0.04 | 1.67 | −1.37 |
| 100 | 0.06 | 0.04 | 1.20 | −0.96 |
| 200 | 0.05 | 0.03 | 0.87 | −0.67 |
| 400 | 0.04 | 0.02 | 0.63 | −0.46 |
| 750 | 0.03 | 0.02 | 0.46 | −0.31 |
| 1550 | 0.03 | 0.01 | 0.31 | −0.19 |
| 3050 | 0.02 | 0.01 | 0.20 | −0.10 |
| 6100 | 0.02 | 0 | 0.11 | −0.05 |
| 12,200 | 0.02 | 0 | 0.05 | −0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watt, M.S.; Jayathunga, S.; Hartley, R.J.L.; Pearse, G.D.; Massam, P.D.; Cajes, D.; Steer, B.S.C.; Estarija, H.J.C. Use of a Consumer-Grade UAV Laser Scanner to Identify Trees and Estimate Key Tree Attributes across a Point Density Range. Forests 2024, 15, 899. https://doi.org/10.3390/f15060899
Watt MS, Jayathunga S, Hartley RJL, Pearse GD, Massam PD, Cajes D, Steer BSC, Estarija HJC. Use of a Consumer-Grade UAV Laser Scanner to Identify Trees and Estimate Key Tree Attributes across a Point Density Range. Forests. 2024; 15(6):899. https://doi.org/10.3390/f15060899
Chicago/Turabian StyleWatt, Michael S., Sadeepa Jayathunga, Robin J. L. Hartley, Grant D. Pearse, Peter D. Massam, David Cajes, Benjamin S. C. Steer, and Honey Jane C. Estarija. 2024. "Use of a Consumer-Grade UAV Laser Scanner to Identify Trees and Estimate Key Tree Attributes across a Point Density Range" Forests 15, no. 6: 899. https://doi.org/10.3390/f15060899
APA StyleWatt, M. S., Jayathunga, S., Hartley, R. J. L., Pearse, G. D., Massam, P. D., Cajes, D., Steer, B. S. C., & Estarija, H. J. C. (2024). Use of a Consumer-Grade UAV Laser Scanner to Identify Trees and Estimate Key Tree Attributes across a Point Density Range. Forests, 15(6), 899. https://doi.org/10.3390/f15060899






