Tracking Community Timing: Pattern and Determinants of Seasonality in Culicoides (Diptera: Ceratopogonidae) in Northern Florida
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect Collections
2.2. Environmental Data
2.3. Culicoides Data
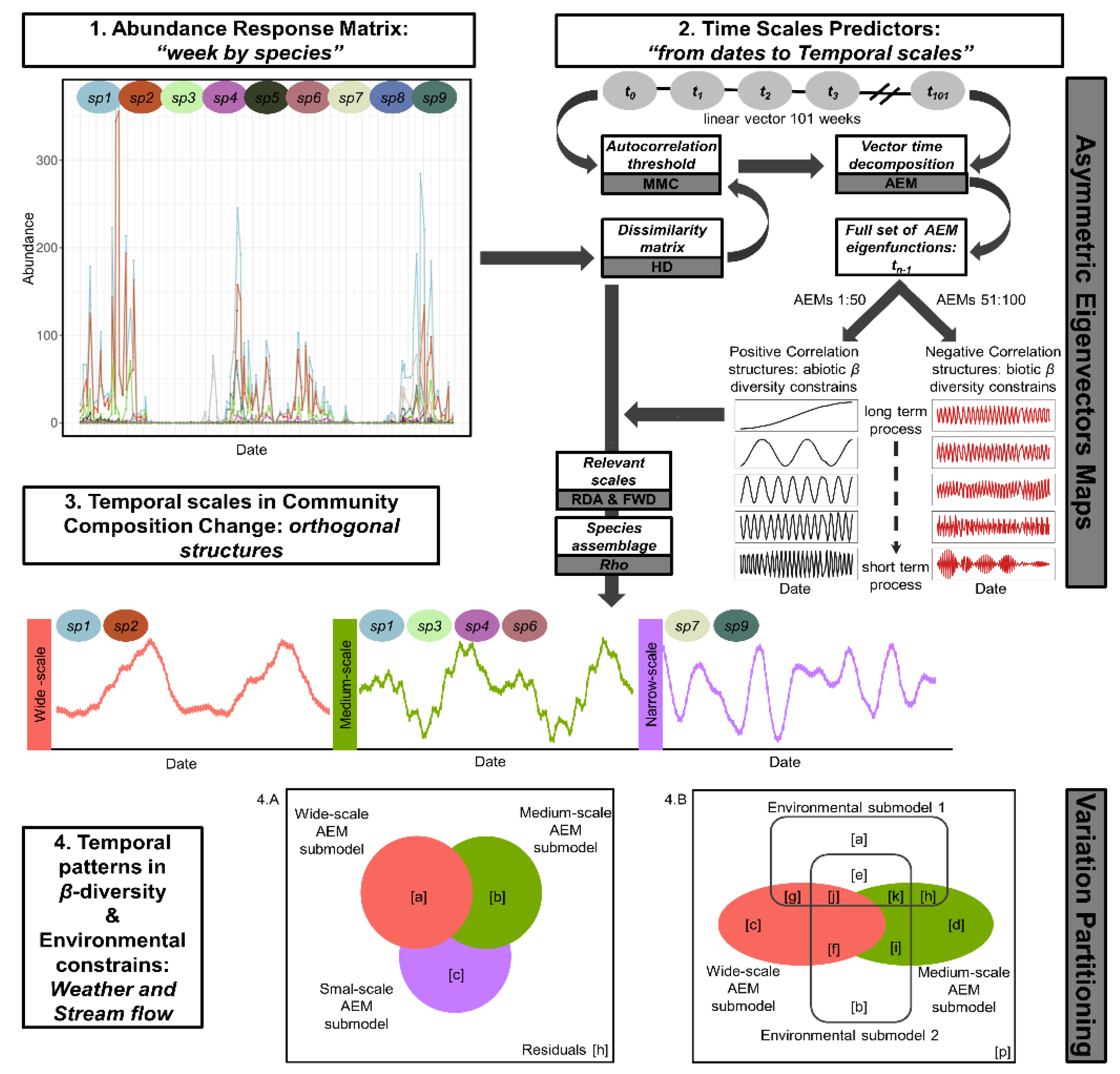
2.4. Culicoides β-Diversity Temporal Profile
2.5. Seasonality in Species Recruitment, Weekly Occurrence and Co-Occurrence Structure
2.6. Environmental Thresholds in the Temporal Dynamics of the Vector Assemblage
3. Results
3.1. Insect Collections
3.2. Culicoides β-Diversity Temporal Profile
3.3. Seasonality in Species Recruitment, Weekly Occurrence and Co-Occurrence Structure
3.4. Environmental Thresholds in the Temporal Dynamic of the Vector Assemblage
4. Discussion
4.1. Seasonality of Culicoides β-Diversity
4.2. Seasonal Culicoides Diversity Structure
4.3. Persistence of Vector-Borne Pathogen in the Ecosystems Depends on Environmental Stability
4.4. Is Culicoides β-Diversity Operating under Wider Temporal Scales?
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fenton, A.; Pedersen, A.B. Community epidemiology framework for classifying disease threats. Emerg. Infect. Dis. 2005, 11, 1815. [Google Scholar] [CrossRef] [PubMed]
- Begon, M. Effects of host diversity on disease dynamics. In Infectious Disease Ecology: Effects of Ecosystems on Disease and of Disease on Ecosystems, 1st ed; Ostfeld, R., Keesing, F., Eviner, V., Eds.; Princeton University Press: Oxford, UK, 2008; pp. 12–29. [Google Scholar]
- Smith, K.F.; Behrens, M.D.; Sax, D.F. Local scale effects of disease on biodiversity. Ecohealth 2009, 6, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Paull, S.H.; Johnson, P.T.J. Experimental warming drives a seasonal shift in the timing of host-parasite dynamics with consequences for disease risk. Ecol. Lett. 2014, 17, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Viana, M.; Mancy, R.; Biek, R.; Cleaveland, S.; Cross, P.C.; Lloyd-Smith, J.O.; Haydon, D.T. Assembling evidence for identifying reservoirs of infection. Trends Ecol. Evol. 2014, 29, 270–279. [Google Scholar] [CrossRef] [Green Version]
- Johnson, P.T.J.; Ostfeld, R.S.; Keesing, F. Frontiers in research on biodiversity and disease. Ecol. Lett. 2015, 18, 1119–1133. [Google Scholar] [CrossRef] [Green Version]
- Johnson, P.T.J.; De Roode, J.C.; Fenton, A. Why infectious disease research needs community ecology. Science 2015, 349. [Google Scholar] [CrossRef] [Green Version]
- Altizer, S.; Dobson, A.; Hosseini, P.; Hudson, P.; Pascual, M.; Rohani, P. Seasonality and the dynamics of infectious diseases. Ecol. Lett. 2006, 9, 467–484. [Google Scholar] [CrossRef] [Green Version]
- Angeler, D.G.; Göthe, E.; Johnson, R.K. Hierarchical Dynamics of Ecological Communities: Do Scales of Space and Time Match? PLoS ONE 2013, 8, e69174. [Google Scholar] [CrossRef]
- Legendre, P.; Gauthier, O. Statistical methods for temporal and space-time analysis of community composition data. Proc. R. Soc. B Biol. Sci. 2014, 281, 20132728. [Google Scholar] [CrossRef] [Green Version]
- Baho, D.L.; Futter, M.N.; Johnson, R.K.; Angeler, D.G. Assessing temporal scales and patterns in time series: Comparing methods based on redundancy analysis. Ecol. Complex. 2015, 22, 162–168. [Google Scholar] [CrossRef] [Green Version]
- Tabachnick, W.J. Challenges in predicting climate and environmental effects on vector-borne disease episystems in a changing world. J. Exp. Biol. 2010, 213, 946–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simón, F.; González-Miguel, J.; Diosdado, A.; Gómez, P.J.; Morchón, R.; Kartashev, V. The Complexity of Zoonotic Filariasis Episystem and Its Consequences: A Multidisciplinary View. Biomed Res. Int. 2017, 2017, 6436130. [Google Scholar] [CrossRef] [Green Version]
- Holyoak, M.; Leibold, M.A.; Holt, R.D. Metacommunities: Spatial Dynamics and Ecological Communities; University of Chicago Press: Chicago, IL, USA, 2005; ISBN 0226350649. [Google Scholar]
- Massol, F.; Gravel, D.; Mouquet, N.; Cadotte, M.W.; Fukami, T.; Leibold, M.A. Linking community and ecosystem dynamics through spatial ecology. Ecol. Lett. 2011, 14, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Dodd, R.S.; Hüberli, D.; Mayer, W.; Harnik, T.Y.; Afzal-Rafii, Z.; Garbelotto, M. Evidence for the role of synchronicity between host phenology and pathogen activity in the distribution of sudden oak death canker disease. New Phytol. 2008, 179, 505–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grulke, N.E.; The, S.; Phytologist, N.; January, N.; Government, S.; Bos, C. The nexus of host and pathogen phenology: Understanding the disease triangle with climate change. New Phytol. 2011, 189, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Kuno, G.; Chang, G.J. Biological Transmission of Arboviruses: Reexamination of and New Insights into Components, Mechanisms, and Unique Traits as Well as Their Evolutionary Trends. Clin. Microbiol. Rev. 2005, 18, 608–637. [Google Scholar] [CrossRef] [Green Version]
- Unnasch, R.S.; Cupp, E.W.; Unnasch, T.R. Host selection and its role in transmission of arboviral encephalitides. Dis. Ecol. Community Struct. Pathog. Dyn. 2006, 73–89. [Google Scholar]
- Takken, W.; Verhulst, N.O. Host Preferences of Blood-Feeding Mosquitoes. Annu. Rev. Entomol. 2013, 58, 433–453. [Google Scholar] [CrossRef] [Green Version]
- Hamer, S.A.; Hickling, G.J.; Sidge, J.L.; Walker, E.D.; Tsao, J.I. Synchronous phenology of juvenile Ixodes scapularis, vertebrate host relationships, and associated patterns of Borrelia burgdorferi ribotypes in the midwestern United States. Ticks Tick. Borne. Dis. 2012, 3, 65–74. [Google Scholar] [CrossRef]
- The Subgeneric Classification of Species of Culicoides—Thoughts and a warning; Last revised: May 16, 2016. Available online: https://www.inhs.illinois.edu/files/5014/6532/8290/CulicoidesSubgenera.pdf (accessed on 24 August 2020).
- Carpenter, S.; Veronesi, E.; Mullens, B.; Venter, G. Vector competence of Culicoides for arboviruses: Three major periods of research, their influence on current studies and future directions. OIE Rev. Sci. Tech. 2015, 34, 97–112. [Google Scholar] [CrossRef]
- Purse, B.V.; Carpenter, S.; Venter, G.J.; Bellis, G.; Mullens, B.A. Bionomics of Temperate and Tropical Culicoides Midges: Knowledge Gaps and Consequences for Transmission of Culicoides-Borne Viruses. Annu. Rev. Entomol. 2015, 60, 373–392. [Google Scholar] [CrossRef] [PubMed]
- Berry, B.S.; Magori, K.; Perofsky, A.C.; Stallknecht, D.E.; Park, A.W. Wetland cover dynamics drive hemorrhagic disease patterns in white-tailed deer in the United States. J. Wildl. Dis. 2013, 49, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Ruder, M.G.; Lysyk, T.J.; Stallknecht, D.E.; Foil, L.D.; Johnson, D.J.; Chase, C.C.; Dargatz, D.A.; Gibbs, E.P.J. Transmission and Epidemiology of Bluetongue and Epizootic Hemorrhagic Disease in North America: Current Perspectives, Research Gaps, and Future Directions. Vector-Borne Zoonotic Dis. 2015, 15, 348–363. [Google Scholar] [CrossRef] [PubMed]
- Cauvin, A.; Dinh, E.T.N.; Orange, J.P.; Shuman, R.M.; Blackburn, J.K.; Wisely, S.M. Antibodies to Epizootic Hemorrhagic Disease Virus (EHDV) in Farmed and Wild Florida White-Tailed Deer (Odocoileus virginianus). J. Wildl. Dis. 2020, 56, 208–213. [Google Scholar] [CrossRef]
- Boughton, R.K.; Wight, B.R.; Wisely, S.; Hood, K.; Main, M.B. White-tailed Deer of Florida. Edis 2020, 2020, 12. [Google Scholar] [CrossRef]
- Becker, M.E.; Roberts, J.; Schroeder, M.E.; Gentry, G.; Foil, L.D. Prospective Study of Epizootic Hemorrhagic Disease Virus and Bluetongue Virus Transmission in Captive Ruminants. J. Med. Entomol. 2020, 57, 1277–1285. [Google Scholar] [CrossRef]
- Blanton, F.S.; Wirth, W.W. The sand flies (Culicoides) of Florida (Diptera: Ceratopogonidae). Arthropods Florida Neighboring Land Areas 1979, 10, 1–204. [Google Scholar]
- Conte, A.; Giovannini, A.; Savini, L.; Goffredo, M.; Calistri, P.; Meiswinkel, R. The effect of climate on the presence of Culicoides imicola in Italy. J. Vet. Med. Ser. B 2003, 50, 139–147. [Google Scholar] [CrossRef]
- Conte, A.; Ippoliti, C.; Savini, L.; Goffredo, M.; Meiswinkel, R. Novel environmental factors influencing the distribution and abundance of Culicoides imicola and the Obsoletus Complex in Italy. Vet. Ital. 2007, 43, 571–580. [Google Scholar]
- Purse, B.V.; Tatem, A.J.; Caracappa, S.; Rogers, D.J.; Mellor, P.S.; Baylis, M.; Torina, A. Modelling the ditributions of Culicoides bluetongue virus vectors in Sicily in relation to satellite-derived climate variables. Med. Vet. Entomol. 2004, 18, 90–101. [Google Scholar] [CrossRef]
- Purse, B.V.; Falconer, D.; Sullivan, M.J.; Carpenter, S.; Mellor, P.S.; Piertney, S.B.; Mordue Luntz, A.J.; Albon, S.; Gunn, G.J.; Blackwell, A. Impacts of climate, host and landscape factors on Culicoides species in Scotland. Med. Vet. Entomol. 2012, 26, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, P.; Ruiz-Fons, F.; Estrada, R.; Márquez, A.L.; Miranda, M.A.; Gortázar, C.; Lucientes, J. A broad assessment of factors determining Culicoides imicola abundance: Modelling the present and forecasting its future in climate change scenarios. PLoS ONE 2010, 5, e14236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, C.J.; Shortall, C.R.; Gubbins, S.; Burgin, L.; Gloster, J.; Harrington, R.; Reynolds, D.R.; Mellor, P.S.; Carpenter, S. Influence of season and meteorological parameters on flight activity of Culicoides biting midges. J. Appl. Ecol. 2011, 48, 1355–1364. [Google Scholar] [CrossRef]
- Peters, J.; Conte, A.; Van doninck, J.; Verhoest, N.E.C.; De Clercq, E.; Goffredo, M.; De Baets, B.; Hendrickx, G.; Ducheyne, E. On the relation between soil moisture dynamics and the geographical distribution of Culicoides imicola. Ecohydrology 2014, 7, 622–632. [Google Scholar] [CrossRef]
- Sloyer, K.E.; Burkett-Cadena, N.D.; Yang, A.; Corn, J.L.; Vigil, S.L.; McGregor, B.L.; Wisely, S.M.; Blackburn, J.K. Ecological niche modeling the potential geographic distribution of four Culicoides species of veterinary significance in florida, USA. PLoS ONE 2019, 14, e0206648. [Google Scholar] [CrossRef] [Green Version]
- Grimaud, Y.; Guis, H.; Chiroleu, F.; Boucher, F.; Tran, A.; Rakotoarivony, I.; Duhayon, M.; Cêtre-Sossah, C.; Esnault, O.; Cardinale, E.; et al. Modelling temporal dynamics of Culicoides Latreille (Diptera: Ceratopogonidae) populations on Reunion Island (Indian Ocean), vectors of viruses of veterinary importance. Parasit. Vectors 2019, 12, 562. [Google Scholar] [CrossRef]
- Hubbell, S.P. The Unified Neutral Theory of Biodiversity and Biogeography (MPB-32); Princeton University Press: Princeton, NJ, USA, 2001; ISBN 0691021287. [Google Scholar]
- Gravel, D.; Canham, C.D.; Beaudet, M.; Messier, C. Reconciling niche and neutrality: The continuum hypothesis. Ecol. Lett. 2006, 9, 399–409. [Google Scholar] [CrossRef] [Green Version]
- Ruokolainen, L.; Ranta, E.; Kaitala, V.; Fowler, M.S. When can we distinguish between neutral and non-neutral processes in community dynamics under ecological drift? Ecol. Lett. 2009, 12, 909–919. [Google Scholar] [CrossRef]
- Vellend, M. Conceptual synthesis in community ecology. Q. Rev. Biol. 2010, 85, 183–206. [Google Scholar] [CrossRef] [Green Version]
- Rosindell, J.; Hubbell, S.P.; He, F.; Harmon, L.J.; Etienne, R.S. The case for ecological neutral theory. Trends Ecol. Evol. 2012, 27, 203–208. [Google Scholar] [CrossRef]
- Kramer, W.L.; Greiner, E.C.; Gibbs, E.P.J. A survey of Culicoides midges (Diptera: Ceratopogonidae) associated with cattle operations in Florida, USA. J. Med. Entomol. 1985, 22, 153–162. [Google Scholar] [CrossRef]
- Kramer, W.L.; Greiner, E.C.; Gibbs, E.P. Seasonal variations in population size, fecundity, and parity rates of Culicoides insignis (Diptera: Ceratopogonidae) in Florida, USA. J. Med. Entomol. 1985, 22, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Baylis, M.; El Hasnaoui, H.; Bouayoune, H.; Touti, J.; Mellor, P.S. The spatial and seasonal distribution of African horse sickness and its potential Culicoides vectors in Morocco. Med. Vet. Entomol. 1997, 11, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Meiswinkel, R.; Scolamacchia, F.; Dik, M.; Mudde, J.; Dijkstra, E.; Van Der Ven, I.J.K.; Elbers, A.R.W. The Mondrian matrix: Culicoides biting midge abundance and seasonal incidence during the 2006-2008 epidemic of bluetongue in the Netherlands. Med. Vet. Entomol. 2014, 28, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Sohier, C.; Deblauwe, I.; De Deken, R.; Madder, M.; Fassotte, C.; Losson, B.; De Regge, N. Longitudinal monitoring of Culicoides in Belgium between 2007 and 2011: Local variation in population dynamics parameters warrant cautious use of monitoring data. Parasit. Vectors 2018, 11, 512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, C.J.; Shortall, C.R.; England, M.; Harrington, R.; Purse, B.; Burgin, L.; Carpenter, S.; Gubbins, S. Long-term shifts in the seasonal abundance of adult Culicoides biting midges and their impact on potential arbovirus outbreaks. J. Appl. Ecol. 2019, 56, 1649–1660. [Google Scholar] [CrossRef] [Green Version]
- Wolfe, S.H.; Reidenauer, J.A.; Means, D.B. An Ecological Characterization of the Florida Panhandle; US Fish and Wildlife Service: Ft. Collins, CO, USA, 1988.
- Foster, N.M.; Breckon, R.D.; Luedke, A.J.; Jones, R.H.; Metcalf, H.E. Transmission of two strains of epizootic hemorrhagic disease virus in deer by Culicoides variipennis. J. Wildl. Dis. 1977, 13, 9–16. [Google Scholar] [CrossRef]
- Jones, R.H.; Roughton, R.D.; Foster, N.M.; Bando, B.M. Culicoides, the vector of epizootic hemorrhagic disease in white-tailed deer in Kentucky in 1971. J. Wildl. Dis. 1977, 13, 2–8. [Google Scholar] [CrossRef] [Green Version]
- Tanya, V.N.; Greiner, E.C.; Gibbs, E.P.J. Evaluation of Culicoides insignis (Diptera: Ceratopogonidae) as a vector of bluetongue virus. Vet. Microbiol. 1992, 32, 1–14. [Google Scholar] [CrossRef]
- Vigil, S.L.; Wlodkowski, J.C.; Parris, J.; Edwards de Vargas, S.; Shaw, D.; Cleveland, C.; Grogan Jr, W.L.; Corn, J.L. New Records of Biting Midges of the Genus Culicoides Latreille from the Southeastern United States (Diptera: Ceratopogonidae). Insecta Mundi 2014, 0394, 1–14. [Google Scholar]
- Vigil, S.L.; Ruder, M.G.; Shaw, D.; Wlodkowski, J.; Garrett, K.; Walter, M.; Corn, J.L. Apparent range expansion of Culicoides (Hoffmania) insignis (Diptera: Ceratopogonidae) in the Southeastern United States. J. Med. Entomol. 2018, 55, 1043–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mullen, G.R.; Hayes, M.E.; Nusbaum, K.E. Potential vectors of bluetongue and epizootic hemorrhagic disease viruses of cattle and white-tailed deer in Alabama. Prog. Clin. Biol. Res. 1985, 178, 201–206. [Google Scholar] [PubMed]
- Gerhardt, R.R. Culicoides spp. Attracted to Ruminants in the Great Smoky Mountains National Park, Tennessee. J. Agric. Entomol. 1986, 3, 192–197. [Google Scholar]
- Smith, K.E.; Stallknecht, D.E.; Nettles, V.F. Experimental Infection of Culicoides lahillei (Diptera: Ceratopogonidae) with Epizootic Hemorrhagic Disease Virus Serotype 2 (Orbivirus: Reoviridae). J. Med. Entomol. 1996, 33, 117–122. [Google Scholar] [CrossRef]
- Smith, K.E.; Stalknecht, D.E.; Sewell, C.T.; Rollor, E.A.; Mullen, G.R.; Anderson, R.R. Monitoring of Culicoides spp. at a site enzootic for hemorrhagic disease in white-tailed deer in Georgia, USA. J. Wildl. Dis. 1996, 32, 627–642. [Google Scholar] [CrossRef]
- McGregor, B.L.; Sloyer, K.E.; Sayler, K.A.; Goodfriend, O.; Krauer, J.M.C.; Acevedo, C.; Zhang, X.; Mathias, D.; Wisely, S.M.; Burkett-Cadena, N.D. Field data implicating Culicoides stellifer and Culicoides venustus (Diptera: Ceratopogonidae) as vectors of epizootic hemorrhagic disease virus. Parasit. Vectors 2019, 12, 258. [Google Scholar] [CrossRef]
- McGregor, B.L.; Stenn, T.; Sayler, K.A.; Blosser, E.M.; Blackburn, J.K.; Wisely, S.M.; Burkett-Cadena, N.D. Host use patterns of Culicoides spp. biting midges at a big game preserve in Florida, U.S.A., and implications for the transmission of orbiviruses. Med. Vet. Entomol. 2019, 33, 110–120. [Google Scholar] [CrossRef] [Green Version]
- Sloyer, K.E.; Acevedo, C.; Runkel, A.E.; Burkett-Cadena, N.D. Host Associations of Biting Midges (Diptera: Ceratopogonidae: Culicoides) Near Sentinel Chicken Surveillance Locations in Florida, USA. J. Am. Mosq. Control Assoc. 2019, 35, 200–206. [Google Scholar] [CrossRef]
- Greiner, E.C.; Barber, T.L.; Pearson, J.E.; Kramer, W.L.; Gibbs, E.P. Orbiviruses from Culicoides in Florida. Prog. Clin. Biol. Res. 1985, 178, 195. [Google Scholar]
- Becker, M.E.; Reeves, W.K.; Dejean, S.K.; Emery, M.P.; Ostlund, E.N.; Foil, L.D. Detection of Bluetongue Virus RNA in Field-Collected Culicoides spp. (Diptera: Ceratopogonidae) Following the Discovery of Bluetongue Virus Serotype 1 in White-Tailed Deer and Cattle in Louisiana. J. Med. Entomol. 2010, 47, 269–273. [Google Scholar] [CrossRef]
- Jones, R.H.; Schmidtmann, E.T.; Foster, N.M. Vector-competence studies for bluetongue and epizootic hemorrhagic disease viruses with Culicoides venustus (Ceratopogonidae). Mosq. News 1983, 43, 184–186. [Google Scholar]
- Venter, G.J.; Paweska, J.T.; Lunt, H.; Mellor, P.S.; Carpenter, S. An alternative method of blood-feeding Culicoides imicola and other haematophagous Culicoides species for vector competence studies. Vet. Parasitol. 2005, 131, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Couvillion, C.E.; Nettles, V.F.; Davidson, W.R.; Pearson, J.E.; Gustafson, G.A. Hemorrhagic disease among white-tailed deer in the Southeast from 1971 through 1980. In Proceedings of the 85th annual meeting of the United States Animal Health Association, St. Louis, MO, USA, 11–16 October 1981; pp. 522–537. [Google Scholar]
- Stallknecht, D.E.; Howerth, E.W.; Gaydos, J.K. Hemorrhagic disease in white-tailed deer: Our current understanding of risk. In Proceedings of the Transactions of the North American Wildlife and Natural Resources Conference, Dallas, TX, USA, 3–7 April 2002; Volume 67, pp. 75–86. [Google Scholar]
- Sayler, K.; Blosser, E.; McGregor, B.; Burkett-Cadena, N.; Wisely, S.M. Overwintering of epizootic hemorrhagic disease virus in white-tailed deer in Florida, USA: Unanticipated seroconversion and the case for alternative vectors. Int. J. Infect. Dis. 2016, 53, 65–66. [Google Scholar] [CrossRef] [Green Version]
- Khalaf, K.T. Distribution and phenology of Culicoides (Diptera: Ceratopogonidae) along the Gulf of Mexico. J. Econ. Entomol. 1969, 62, 1153–1161. [Google Scholar] [CrossRef]
- Zimmerman, R.H.; Turner, E.C., Jr. Seasonal abundance and parity of common Culicoides collected in blacklight traps in Virginia pastures. Mosq. News 1983, 43, 63–69. [Google Scholar]
- Pfannenstiel, R.S.; Mullens, B.A.; Ruder, M.G.; Zurek, L.; Cohnstaedt, L.W.; Nayduch, D. Management of North American Culicoides Biting Midges: Current Knowledge and Research Needs. Vector-Borne Zoonotic Dis. 2015, 15, 374–384. [Google Scholar] [CrossRef]
- Drolet, B.S.; Van Rijn, P.; Howerth, E.W.; Beer, M.; Mertens, P.P. A Review of Knowledge Gaps and Tools for Orbivirus Research. Vector-Borne Zoonotic Dis. 2015, 15, 339–347. [Google Scholar] [CrossRef]
- McGregor, B.L.; Runkel, A.E.; Wisely, S.M.; Burkett-Cadena, N.D. Vertical stratification of Culicoides biting midges at a Florida big game preserve. Parasit. Vectors 2018, 11, 505. [Google Scholar] [CrossRef] [Green Version]
- Dinh, E.T.N.; Cauvin, A.; Orange, J.P.; Shuman, R.M.; Wisely, S.M.; Blackburn, J.K. Living la Vida T-LoCoH: Site fidelity of Florida ranched and wild white-tailed deer (Odocoileus virginianus) during the epizootic hemorrhagic disease virus (EHDV) transmission period. Mov. Ecol. 2020, 8, 1–9. [Google Scholar] [CrossRef]
- Sloyer, K.E.; Wisely, S.M.; Burkett-Cadena, N.D. Effects of ultraviolet LED versus incandescent bulb and carbon dioxide for sampling abundance and diversity of Culicoides in Florida. J. Med. Entomol. 2019, 56, 353–361. [Google Scholar] [CrossRef]
- Dyce, A.L. the Recognition of Nulliparous and Parous Culicoides (Diptera: Ceratopogonidae) Without Dissection. Aust. J. Entomol. 1969, 8, 11–15. [Google Scholar] [CrossRef]
- Akey, D.H.; Potter, H.W. Pigmentation associated with oogenesis in the biting fly Culicoides variipennis (Diptera: Ceratopogonidae): Determination of parity. J. Med. Entomol. 1979, 16, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Data Access National Centers for Environmental Information (NCEI) Formerly Known as National Climatic Data Center (NCDC). Available online: https://www.ncdc.noaa.gov/data-access (accessed on 19 June 2020).
- USGS Surface-Water Data for the Nation. Available online: https://waterdata.usgs.gov/nwis/sw (accessed on 19 June 2020).
- Peres-Neto, P.R.; Legendre, P.; Dray, S.; Borcard, D. Variation partitioning of species data matrices: Estimation and comparison of fractions. Ecology 2006, 87, 2614–2625. [Google Scholar] [CrossRef]
- Legendre, P.; Borcard, D.; Roberts, D.W. Variation partitioning involving orthogonal spatial eigenfunction submodels. Ecology 2012, 93, 1234–1240. [Google Scholar] [CrossRef] [Green Version]
- Legendre, P. Variation partitioning of a community matrix by 2, 3, or 4 explanatory matrices containing environmental variables and spatial eigenfunction submodels (R Doc in supplement 1). Ecology 2012, 93, 1234–1240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanchet, F.G.; Legendre, P.; Borcard, D. Modelling directional spatial processes in ecological data. Ecol. Modell. 2008, 215, 325–336. [Google Scholar] [CrossRef]
- Dray, S.; Pélissier, R.; Couteron, P.; Fortin, M.J.; Legendre, P.; Peres-Neto, P.R.; Bellier, E.; Bivand, R.; Blanchet, F.G.; De Cáceres, M.; et al. Community ecology in the age of multivariate multiscale spatial analysis. Ecol. Monogr. 2012, 82, 257–275. [Google Scholar] [CrossRef]
- Borcard, D.; Gillet, F.; Legendre, P. Spatial analysis of ecological data. In Numerical Ecology with R; Springer: Berlin/Heidelberg, Germany, 2018; pp. 299–367. [Google Scholar]
- Blanchet, F.G.; Legendre, P.; Borcard, D. Forward selection of explanatory variables. Ecology 2008, 89, 2623–2632. [Google Scholar] [CrossRef]
- Legendre, P.; De Cáceres, M. Beta diversity as the variance of community data: Dissimilarity coefficients and partitioning. Ecol. Lett. 2013, 16, 951–963. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package. R Package Version 1.17-4. 2010. Available online: http//CRAN.R-project.org/package=vegan (accessed on 10 March 2020).
- Dray, S.; Blanchet, G.; Borcard, D.; Clappe, S.; Guenard, G.; Jombart, T. Adespatial: Multivariate multiscale spatial analysis. R package ver. 0.1-1 2018. Available online: https://cran.r-project.org/web/packages/adespatial/index.html (accessed on 10 March 2020).
- Chao, A.; Gotelli, N.J.; Hsieh, T.C.; Sander, E.L.; Ma, K.H.; Colwell, R.K.; Ellison, A.M. Rarefaction and extrapolation with Hill numbers: A framework for sampling and estimation in species diversity studies. Ecol. Monogr. 2014, 84, 45–67. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, T.C.; Ma, K.H.; Chao, A. iNEXT: An R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evol. 2016, 7, 1451–1456. [Google Scholar] [CrossRef]
- Ulrich, W.; Almeida-Neto, M.; Gotelli, N.J. A consumer’s guide to nestedness analysis. Oikos 2009, 118, 3–17. [Google Scholar] [CrossRef]
- Patterson, B.D.; Atmar, W. Nested subsets and the structure of insular mammalian faunas and archipelagos. Biol. J. Linn. Soc. 1986, 28, 65–82. [Google Scholar] [CrossRef]
- Almeida-Neto, M.; Guimarães, P.; Guimarães, P.R.; Loyola, R.D.; Ulrich, W. A consistent metric for nestedness analysis in ecological systems: Reconciling concept and measurement. Oikos 2008, 117, 1227–1239. [Google Scholar] [CrossRef]
- Lane, P.W.; Lindenmayer, D.B.; Barton, P.S.; Blanchard, W.; Westgate, M.J. Visualization of species pairwise associations: A case study of surrogacy in bird assemblages. Ecol. Evol. 2014, 4, 3279–3289. [Google Scholar] [CrossRef]
- Westgate, M.J.; Lane, P.W. Sppairs: Species pairwise association analysis in R.–Version 0.2 2015. Available online: https://github.com/mjwestgate/sppairs (accessed on 10 March 2020).
- Newman, M.E.J. Modularity and community structure in networks. Proc. Natl. Acad. Sci. USA 2006, 103, 8577–8582. [Google Scholar] [CrossRef] [Green Version]
- Newman, M.E.J.; Girvan, M. Finding and evaluating community structure in networks. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2004, 69, 026113. [Google Scholar] [CrossRef] [Green Version]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. InterJ. Complex Syst. 2006, 1695. [Google Scholar]
- Guimerà, R.; Amaral, L.A.N. Functional cartography of complex metabolic networks. Nature 2005, 433, 895–900. [Google Scholar] [CrossRef] [Green Version]
- Dormann, C.; Gruber, B.; Fründ, J. Introducing the bipartite package: Analysing ecological networks. Interaction 2008, 8, 8–11. [Google Scholar]
- Dáttilo, W.; Lara-Rodríguez, N.; Jordano, P.; Guimarães, P.R.; Thompson, J.N.; Marquis, R.J.; Medeiros, L.P.; Ortiz-Pulido, R.; Marcos-García, M.A.; Rico-Gray, V. Unravelling darwin’s entangled bank: Architecture and robustness of mutualistic networks with multiple interaction types. Proc. R. Soc. B Biol. Sci. 2016, 283, 20161564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De’ath, G. Multivariate Regression Trees: A New Technique for Modelling. Ecology 2002, 83, 1105–1117. [Google Scholar] [CrossRef]
- De’ath, G.; Therneau, T.M.; Atkinson, B.; Ripley, B.; Oksanen, J. Mvpart: Multivariate partitioning. R Package. version 1.6-1. 2013. Available online: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.398.6589&rep=rep1&type=pdf (accessed on 10 March 2020).
- Dufrêne, M.; Legendre, P. Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol. Monogr. 1997, 67, 345–366. [Google Scholar] [CrossRef]
- Westgate, M.J. Circleplot: Circular plots of distance and association matrices. R Package. version 0.4. 2016. Available online: https://github.com/mjwestgate/circleplot (accessed on 10 March 2020).
- Aybar, C.A.V.; Juri, M.J.D.; de Grosso, M.S.L.; Spinelli, G.R. Spatial and Temporal Distribution of Culicoides insignis and Culicoides paraensis in the Subtropical Mountain Forest of Tucumán, Northwestern Argentina. Fla. Entomol. 2011, 94, 1018–1025. [Google Scholar] [CrossRef]
- Silva, F.S.; Carvalho, L.P.C. A Population Study of the Culicoides Biting Midges (Diptera: Ceratopogonidae) in Urban, Rural, and Forested Sites in a Cerrado Area of Northeastern Brazil. Ann. Entomol. Soc. Am. 2013, 106, 463–470. [Google Scholar] [CrossRef]
- Diarra, M.; Fall, M.; Fall, A.G.; Diop, A.; Seck, M.T.; Garros, C.; Balenghien, T.; Allène, X.; Rakotoarivony, I.; Lancelot, R.; et al. Seasonal dynamics of Culicoides (Diptera: Ceratopogonidae) biting midges, potential vectors of African horse sickness and bluetongue viruses in the Niayes area of Senegal. Parasit. Vectors 2014, 7, 147. [Google Scholar] [CrossRef] [Green Version]
- Kraft, N.J.B.; Adler, P.B.; Godoy, O.; James, E.C.; Fuller, S.; Levine, J.M. Community assembly, coexistence and the environmental filtering metaphor. Funct. Ecol. 2015, 29, 592–599. [Google Scholar] [CrossRef]
- Gerry, A.C.; Mullens, B.A. Seasonal Abundance and Survivorship of Culicoides sonorensis (Diptera: Ceratopogonidae) at a Southern California Dairy, with Reference to Potential Bluetongue Virus Transmission and Persistence. J. Med. Entomol. 2009, 37, 675–688. [Google Scholar] [CrossRef]
- Scolamacchia, F.; Van Den Broek, J.; Meiswinkel, R.; Heesterbeek, J.A.P.; Elbers, A.R.W. Principal climatic and edaphic determinants of Culicoides biting midge abundance during the 2007-2008 bluetongue epidemic in the Netherlands, based on OVI light trap data. Med. Vet. Entomol. 2014, 28, 143–156. [Google Scholar] [CrossRef]
- Diarra, M.; Fall, M.; Lancelot, R.; Diop, A.; Fall, A.G.; Dicko, A.; Seck, M.T.; Garros, C.; Allène, X.; Rakotoarivony, I.; et al. Modelling the abundances of two major Culicoides (Diptera: Ceratopogonidae) species in the Niayes area of senegal. PLoS ONE 2015, 10, e0131021. [Google Scholar] [CrossRef] [Green Version]
- Lysyk, T.J.; Danyk, T. Effect of Temperature on Life History Parameters of Adult Culicoides sonorensis (Diptera: Ceratopogonidae) in Relation to Geographic Origin and Vectorial Capacity for Bluetongue Virus. J. Med. Entomol. 2007, 44, 741–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veronesi, E.; Venter, G.J.; Labuschagne, K.; Mellor, P.S.; Carpenter, S. Life-history parameters of Culicoides (Avaritia) imicola Kieffer in the laboratory at different rearing temperatures. Vet. Parasitol. 2009, 163, 370–373. [Google Scholar] [CrossRef] [PubMed]
- Verhoef, F.A.; Venter, G.J.; Weldon, C.W. Thermal limits of two biting midges, Culicoides imicola Kieffer and C. bolitinos Meiswinkel (Diptera: Ceratopogonidae). Parasit. Vectors 2014, 7, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wittmann, E.J.; Mellor, P.S.; Baylis, M. Effect of temperature on the transmission of orbiviruses by the biting midge, Culicoides sonorensis. Med. Vet. Entomol. 2002, 16, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Mullens, B.A.; Gerry, A.C.; Lysyk, T.J.; Schmidtmann, E. Environmental effects on vector competence and virogenesis of bluetongue virus in Culicoides: Interpreting laboratory data in a field context. Vet. Ital. 2004, 40, 160. [Google Scholar] [PubMed]
- Gubbins, S.; Carpenter, S.; Baylis, M.; Wood, J.L.N.; Mellor, P.S. Assessing the risk of bluetongue to UK livestock: Uncertainty and sensitivity analyses of a temperature-dependent model for the basic reproduction number. J. R. Soc. Interface 2008, 5, 363–371. [Google Scholar] [CrossRef] [Green Version]
- Ruder, M.G.; Stallknecht, D.E.; Howerth, E.W.; Carter, D.L.; Pfannenstiel, R.S.; Allison, A.B.; Mead, D.G. Effect of Temperature on Replication of Epizootic Hemorrhagic Disease Viruses in Culicoides sonorensis (Diptera: Ceratopogonidae). J. Med. Entomol. 2015, 52, 1050–1059. [Google Scholar] [CrossRef] [Green Version]
- Blackwell, A.; Lock, K.A.; Marshall, B.; Boag, B.; Gordon, S.C. The spatial distribution of larvae of Culicoides impunctatus biting midges. Med. Vet. Entomol. 1999, 13, 362–371. [Google Scholar] [CrossRef]
- Harrup, L.E.; Purse, B.V.; Golding, N.; Mellor, P.S.; Carpenter, S. Larval development and emergence sites of farm-associated Culicoides in the United Kingdom. Med. Vet. Entomol. 2013, 27, 441–449. [Google Scholar] [CrossRef]
- Erram, D.; Burkett-Cadena, N. Laboratory Rearing of Culicoides stellifer (Diptera: Ceratopogonidae), a Suspected Vector of Orbiviruses in the United States. J. Med. Entomol. 2020, 57, 25–32. [Google Scholar] [CrossRef]
- Erram, D.; Blosser, E.M.; Burkett-Cadena, N. Habitat associations of Culicoides species (Diptera: Ceratopogonidae) abundant on a commercial cervid farm in Florida, USA. Parasit. Vectors 2019, 12, 367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mellor, P.S.; Boorman, J.; Baylis, M. Culicoides Biting Midges: Their Role as Arbovirus Vectors. Annu. Rev. Entomol. 2000, 45, 307–340. [Google Scholar] [CrossRef] [PubMed]
- Veggiani Aybar, C.A.; Dantur Juri, M.J.; Lizarralde De Grosso, M.S.; Spinelli, G.R. Species diversity and seasonal abundance of Culicoides biting midges in northwestern Argentina. Med. Vet. Entomol. 2010, 24, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Searle, K.R.; Barber, J.; Stubbins, F.; Labuschagne, K.; Carpenter, S.; Butler, A.; Denison, E.; Sers, C.; Mellor, P.S.; Wilson, A.; et al. Environmental drivers of Culicoides phenology: How important is species-specific variation when determining disease policy? PLoS ONE 2014, 9, e111876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourquia, M.; Garros, C.; Rakotoarivony, I.; Boukhari, I.; Chakrani, M.; Huber, K.; Gardès, L.; Wint, W.; Baldet, T.; Khallaayoune, K.; et al. Composition and seasonality of Culicoides in three host environments in Rabat region (Morocco). Rev. D’élevage Médecine Vétérinaire des Pays Trop. 2020, 73, 37–46. [Google Scholar] [CrossRef]
- Cuéllar, A.C.; Kjær, L.J.; Baum, A.; Stockmarr, A.; Skovgard, H.; Nielsen, S.A.; Andersson, M.G.; Lindström, A.; Chirico, J.; Lühken, R.; et al. Modelling the monthly abundance of Culicoides biting midges in nine European countries using Random Forests machine learning. Parasit. Vectors 2020, 13, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Christensen, S.A.; Ruder, M.G.; Williams, D.M.; Porter, W.F.; Stallknecht, D.E. The role of drought as a determinant of hemorrhagic disease in the eastern United States. Glob. Chang. Biol. 2020, 26, 3799–3808. [Google Scholar] [CrossRef]
- Xu, B.; Madden, M.; Stallknecht, D.E.; Hodler, T.W.; Parker, K.C. Spatial-temporal model of haemorrhagic disease in white-tailed deer in south-east USA, 1983 to 2000. Vet. Rec. 2012, 170, 288. [Google Scholar] [CrossRef]
- Kurushima, H.; Yoshimura, J.; Kim, J.K.; Kim, J.K.; Nishimoto, Y.; Sayama, K.; Kato, M.; Watanabe, K.; Hasegawa, E.; Roff, D.A.; et al. Co-occurrence of ecologically equivalent cryptic species of spider wasps. R. Soc. Open Sci. 2016, 3, 160119. [Google Scholar] [CrossRef] [Green Version]
- Gerry, A.C.; Monteys, V.S.I.; Vidal, J.-O.M.; Francino, O.; Mullens, B.A. Biting Rates of Culicoides Midges (Diptera: Ceratopogonidae) on Sheep in Northeastern Spain in Relation to Midge Capture Using UV Light and Carbon Dioxide-Baited Traps. J. Med. Entomol. 2009, 46, 615–624. [Google Scholar] [CrossRef]
- Mayo, C.E.; Mullens, B.A.; Reisen, W.K.; Osborne, C.J.; Gibbs, E.P.J.; Gardner, I.A.; MacLachlan, N.J. Seasonal and interseasonal dynamics of bluetongue virus infection of dairy cattle and Culicoides sonorensis midges in northern California—Implications for virus overwintering in temperate zones. PLoS ONE 2014, 9, e106975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mullens, B.A.; McDermott, E.G.; Gerry, A.C. Progressi e lacune nella conoscenza dell’ecologia di Culicoides e del relativo controllo. Vet. Ital. 2015, 51, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.; Darpel, K.; Mellor, P.S. Where does bluetongue virus sleep in the winter? PLoS Biol. 2008, 6, e210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruder, M.G.; Allison, A.B.; Stallknecht, D.E.; Mead, D.G.; Mcgraw, S.M.; Carter, D.L.; Kubiski, S.V.; Batten, C.A.; Klement, E.; Howerth, E.W. Susceptibility of white-tailed deer (Odocoileus virginianus) to experimental infection with epizootic hemorrhagic disease virus serotype 7. J. Wildl. Dis. 2012, 48, 676–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, L.L.; Ruder, M.G.; Kienzle-Dean, C.; Carter, D.; Stallknecht, D.; Howerth, E.W. Experimental infection of white-tailed deer (Odocoileus Virginianus) with bluetongue virus serotype 3. J. Wildl. Dis. 2019, 55, 627–636. [Google Scholar] [CrossRef]
- CPC—Climate Weather Linkage: El Niño Southern Oscillation. Available online: https://www.cpc.ncep.noaa.gov/products/precip/CWlink/MJO/enso.shtml#history (accessed on 19 June 2020).
- Baylis, M.; Mellor, P.S.; Meiswinkel, R. Horse sickness and ENSO in South Africa. Nature 1999, 397, 574. [Google Scholar] [CrossRef] [PubMed]
- Baygents, G.; Bani-Yaghoub, M. Cluster analysis of hemorrhagic disease in Missouri’s white-tailed deer population: 1980–2013. BMC Ecol. 2018, 18, 35. [Google Scholar] [CrossRef] [Green Version]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quaglia, A.I.; Blosser, E.M.; McGregor, B.L.; Runkel, A.E., IV; Sloyer, K.E.; Erram, D.; Wisely, S.M.; Burkett-Cadena, N.D. Tracking Community Timing: Pattern and Determinants of Seasonality in Culicoides (Diptera: Ceratopogonidae) in Northern Florida. Viruses 2020, 12, 931. https://doi.org/10.3390/v12090931
Quaglia AI, Blosser EM, McGregor BL, Runkel AE IV, Sloyer KE, Erram D, Wisely SM, Burkett-Cadena ND. Tracking Community Timing: Pattern and Determinants of Seasonality in Culicoides (Diptera: Ceratopogonidae) in Northern Florida. Viruses. 2020; 12(9):931. https://doi.org/10.3390/v12090931
Chicago/Turabian StyleQuaglia, Agustin I., Erik M. Blosser, Bethany L. McGregor, Alfred E. Runkel, IV, Kristin E. Sloyer, Dinesh Erram, Samantha M. Wisely, and Nathan D. Burkett-Cadena. 2020. "Tracking Community Timing: Pattern and Determinants of Seasonality in Culicoides (Diptera: Ceratopogonidae) in Northern Florida" Viruses 12, no. 9: 931. https://doi.org/10.3390/v12090931






