New Role in the 5-HT Receptor: The Sex Attracting of Bursaphelenchus mucronatus
Abstract
1. Introduction
2. Materials and Methods
2.1. Nematode Samples and Culture Conditions
2.2. Gene Cloning
2.3. Bioinformatic Analysis of Bmu-ser-1
2.4. Quantitative Real-Time PCR
2.5. In Situ Hybridization of Bm-ser-1
2.6. RNAi of Bm-ser-1
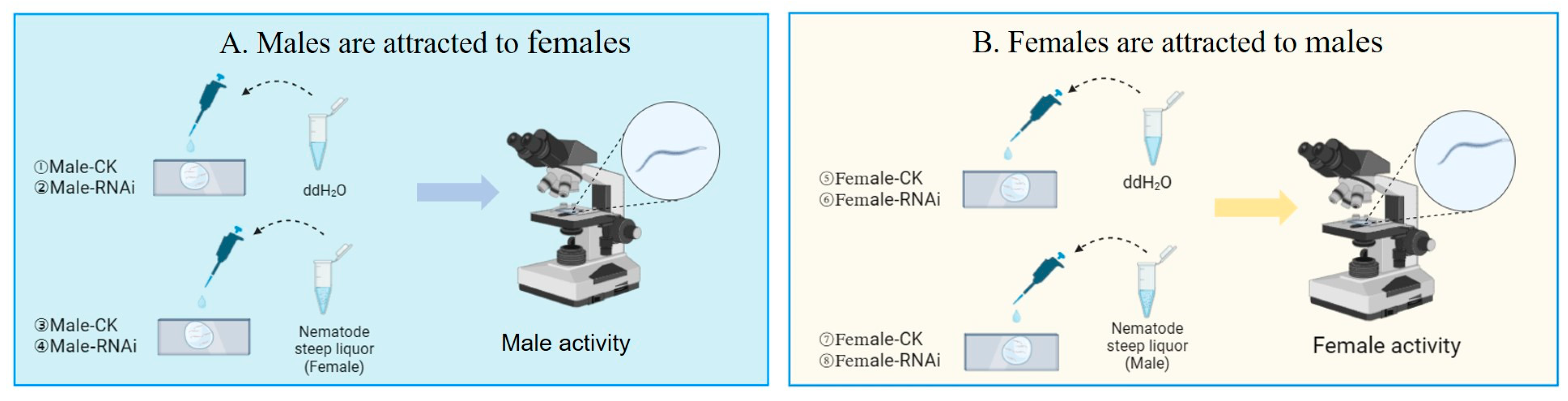
2.7. Sexual Arousal Experiments
2.8. Data Analysis
3. Results
3.1. Sequence Analysis of Bm-ser-1
3.2. Expression Pattern of Bmu-ser-1
3.3. In Situ Hybridization of Bm-ser-1
3.4. RNAi for the Bm-ser-1 Gene
3.5. The Bm-ser-1 Gene Is a Potential Sex Pheromone Receptor Gene
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kikuchi, T.; Cotton, J.A.; Dalzell, J.J.; Hasegawa, K.; Kanzaki, N.; McVeigh, P.; Takanashi, T.; Tsai, I.J.; Assefa, S.A.; Cock, P.J.A.; et al. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLOS Pathog. 2011, 7, e1002219. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, X.; Wei, Y.; Zhou, J.; Zhang, W.; Qin, P.; Chinta, S.; Kong, X.; Liu, Y.; Yu, H.; et al. Ascarosides coordinate the dispersal of a plant-parasitic nematode with the metamorphosis of its vector beetle. Nat. Commun. 2016, 7, 12341. [Google Scholar] [CrossRef]
- Webster, J.M.; Anderson, R.V.; Baillie, D.L.; Beckenbach, K.; Rutherford, T.A. DNA probes for differentiating isolates of the pinewood nematode species complex. Rev. Nématologie 1990, 13, 255–263. [Google Scholar] [CrossRef]
- Mamiya, Y.; Enda, N.J.N. Bursaphelenchus mucronatus n. sp.(Nematoda: Aphelenchoididae) from pine wood and its biology and pathogenicity to pine trees. Nematologica 1979, 25, 353–361. [Google Scholar] [CrossRef]
- Howard, A.D.; McAllister, G.; Feighner, S.D.; Liu, Q.; Nargund, R.P.; Van der Ploeg, L.H.T.; Patchett, A.A. Orphan G-protein-coupled receptors and natural ligand discovery. Trends Pharmacol. Sci. 2001, 22, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.P. Regulation of Behavioral Arousal and Quiescence in C. elegans. Genetics 2016, 199, 523–531. [Google Scholar]
- Bockaert, J.; Pin, J.j. Molecular tinkering of G protein-coupled receptors: An evolutionary success. EMBO J. 1999, 18, 1723–1729. [Google Scholar] [CrossRef] [PubMed]
- Anganathan, R.; Cannon, S.C.; Horvitz, H.R.J.N. MOD-1 is a serotonin-gated chloride channel that modulates locomotory behaviour in C. elegans. Nature 2000, 408, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Churgin, M.A.; McCloskey, R.J.; Peters, E.; Fang-Yen, C.J.J.o.N. Antagonistic serotonergic and octopaminergic neural circuits mediate food-dependent locomotory behavior in Caenorhabditis elegans. J. Neurosci. 2017, 37, 7811–7823. [Google Scholar] [CrossRef]
- Dernovici, S.; Starc, T.; Dent, J.A.; Ribeiro, P.J.D. The serotonin receptor SER-1 (5HT2ce) contributes to the regulation of locomotion in Caenorhabditis elegans. Dev. Neurobiol. 2007, 67, 189–204. [Google Scholar] [CrossRef]
- Carnell, L.; Illi, J.; Hong, S.W.; McIntire, S.L. The G-protein-coupled serotonin receptor SER-1 regulates egg laying and male mating behaviors in Caenorhabditis elegans. J. Neurosci. 2005, 25, 10671–10681. [Google Scholar] [CrossRef]
- Kiyohara, T. Sexual Attraction in Bursaphelenchus xylophilus. Jpn. J. Nematol. 1982, 11, 7–12. [Google Scholar] [CrossRef]
- Shinya, R.; Chen, A.; Sternberg, P.W. Sex attraction and mating in Bursaphelenchus okinawaensis and B. xylophilus. J. Nematol. 2015, 47, 176–183. [Google Scholar] [CrossRef]
- Meng, J.; Wickham, J.D.; Ren, W.; Zhao, L.; Sun, J. Species displacement facilitated by ascarosides between two sympatric sibling species: A native and invasive nematode. J. Pest Sci. 2020, 93, 1059–1071. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, W.; Bai, L.; Liu, H.; Wang, J.; Ma, X.; Huang, L.; Guo, K.; Yu, H.; Hu, J. Female’s war: A story of the invasion and competitive displacement between two xylophilus group nematode species. J. Pest Sci. 2023, 96, 1301–1311. [Google Scholar] [CrossRef]
- Zhou, L.; Ma, X.; Zhu, N.; Zou, Q.; Guo, K.; Bai, L.; Yu, H.; Hu, J. The role of mab-3 in spermatogenesis and ontogenesis of pinewood nematode, Bursaphelenchus xylophilus. Pest Manag. Sci. 2021, 77, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Urwin, P.E.; Lilley, C.J.; Atkinson, H.J. Ingestion of double-stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol. Plant-Microbe Interact. 2002, 15, 747–752. [Google Scholar] [CrossRef]
- Wang, J.; Liu, H.; Guo, K.; Yu, H.; Ye, J.; Hu, J. Molecular characterization and functional analysis of Bxy-octr-1 in Bursaphelenchus xylophilus. Gene 2022, 823, 146350. [Google Scholar] [CrossRef] [PubMed]
- Saudou, F.; Boschert, U.; Amlaiky, N.; Plassat, J.L.; Hen, R.J.E.J. A family of Drosophila serotonin receptors with distinct intracellular signalling properties and expression patterns. J. Entomol. Soc. Br. 1992, 11, 7–17. [Google Scholar] [CrossRef]
- Cunningham, K.A.; Hua, Z.; Srinivasan, S.; Liu, J.; Lee, B.H.; Edwards, R.H.; Ashrafi, K.J.C.M. AMP-activated kinase links serotonergic signaling to glutamate release for regulation of feeding behavior in C. elegans. Cell Metab. 2012, 16, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Ranawade, A.V.; Cumbo, P.; Gupta, B.P. Caenorhabditis elegans histone deacetylase hda-1 is required for morphogenesis of the vulva and LIN-12/Notch-mediated specification of uterine cell fates. Genes Genomes Genet. 2013, 3, 1363–1374. [Google Scholar] [CrossRef]
- Dufourcq, P.; Victor, M.; Gay, F.; Calvo, D.; Hodgkin, J.; Shi, Y. Functional Requirement for Histone Deacetylase 1 in Caenorhabditis elegans Gonadogenesis. Mol. Cell. Biol. 2002, 22, 3024–3034. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Bai, L.; Schütz, S.A.; Liu, B.; Liu, Z.; Zhang, X.Y.; Yu, H.; Hu, J. Observation and quantification of mating behavior in the pinewood nematode, Bursaphelenchus xylophilus. J. Vis. Exp. 2016, 118, 54842. [Google Scholar] [CrossRef]
- Dempsey, C.M.; Mackenzie, S.M.; Gargus, A.; Blanco, G.; Sze, J.Y. Serotonin (5HT), Fluoxetine, imipramine and dopamine target distinct 5HT receptor signaling to modulate Caenorhabditis elegans egg-laying behavior. Genetics 2005, 169, 1425–1436. [Google Scholar] [CrossRef] [PubMed]
- Aprison, E.Z.; Ruvinsky, I. The roles of several sensory neurons and the feedback from egg laying in regulating the germline response to a sex pheromone in C. elegans hermaphrodites. Micropublication Biol. 2022. [Google Scholar] [CrossRef]
- Park, J.; Oh, H.; Kim, D.Y.; Cheon, Y.; Park, Y.J.; Hwang, H.; Neal, S.J.; Dar, A.R.; Butcher, R.A.; Sengupta, P.; et al. CREB mediates the C. elegans dauer polyphenism through direct and cell-autonomous regulation of TGF-β expression. PLoS Genet. 2021, 17, e1009678. [Google Scholar] [CrossRef]






| Primer | Sequence (5′-3′) | |
|---|---|---|
| Cloning | Bmu-ser-1-C-F | TTAGCGATGGCAGATTTA |
| Bmu-ser-1-C-R | TTCGGGTAGTCAACACTC | |
| RT-qPCR | 18S-F | TTCTGACCGTAAACGA TGCCAACT |
| 18S-R | CCTCACCTCGAACGCCGAATTA | |
| Bmu-ser-1-Q-F | GGACCGACGGCTTCAAAATG | |
| Bmu-ser-1-Q-R | CGGAAACCGGTATAACCAGC | |
| RNAi | T7 promoter | TAATACGACTCACTATAGGG |
| Group | Nematode | Added Solution |
|---|---|---|
| 1 | Male-CK | ddH2O |
| 2 | Male-RNAi | ddH2O |
| 3 | Male-CK | Female steep liquor |
| 4 | Male-RNAi | Female steep liquor |
| 5 | Female-CK | ddH2O |
| 6 | Female-RNAi | ddH2O |
| 7 | Female-CK | Male steep liquor |
| 8 | Female-RNAi | Male steep liquor |
| Group | Mating Rate (%) | Mating Time/min | Wrong Mating Rate (%) | Effective Mating Rate (%) | Brood Size |
|---|---|---|---|---|---|
| CK♀ + CK♂ | 82.29 ± 2.25 a * | 14.34 ± 0.45 a | 5.77 ± 3.12 b | 73.50 ± 1.09 a | 10.24 ± 0.21 a |
| RNAi♀ + CK♂ | 83.33 ± 1.70 a | 13.21 ± 0.44 a | 9.67 ± 4.05 b | 63.73 ± 0.93 a | 5.27 ± 0.42 b |
| CK♀ + RNAi♂ | 78.13 ± 0.47 b | 8.00 ± 0.49 b | 22.10 ± 4.24 a | 34.10 ± 2.41 b | 6.12 ± 0.42 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Ma, R.; Chen, J.; Li, Q.; Guo, K.; Shao, H.; Hu, J. New Role in the 5-HT Receptor: The Sex Attracting of Bursaphelenchus mucronatus. Forests 2024, 15, 1115. https://doi.org/10.3390/f15071115
Liu W, Ma R, Chen J, Li Q, Guo K, Shao H, Hu J. New Role in the 5-HT Receptor: The Sex Attracting of Bursaphelenchus mucronatus. Forests. 2024; 15(7):1115. https://doi.org/10.3390/f15071115
Chicago/Turabian StyleLiu, Wenyi, Rui Ma, Jing Chen, Quan Li, Kai Guo, Hudie Shao, and Jiafu Hu. 2024. "New Role in the 5-HT Receptor: The Sex Attracting of Bursaphelenchus mucronatus" Forests 15, no. 7: 1115. https://doi.org/10.3390/f15071115
APA StyleLiu, W., Ma, R., Chen, J., Li, Q., Guo, K., Shao, H., & Hu, J. (2024). New Role in the 5-HT Receptor: The Sex Attracting of Bursaphelenchus mucronatus. Forests, 15(7), 1115. https://doi.org/10.3390/f15071115






