Soil Fungal Function Centralization Enhances the Decomposition of Fine Roots at Canopy Gap Borders
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description
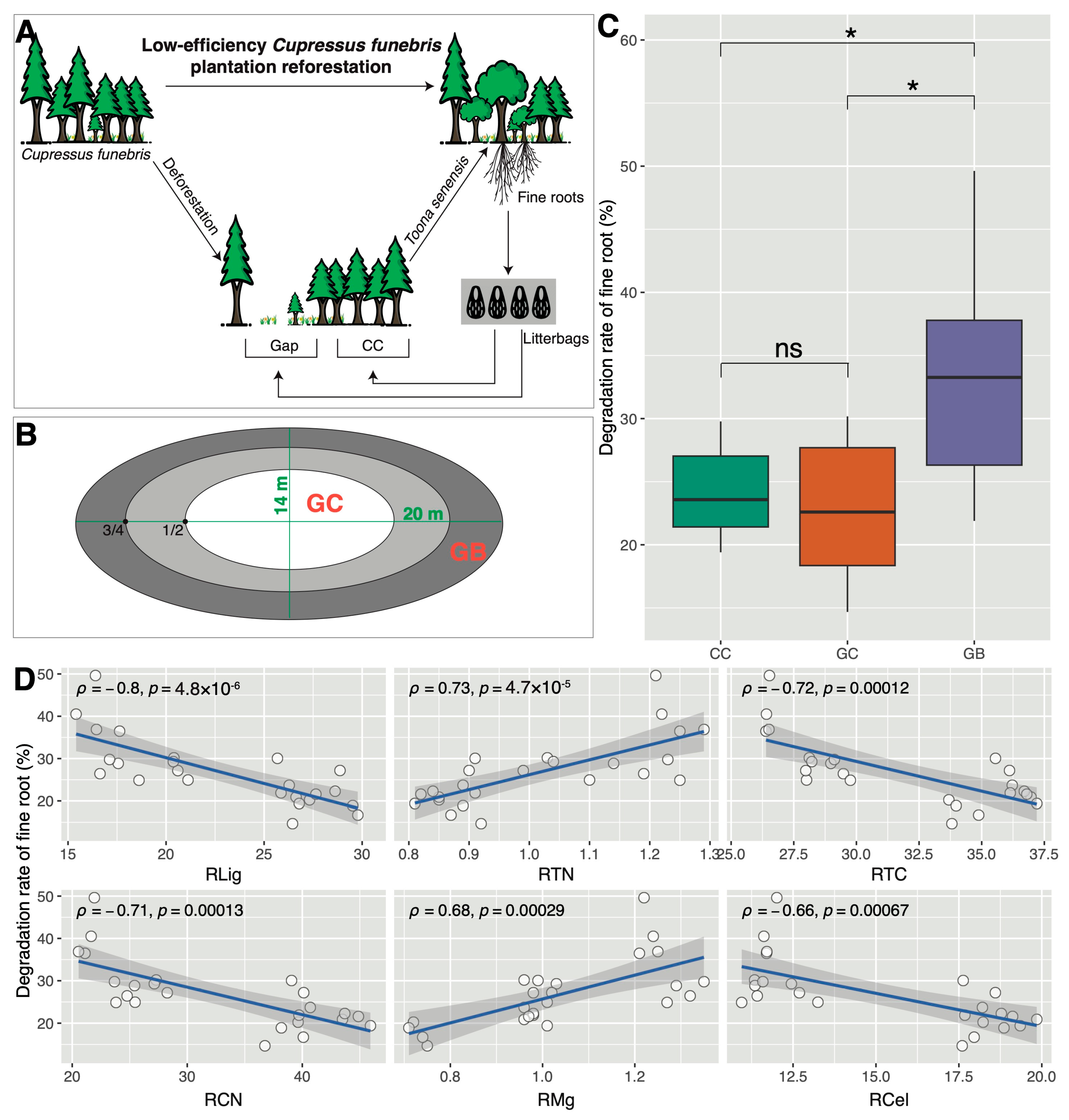
2.2. Experimental Design and Sample Collection
2.3. Soil Property and Root Nutrient Content Measurement
2.4. DNA Extraction, Sequencing, Bioinformatics Analysis, and Ecological Metric Calculation
2.5. Statistical Analysis
3. Results
3.1. Shifts in Soil Properties and Decomposition Rates across Gap Locations
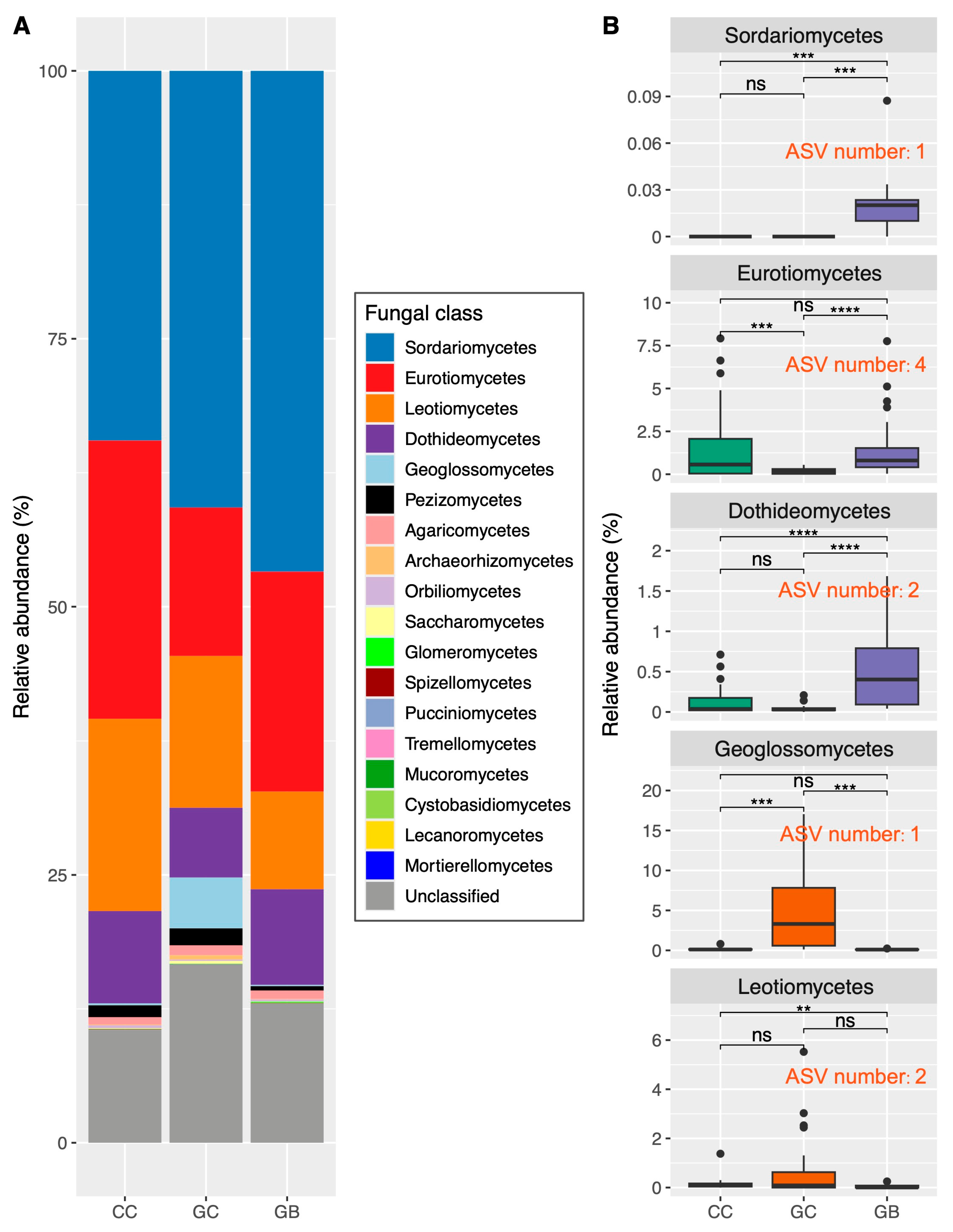
3.2. Shifts in Fungal Taxonomic Composition across Gap Locations and Associated Key Drivers
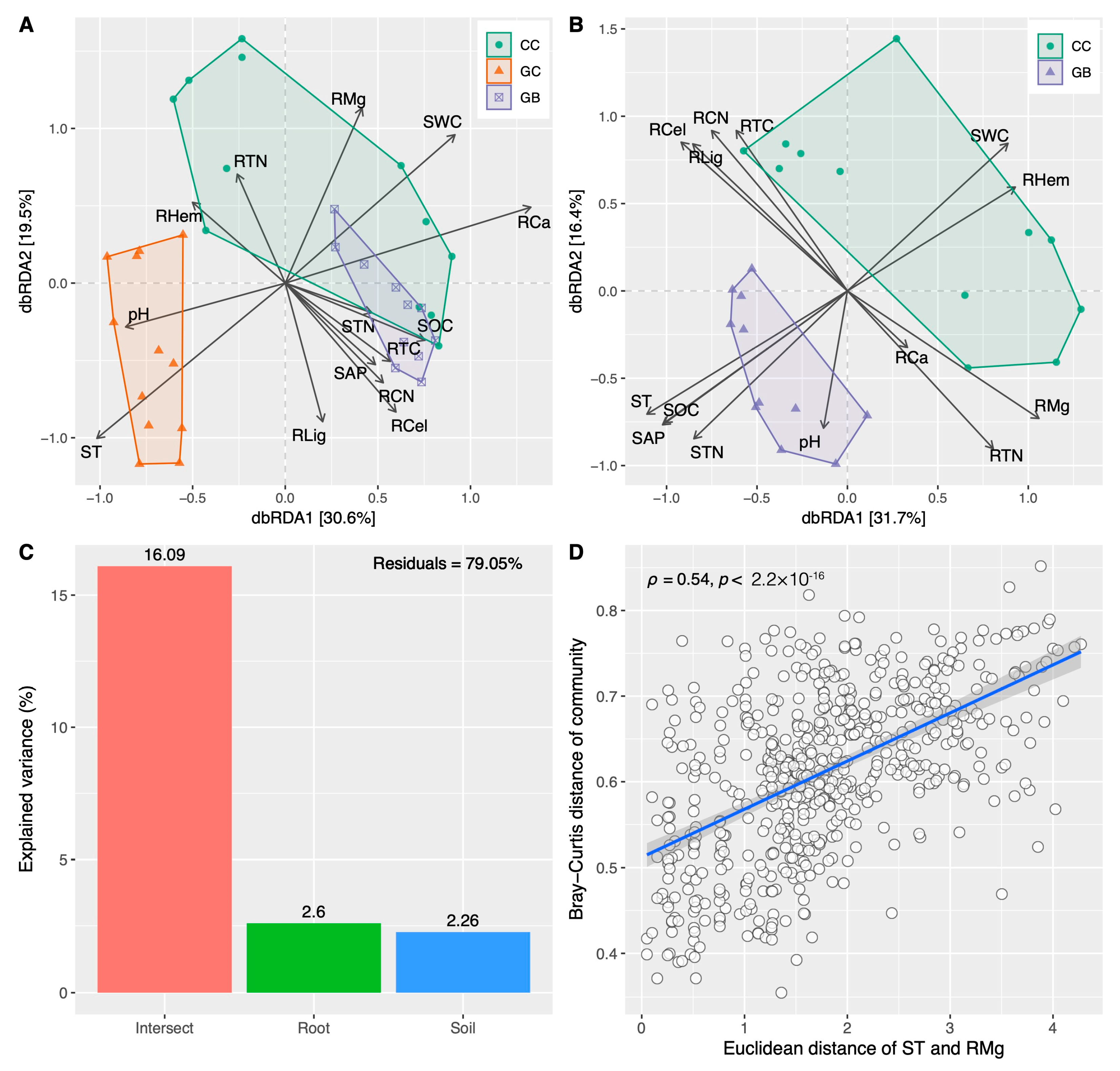
3.3. Shifts in Fungal Diversity and Community across Gap Locations and Their Key Drivers
4. Discussion
4.1. Gap Borders Exhibit the Highest Decomposition Rate of Toona Senensis Fine Roots
4.2. Saprophytic Fungi Rather Than Soil Properties Mainly Determine Fine Root Decomposition
4.3. Phylogenetic Clustering Results in the Centralization of Soil Saprophytic Fungi Functions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muscolo, A.; Bagnato, S.; Sidari, M.; Mercurio, R. A review of the roles of forest canopy gaps. J. For. Res. 2014, 25, 725–736. [Google Scholar] [CrossRef]
- Nygaard, P.H.; Strand, L.T.; Stuanes, A.O. Gap formation and dynamics after long-term steady state in an old-growth Picea abies stand in Norway: Above- and belowground interactions. Ecol. Evol. 2018, 8, 462–476. [Google Scholar] [CrossRef] [PubMed]
- Kelemen, K.; Mihók, B.; Gálhidy, L.; Standovár, T. Dynamic response of herbaceous vegetation to gap opening in a Central European beech stand. Silva Fenn. 2012, 46, 53–65. [Google Scholar] [CrossRef]
- McNab, W.H.; Kilgo, J.C.; Blake, J.I.; Zarnoch, S.J. Effect of gap size on composition and structure of regeneration 19 years after harvest in a southeastern bottomland forest, USA. Can. J. For. Res. 2020, 51, 380–392. [Google Scholar] [CrossRef]
- Fahey, R.T.; Puettmann, K.J. Patterns in spatial extent of gap influence on understory plant communities. For. Ecol. Manag. 2008, 255, 2801–2810. [Google Scholar] [CrossRef]
- Tong, R.; Ji, B.; Wang, G.G.; Lou, C.; Ma, C.; Zhu, N.; Yuan, W.; Wu, T. Canopy gap impacts on soil organic carbon and nutrient dynamic: A meta-analysis. Ann. For. Sci. 2024, 81, 12. [Google Scholar] [CrossRef]
- Saha, S.; Huang, L.; Khoso, M.A.; Wu, H.; Han, D.; Ma, X.; Poudel, T.R.; Li, B.; Zhu, M.; Lan, Q.; et al. Fine root decomposition in forest ecosystems: An ecological perspective. Front. Plant Sci. 2023, 14, 1277510. [Google Scholar] [CrossRef]
- Zhao, R.; Zhu, Y.-j.; Mao, Z.; Gu, W.; Zhang, H.; Long, F.; Sun, T. Predicting fine root decomposition from functional traits in 10 temperate tree species. Forests 2023, 14, 372. [Google Scholar] [CrossRef]
- Fujimaki, R.; Takeda, H.; Wiwatiwitaya, D. Fine root decomposition in tropical dry evergreen and dry deciduous forests in Thailand. J. For. Res. 2008, 13, 338–346. [Google Scholar] [CrossRef]
- Hobbie, S.; Oleksyn, J.; Eissenstat, D.; Reich, P. Fine root decomposition rates do not mirror those of leaf litter among temperate tree species. Oecologia 2009, 162, 505–513. [Google Scholar] [CrossRef]
- Wang, H.; Liu, S.; Mo, J. Correlation between leaf litter and fine root decomposition among subtropical tree species. Plant Soil 2010, 335, 289–298. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, L.; Guan, D.; Chen, Y.; Motelica-Heino, M.; Peng, Y.; Lee, S.Y. The role of mangrove fine root production and decomposition on soil organic carbon component ratios. Ecol. Indic. 2021, 125, 107525. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, X.; Zhang, D.-j.; Zhang, J. The effects of gap size and litter species on colonization of soil fauna during litter decomposition in Pinus massoniana plantations. Appl. Soil Ecol. 2020, 155, 103611. [Google Scholar] [CrossRef]
- Chen, J.; Zhu, J.; Wang, Z.; Xing, C.; Chen, B.; Wang, X.; Wei, C.; Liu, J.-f.; He, Z.-s.; Xu, D.-w. Canopy gaps control litter decomposition and nutrient release in subtropical forests. Forests 2023, 14, 673. [Google Scholar] [CrossRef]
- He, L.; Jia, Z.; Li, Q.-X.; Feng, L.; Yang, K. Fine-root decomposition characteristics of four typical shrubs in sandy areas of an arid and semiarid alpine region in western China. Ecol. Evol. 2019, 9, 5407–5419. [Google Scholar] [CrossRef]
- Zhao, B.; Xing, P.; Wu, Q.L. Interactions between bacteria and fungi in macrophyte leaf litter decomposition. Environ. Microbiol. 2020, 23, 1130–1144. [Google Scholar] [CrossRef]
- Li, D.; Li, X.; Su, Y.; Li, X.; Yin, H.; Li, X.; Guo, M.; He, Y. Forest gaps influence fungal community assembly in a weeping cypress forest. Appl. Microbiol. Biotechnol. 2019, 103, 3215–3224. [Google Scholar] [CrossRef]
- Bonfante, P.; Genre, A. Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat. Commun. 2010, 1, 48. [Google Scholar] [CrossRef] [PubMed]
- Schädler, M.; Brandl, R.; Kempel, A. “Afterlife” effects of mycorrhization on the decomposition of plant residues. Soil Biol. Biochem. 2010, 42, 521–523. [Google Scholar] [CrossRef]
- Argiroff, W.A.; Zak, D.; Upchurch, R.; Pellitier, P.; Belke, J.P. Fungal community composition and genetic potential regulate fine root decay in northern temperate forests. Mol. Ecol. 2023, 32, 2005–2021. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Jiang, S.; El-Naggar, A.; Tang, Y.; Liu, X.; Zhang, J. Dynamics of fine root decomposition in different vegetation types: Investigating the impact of soil fungal communities and enzyme activities. Forests 2023, 14, 1321. [Google Scholar] [CrossRef]
- Fahey, T.; Fahey, C.; Goebel, M.; Antunes, P.M.; Yavitt, J. Fine root decomposition and macronutrient release in temperate forests: Examining the litter bag and intact soil core approaches and their relationship with fungal community composition. Appl. Soil Ecol. 2024, 193, 105147. [Google Scholar] [CrossRef]
- Fisk, M.; Fahey, T.; Sobieraj, J.; Staniec, A.C.; Crist, T.O. Rhizosphere disturbance influences fungal colonization and community development on dead fine roots. Plant Soil 2011, 341, 279–293. [Google Scholar] [CrossRef]
- Gray, L.; Kernaghan, G. Fungal succession during the decomposition of ectomycorrhizal fine roots. Microb. Ecol. 2019, 79, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Kohout, P.; Charvátová, M.; Štursová, M.; Mašínová, T.; Tomšovský, M.; Baldrian, P. Clearcutting alters decomposition processes and initiates complex restructuring of fungal communities in soil and tree roots. ISME J. 2018, 12, 692–703. [Google Scholar] [CrossRef]
- Kohout, P.; Sudová, R.; Brabcová, V.; Vosolsobě, S.; Baldrian, P.; Albrechtová, J. Forest microhabitat affects succession of fungal communities on decomposing fine tree roots. Front. Microbiol. 2021, 12, 541583. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Fahey, T.; Pawlowska, T.; Fisk, M.; Burtis, J. Fine root decomposition, nutrient mobilization and fungal communities in a pine forest ecosystem. Soil Biol. Biochem. 2015, 83, 76–83. [Google Scholar] [CrossRef]
- Baldrian, P. Forest microbiome: Diversity, complexity and dynamics. FEMS Microbiol. Rev. 2016, 41, 109–130. [Google Scholar] [CrossRef] [PubMed]
- Purahong, W.; Wubet, T.; Lentendu, G.; Schloter, M.; Pecyna, M.; Kapturska, D.; Hofrichter, M.; Krüger, D.; Buscot, F. Life in leaf litter: Novel insights into community dynamics of bacteria and fungi during litter decomposition. Mol. Ecol. 2016, 25, 4059–4074. [Google Scholar] [CrossRef]
- Zhang, Q.; Zak, J.C. Effects of gap size on litter decomposition and microbial activity in a subtropical forest. Ecology 1995, 76, 2196–2204. [Google Scholar] [CrossRef]
- Xu, J.; Xue, L.; Su, Z. Impacts of forest gaps on soil properties after a severe ice storm in a Cunninghamia lanceolata Stand. Pedosphere 2016, 26, 408–416. [Google Scholar] [CrossRef]
- Sariyildiz, T. Effects of gap-size classes on long-term litter decomposition rates of beech, oak and chestnut species at high elevations in Northeast Turkey. Ecosystems 2008, 11, 841–853. [Google Scholar] [CrossRef]
- Ritter, E. Litter decomposition and nitrogen mineralization in newly formed gaps in a Danish beech (Fagus sylvatica) forest. Soil Biol. Biochem. 2005, 37, 1237–1247. [Google Scholar] [CrossRef]
- Prescott, C.E.; Hope, G.D.; Blevins, L.L. Effect of gap size on litter decomposition and soil nitrate concentrations in a high-elevation spruce fir forest. Can. J. For. Res. 2003, 33, 2210–2220. [Google Scholar] [CrossRef]
- Muscolo, A.; Sidari, M.; Mercurio, R. Influence of gap size on organic matter decomposition, microbial biomass and nutrient cycle in Calabrian pine (Pinus laricio, Poiret) stands. For. Ecol. Manag. 2007, 242, 412–418. [Google Scholar] [CrossRef]
- Han, M.; Tang, M.; Shi, B.; Jin, G. Effect of canopy gap size on soil respiration in a mixed broadleaved-Korean pine forest: Evidence from biotic and abiotic factors. Eur. J. Soil Biol. 2020, 99, 103194. [Google Scholar] [CrossRef]
- Denslow, J.; Ellison, A.; Sanford, R.E. Treefall gap size effects on above- and below-ground processes in a tropical wet forest. J. Ecol. 1998, 86, 597–609. [Google Scholar] [CrossRef]
- Scharenbroch, B.; Bockheim, J. Impacts of forest gaps on soil properties and processes in old growth northern hardwood-hemlock forests. Plant Soil 2007, 294, 219–233. [Google Scholar] [CrossRef]
- Ni, X.; Yang, W.; Tan, B.; Li, H.; He, J.; Xu, L.; Wu, F. Forest gaps slow the sequestration of soil organic matter: A humification experiment with six foliar litters in an alpine forest. Sci. Rep. 2016, 6, 19744. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Chen, H.Y. Fine root dynamics with stand development in the boreal forest. Funct. Ecol. 2012, 26, 991–998. [Google Scholar] [CrossRef]
- Wang, G.; Liu, F. The influence of gap creation on the regeneration of Pinus tabuliformis planted forest and its role in the near-natural cultivation strategy for planted forest management. For. Ecol. Manag. 2011, 262, 413–423. [Google Scholar] [CrossRef]
- Pregitzer, K.S.; DeForest, J.L.; Burton, A.J.; Allen, M.F.; Ruess, R.W.; Hendrick, R.L. Fine root architecture of nine north American trees. Ecol. Monogr. 2002, 72, 293–309. [Google Scholar] [CrossRef]
- Li, C.; Liao, H.; Li, D.; Jing, Y. Abundant species govern the altitude patterns of bacterial community in natural and disturbed subalpine forest soils. Diversity 2024, 16, 242. [Google Scholar] [CrossRef]
- Olsen, S.R. Estimation of Available Phosphorus in Soils by Extraction with Sodium Bicarbonate; US Department of Agriculture: Washington, DC, USA, 1954.
- Brändli, R.C.; Bergsli, A.; Ghosh, U.; Hartnik, T.; Breedveld, G.D.; Cornelissen, G. Quantification of activated carbon contents in soils and sediments using chemothermal and wet oxidation methods. Environ. Pollut. 2009, 157, 3465–3470. [Google Scholar] [CrossRef]
- Bremner, J. Total nitrogen. In Methods of Soil Analysis: Part 2 Chemical and Microbiological Properties; American Society of Agronomy, Soil Science Society of America: Madison, WI, USA, 1965; Volume 9, pp. 1149–1178. [Google Scholar]
- Rowland, A.; Roberts, J.D. Lignin and cellulose fractionation in decomposition studies using acid-detergent fibre methods. Commun. Soil Sci. Plant Anal. 1994, 25, 269–277. [Google Scholar] [CrossRef]
- Blumenkrantz, N.; Asboe-Hansen, G. New method for quantitative determination of uronic acids. Anal. Biochem. 1973, 54, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liao, H.; Li, D.; Jing, Y. The fungal functional guilds at the early-stage restoration of subalpine forest soils disrupted by highway construction in Southwest China. Forests 2024, 15, 636. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.; Schigel, D.; Kennedy, P.; Picard, K.T.; Glöckner, F.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2018, 47, D259–D264. [Google Scholar] [CrossRef]
- Li, C.; Liu, C.; Li, H.; Liao, H.; Xu, L.; Yao, M.; Li, X. The microgeo: An R package rapidly displays the biogeography of soil microbial community traits on maps. FEMS Microbiol. Ecol. 2024, 100, fiae087. [Google Scholar] [CrossRef]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, B.M.; Stegen, J.C.; Kim, M.; Dong, K.; Adams, J.M.; Lee, Y.K. Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 2018, 12, 1072–1083. [Google Scholar] [CrossRef] [PubMed]
- Ning, D.L.; Yuan, M.T.; Wu, L.W.; Zhang, Y.; Guo, X.; Zhou, X.S.; Yang, Y.F.; Arkin, A.P.; Firestone, M.K.; Zhou, J.Z. A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming. Nat. Commun. 2020, 11, e4717. [Google Scholar] [CrossRef] [PubMed]
- Stegen, J.C.; Lin, X.; Konopka, A.E.; Fredrickson, J.K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012, 6, 1653–1664. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef] [PubMed]
- Põlme, S.; Abarenkov, K.; Henrik Nilsson, R.; Lindahl, B.D.; Clemmensen, K.E.; Kauserud, H.; Nguyen, N.; Kjøller, R.; Bates, S.T.; Baldrian, P.; et al. FungalTraits: A user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 2020, 105, 1–16. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Remy, E.; Wuyts, K.; Nevel, L.v.; Smedt, P.D.; Boeckx, P.; Verheyen, K. Driving factors behind litter decomposition and nutrient release at temperate forest edges. Ecosystems 2017, 21, 755–771. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, D.J.; Li, X.; Liu, H.; Zhang, M.J.; Yang, W.Q.; Zhang, J. Edge effects of forest gap in Pinus massoniana plantations on the decomposition of leaf litter recalcitrant components of Cinnamomum camphora and Toona ciliata. Ying Yong Sheng Tai Xue Bao 2016, 27, 1116–1124. [Google Scholar]
- Li, H.; Wu, F.; Yang, W.; Xu, L.; Ni, X.; He, J.; Tan, B.; Hu, Y. Effects of forest gaps on litter lignin and cellulose dynamics vary seasonally in an alpine forest. Forests 2016, 7, 27. [Google Scholar] [CrossRef]
- He, W.; Wu, F.; Yang, W.; Zhang, D.; Xu, Z.; Tan, B.; Zhao, Y.; Justine, M.F. Gap locations influence the release of carbon, nitrogen and phosphorus in two shrub foliar litter in an alpine fir forest. Sci. Rep. 2016, 6, 22014. [Google Scholar] [CrossRef]
- Ritter, E.; Dalsgaard, L.; Einhorn, K. Light, temperature and soil moisture regimes following gap formation in a semi-natural beech-dominated forest in Denmark. For. Ecol. Manag. 2005, 206, 15–33. [Google Scholar] [CrossRef]
- Zhu, J.; Matsuzaki, T.; Lee, F.-q.; Gonda, Y. Effect of gap size created by thinning on seedling emergency, survival and establishment in a coastal pine forest. For. Ecol. Manag. 2003, 182, 339–354. [Google Scholar] [CrossRef]
- Li, S.-P.; Cadotte, M.W.; Meiners, S.J.; Hua, Z.-S.; Jiang, L.; Shu, W.-S. Species colonisation, not competitive exclusion, drives community overdispersion over long-term succession. Ecol. Lett. 2015, 18, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Chen, X.; Kennedy, D.W.; Murray, C.J.; Rockhold, M.L.; Konopka, A. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013, 7, 2069–2079. [Google Scholar] [CrossRef] [PubMed]
- Žifčáková, L.; Větrovský, T.; Howe, A.; Baldrian, P. Microbial activity in forest soil reflects the changes in ecosystem properties between summer and winter. Environ. Microbiol. 2015, 18, 288–301. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; TripathI, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, H.; Li, C.; Han, Z.; Luo, G.; Yang, Y.; Wu, Q.; An, A.; Li, X.; Li, D. Soil Fungal Function Centralization Enhances the Decomposition of Fine Roots at Canopy Gap Borders. Forests 2024, 15, 1293. https://doi.org/10.3390/f15081293
Liao H, Li C, Han Z, Luo G, Yang Y, Wu Q, An A, Li X, Li D. Soil Fungal Function Centralization Enhances the Decomposition of Fine Roots at Canopy Gap Borders. Forests. 2024; 15(8):1293. https://doi.org/10.3390/f15081293
Chicago/Turabian StyleLiao, Haijun, Chaonan Li, Zhoulin Han, Guorong Luo, Yulian Yang, Qinggui Wu, Aluo An, Xianwei Li, and Dehui Li. 2024. "Soil Fungal Function Centralization Enhances the Decomposition of Fine Roots at Canopy Gap Borders" Forests 15, no. 8: 1293. https://doi.org/10.3390/f15081293
APA StyleLiao, H., Li, C., Han, Z., Luo, G., Yang, Y., Wu, Q., An, A., Li, X., & Li, D. (2024). Soil Fungal Function Centralization Enhances the Decomposition of Fine Roots at Canopy Gap Borders. Forests, 15(8), 1293. https://doi.org/10.3390/f15081293




