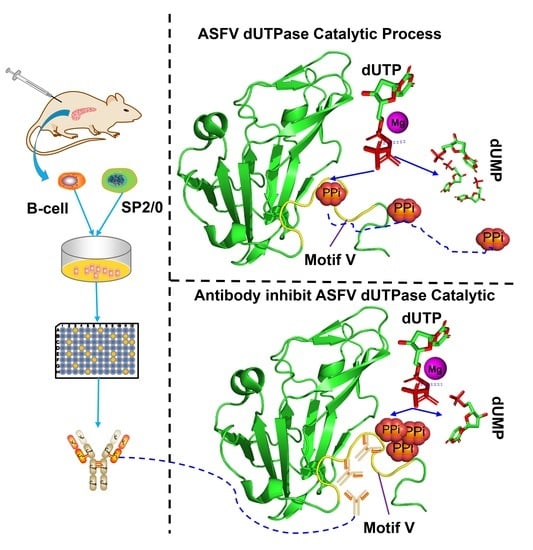
Identification and Characterization of a Novel Epitope of ASFV-Encoded dUTPase by Monoclonal Antibodies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Recombinant Plasmid Constructs for Protein Expression and Purification
2.2. Enzyme Kinetics Measurement
2.3. mAbs Production and Purification
2.4. Indirect ELISAs
2.5. Western Blotting Assay
2.6. Immunofluorescence Assay (IFA)
2.7. Epitope Mapping of E165R
2.8. Testing of Inhibitory Antibody
2.9. Monoclonal Antibody Isotyping
2.10. Sequence Alignment of Classical dUTPases
2.11. Structure Analysis and Specificity Verification of Inhibitory Antibody-Recognized Epitope
2.12. Data Analysis
3. Results
3.1. Expression and Purification of the Recombinant E165R Protein
3.2. Production and Characterization of Anti-E165R mAbs
3.3. Identification of Epitopes Recognized by the mAbs
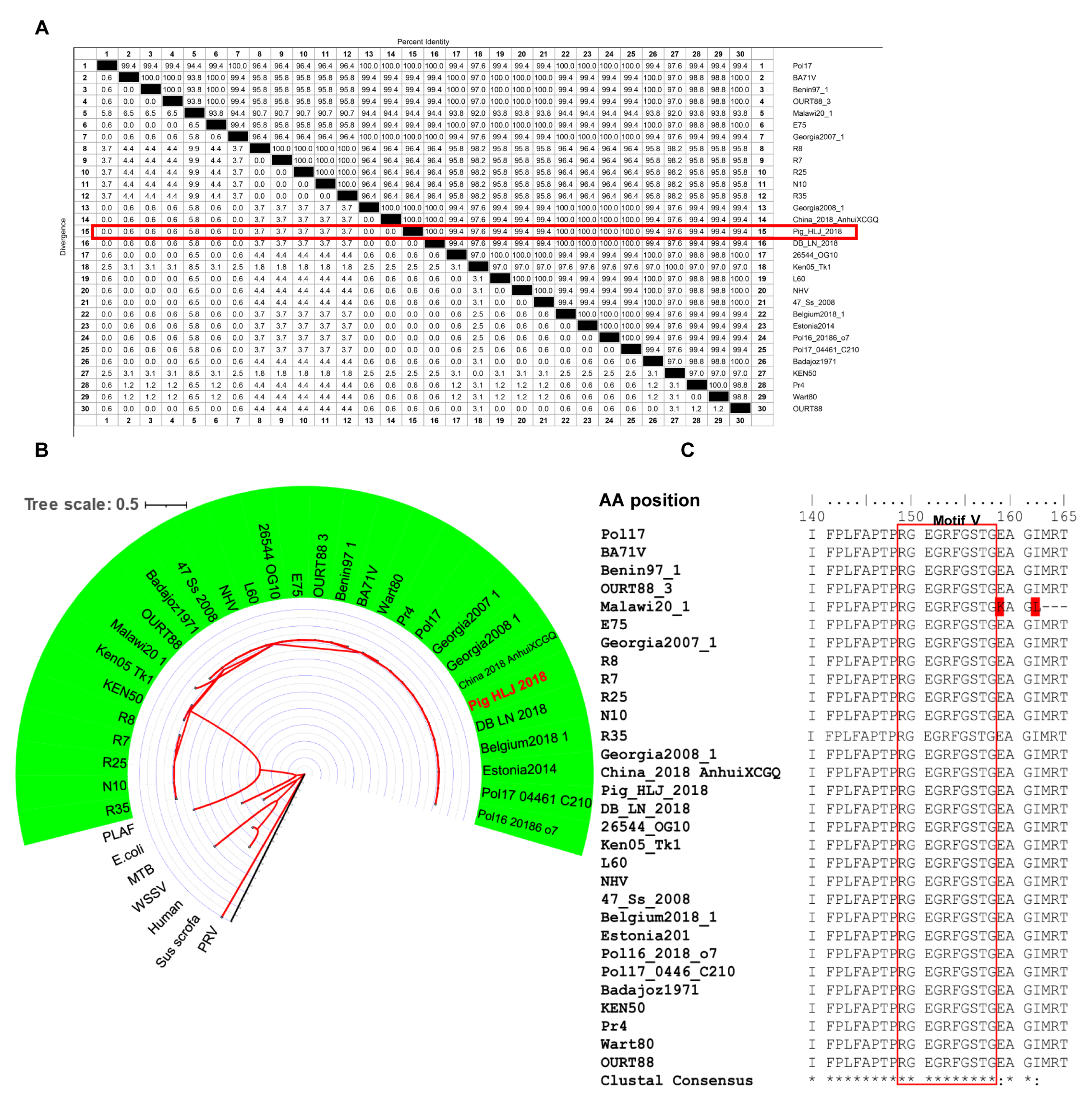
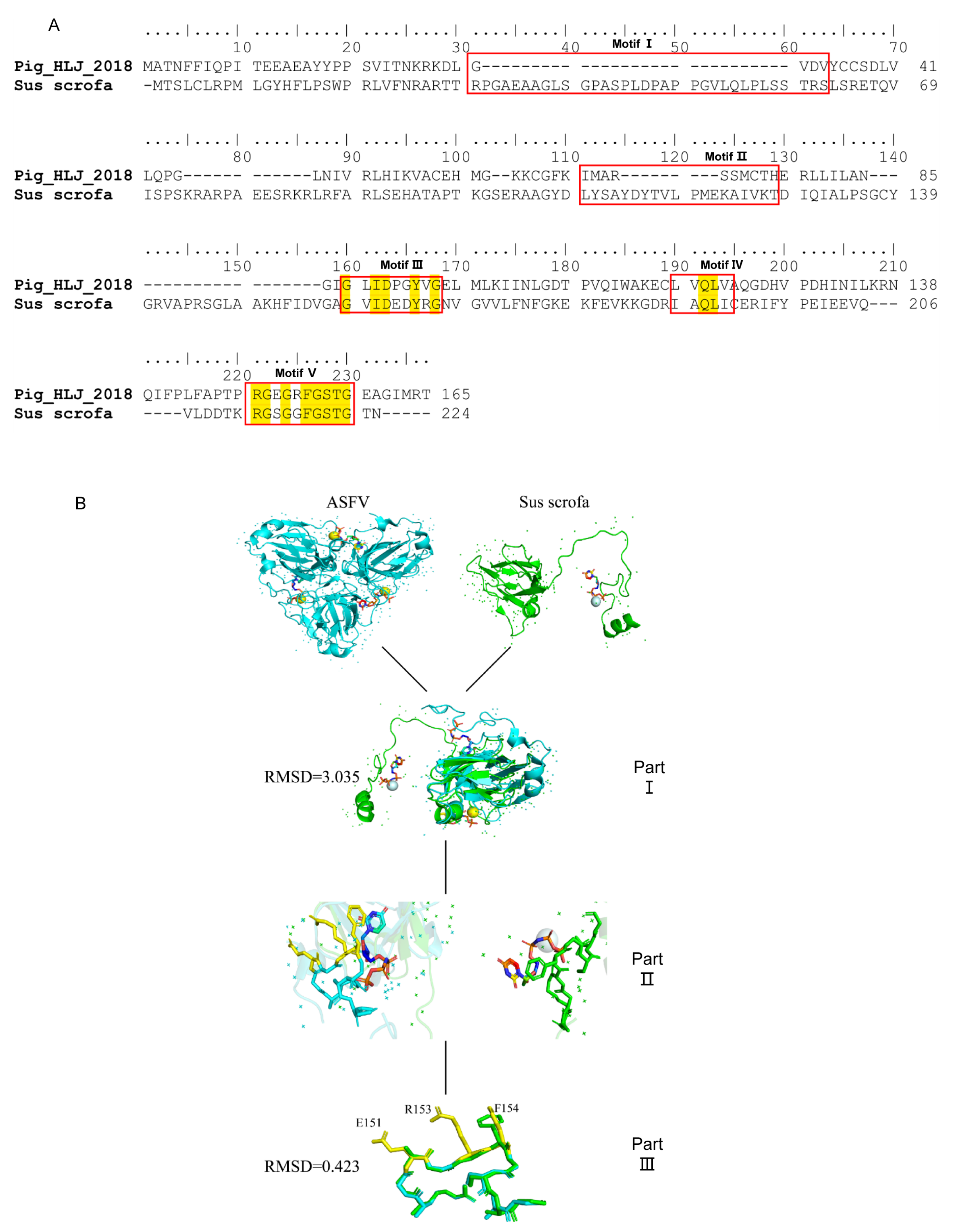
3.4. Conservation of Epitopes Recognized by mAbs
3.5. The Epitope on ASFV dUTPase Was Specifically Recognized by 8F9 mAb
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karger, A.; Perez-Nunez, D.; Urquiza, J.; Hinojar, P.; Alonso, C.; Freitas, F.B.; Revilla, Y.; Le Potier, M.F.; Montoya, M. An Update on African Swine Fever Virology. Viruses 2019, 11, 864. [Google Scholar] [CrossRef] [Green Version]
- Ge, S.; Li, J.; Fan, X.; Liu, F.; Li, L.; Wang, Q.; Ren, W.; Bao, J.; Liu, C.; Wang, H. Molecular Characterization of African Swine Fever Virus, China, 2018. Emerg. Infect. Dis. 2018, 24, 2131–2133. [Google Scholar] [CrossRef] [Green Version]
- Teklue, T.; Sun, Y.; Abid, M.; Luo, Y.; Qiu, H.J. Current status and evolving approaches to African swine fever vaccine de-velopment. Transbound. Emerg. Dis. 2020, 67, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Dixon, L.K.; Sun, H.; Roberts, H. African swine fever. Antivir. Res. 2019, 165, 34–41. [Google Scholar] [CrossRef]
- Wang, N.; Zhao, D.; Wang, J.; Zhang, Y.; Wang, M.; Gao, Y.; Li, F.; Wang, J.; Bu, Z.; Rao, Z. Architecture of African swine fever virus and implications for viral assembly. Science 2019, 366, 640–644. [Google Scholar] [CrossRef]
- Liu, S.; Luo, Y.; Wang, Y.; Li, S.; Zhao, Z.; Bi, Y.; Sun, J.; Peng, R.; Song, H.; Zhu, D. Cryo-EM Structure of the African Swine Fever Virus. Cell Host Microbe 2019, 26, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Wang, Q.; Lin, P.; Liu, C.; Li, L.; Wu, X.; Chi, T.; Xu, T.; Ge, S.; Liu, Y. Genome comparison of African swine fever virus China/2018/AnhuiXCGQ strain and related European p72 GenotypeII strains. Transbound. Emerg. Dis. 2019, 66, 1167–1176. [Google Scholar] [CrossRef]
- Alejo, A.; Matamoros, T.; Guerra, M.; Andrés, G. A Proteomic Atlas of the African Swine Fever Virus Particle. J. Virol. 2018, 92, e01293-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Chai, Y.; Song, H.; Weng, C.; Qi, J.; Sun, Y.; Gao, G.F. Crystal Structure of African Swine Fever Virus dUTPase Reveals a Potential Drug Target. mBio 2019, 10, e02483-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Wang, C.; Yang, M.; Cao, L.; Fu, D.; Liu, X.; Sun, D.; Chen, C.; Wang, Y.; Jia, Z. Structural Insight into African Swine Fever Virus dUTPase Reveals a Novel Folding Pattern in the dUTPase Family. J. Virol. 2020, 94, e01698-19. [Google Scholar] [CrossRef]
- Oliveros, M.; Garcia-Escudero, R.; Alejo, A.; Vinuela, E.; Salas, M.L.; Salas, J. African swine fever virus dUTPase is a highly specific enzyme required for efficient replication in swine macrophages. J. Virol. 1999, 73, 8934–8943. [Google Scholar] [CrossRef] [Green Version]
- Mol, C.D.; Harris, J.M.; McIntosh, E.M.; Tainer, J.A. Human dUTP pyrophosphatase: Uracil recognition by a beta hairpin and active sites formed by three separate subunits. Structure 1996, 4, 1077–1092. [Google Scholar] [CrossRef] [Green Version]
- Cedergren-Zeppezauer, E.S.; Larsson, G.; Nyman, P.O.; Dauter, Z.; Wilson, K.S. Crystal structure of a dUTPase. Nature 1992, 355, 740–743. [Google Scholar] [CrossRef]
- Prasad, G.S.; Stura, E.A.; McRee, D.E.; Laco, G.S.; Hasselkus-Light, C.; Elder, J.H.; Stout, C.D. Crystal structure of dUTP pyrophosphatase from feline immunodeficiency virus. Protein Sci. 1996, 5, 2429–2437. [Google Scholar] [CrossRef] [Green Version]
- Dauter, Z.; Persson, R.; Rosengren, A.M.; Nyman, P.O.; Wilson, K.S.; Cedergren-Zeppezauer, E.S. Crystal structure of dUTPase from equine infectious anaemia virus; active site metal binding in a substrate analogue complex. J. Mol. Biol. 1999, 285, 655–673. [Google Scholar] [CrossRef] [Green Version]
- Chan, S.; Segelke, B.; Lekin, T.; Krupka, H.; Cho, U.S.; Kim, M.Y.; So, M.; Kim, C.Y.; Naranjo, C.M.; Rogers, Y.C. Crystal structure of the Mycobacterium tuberculosis dUTPase: Insights into the catalytic mechanism. J. Mol. Biol. 2004, 341, 503–517. [Google Scholar] [CrossRef]
- Zang, K.; Li, F.; Ma, Q. The dUTPase of white spot syndrome virus assembles its active sites in a noncanonical manner. J. Biol. Chem. 2018, 293, 1088–1099. [Google Scholar] [CrossRef] [Green Version]
- Liang, R.; Wang, G.; Zhang, D.; Ye, G.; Li, M.; Shi, Y.; Shi, J.; Chen, H.; Peng, G. Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development. J. Biol. Chem. 2020, 296, 100015. [Google Scholar] [CrossRef]
- Vertessy, B.G.; Persson, R.; Rosengren, A.M.; Zeppezauer, M.; Nyman, P.O. Specific derivatization of the active site tyrosine in dUTPase perturbs ligand binding to the active site. Biochem. Biophys. Res. Commun. 1996, 219, 294–300. [Google Scholar] [CrossRef]
- Larsson, G.; Nyman, P.O.; Kvassman, J.O. Kinetic characterization of dUTPase from Escherichia coli. J. Biol. Chem. 1996, 271, 24010–24016. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.K.; Liu, J.M.; Wang, L.X.; Fan, S.Q.; Li, Z.Y.; Li, Y.W.; Yi, L.; Ding, H.X.; Zhao, M.Q.; Chen, J.D. Current State of Global African Swine Fever Vaccine Development under the Prevalence and Transmission of ASF in China. Vaccines 2020, 8, 531. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xu, A.; Qin, C.; Zhang, Q.; Chen, S.; Lang, Y.; Wang, M.; Li, C.; Feng, W.; Zhang, R. Pseudorabies Virus dUTPase UL50 Induces Lysosomal Degradation of Type I Interferon Receptor 1 and Antagonizes the Alpha Interferon Re-sponse. J. Virol. 2017, 91, e01148-17. [Google Scholar] [CrossRef] [Green Version]
- Jons, A.; Mettenleiter, T.C. Identification and characterization of pseudorabies virus dUTPase. J. Virol. 1996, 70, 1242–1245. [Google Scholar] [CrossRef] [Green Version]
- Ariza, M.E.; Glaser, R.; Kaumaya, P.T.; Jones, C.; Williams, M.V. The EBV-encoded dUTPase activates NF-kappa B through the TLR2 and MyD88-dependent signaling pathway. J. Immunol. 2009, 182, 851–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams Ph, D.M.; Cox, B.; Lafuse Ph, D.W.; Ariza, M.E. Epstein-Barr Virus dUTPase Induces Neuroinflammatory Media-tors: Implications for Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Clin. Ther. 2019, 41, 848–863. [Google Scholar] [CrossRef] [Green Version]
- Kato, A.; Adachi, S.; Kawano, S.; Takeshima, K.; Watanabe, M.; Kitazume, S.; Sato, R.; Kusano, H.; Koyanagi, N.; Maruzuru, Y. Identification of a herpes simplex virus 1 gene encoding neurovirulence factor by chemical proteomics. Nat. Commun. 2020, 11, 4894. [Google Scholar] [CrossRef]
- Pavlova, S.P.; Veits, J.; Mettenleiter, T.C.; Fuchs, W. Live vaccination with an H5-hemagglutinin-expressing infectious lar-yngotracheitis virus recombinant protects chickens against different highly pathogenic avian influenza viruses of the H5 subtype. Vaccine 2009, 27, 5085–5090. [Google Scholar] [CrossRef] [PubMed]
- Yokogawa, T.; Yano, W.; Tsukioka, S.; Osada, A.; Wakasa, T.; Ueno, H.; Hoshino, T.; Yamamura, K.; Fujioka, A.; Fukuoka, M. dUTPase inhibition confers susceptibility to a thymidylate synthase inhibitor in DNA-repair-defective human cancer cells. Cancer Sci. 2021, 112, 422–432. [Google Scholar] [CrossRef]
- Yamamoto, N.; Hayashi, H.; Planchard, D.; Moran, T.; Gregorc, V.; Dowell, J.; Sakai, H.; Yoh, K.; Nishio, M.; Cortot, A.B. A randomized, phase 2 study of deoxyuridine triphosphatase inhibitor, TAS-114, in combination with S-1 versus S-1 alone in patients with advanced non-small-cell lung cancer. Investig. New Drugs 2020, 38, 1588–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doi, T.; Yoh, K.; Shitara, K.; Takahashi, H.; Ueno, M.; Kobayashi, S.; Morimoto, M.; Okusaka, T.; Ueno, H.; Morizane, C. First-in-human phase 1 study of novel dUTPase inhibitor TAS-114 in combination with S-1 in Japanese patients with advanced solid tumors. Investig. New Drugs 2019, 37, 507–518. [Google Scholar] [CrossRef] [Green Version]
- Miyakoshi, H.; Miyahara, S.; Yokogawa, T.; Endoh, K.; Muto, T.; Yano, W.; Wakasa, T.; Ueno, H.; Chong, K.T.; Taguchi, J. 1,2,3-Triazole-containing uracil derivatives with excellent pharmacokinetics as a novel class of potent human deoxyuridine triphosphatase inhibitors. J. Med. Chem. 2012, 55, 6427–6437. [Google Scholar] [CrossRef] [PubMed]
- Miyahara, S.; Miyakoshi, H.; Yokogawa, T.; Chong, K.T.; Taguchi, J.; Muto, T.; Endoh, K.; Yano, W.; Wakasa, T.; Ueno, H. Discovery of a novel class of potent human deoxyuridine triphosphatase inhibitors remarkably enhancing the antitumor activity of thymidylate synthase inhibitors. J. Med. Chem. 2012, 55, 2970–2980. [Google Scholar] [CrossRef] [PubMed]
- Kiyonari, S.; Iimori, M.; Matsuoka, K.; Watanabe, S.; Morikawa-Ichinose, T.; Miura, D.; Niimi, S.; Saeki, H.; Tokunaga, E.; Oki, E. The 1,2-Diaminocyclohexane Carrier Ligand in Oxaliplatin Induces p53-Dependent Transcriptional Repression of Factors Involved in Thymidylate Biosynthesis. Mol. Cancer Ther. 2015, 14, 2332–2342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarthy, O.; Musso-Buendia, A.; Kaiser, M.; Brun, R.; Ruiz-Perez, L.M.; Johansson, N.G.; Pacanowska, D.G.; Gilbert, I.H. Design, synthesis and evaluation of novel uracil acetamide derivatives as potential inhibitors of Plasmodium falciparum dUTP nucleotidohydrolase. Eur. J. Med. Chem. 2009, 44, 678–688. [Google Scholar] [CrossRef]
- Baragana, B.; McCarthy, O.; Sanchez, P.; Bosch-Navarrete, C.; Kaiser, M.; Brun, R.; Whittingham, J.L.; Roberts, S.M.; Zhou, X.X.; Wilson, K.S. Beta-branched acyclic nucleoside analogues as inhibitors of Plasmodium falciparum dUTPase. Bioorg. Med. Chem. 2011, 19, 2378–2391. [Google Scholar] [CrossRef]
- Nguyen, C.; Ruda, G.F.; Schipani, A.; Kasinathan, G.; Leal, I.; Musso-Buendia, A.; Kaiser, M.; Brun, R.; Ruiz-Perez, L.M.; Sahlberg, B.L. Acyclic nucleoside analogues as inhibitors of Plasmodium falciparum dUTPase. J. Med. Chem. 2006, 49, 4183–4195. [Google Scholar] [CrossRef]
- Lima, M.N.N.; Melo-Filho, C.C.; Cassiano, G.C.; Neves, B.J.; Alves, V.M.; Braga, R.C.; Cravo, P.V.L.; Muratov, E.N.; Calit, J.; Bargieri, D.Y. QSAR-driven design and discovery of novel compounds with antiplasmodial and transmission blocking activities. Front. Pharm. 2018, 9, 146. [Google Scholar] [CrossRef] [Green Version]
- Hirmondo, R.; Szabo, J.E.; Nyiri, K.; Tarjanyi, S.; Dobrotka, P.; Toth, J.; Vertessy, B.G. Cross-species inhibition of dUTPase via the Staphylococcal Stl protein perturbs dNTP pool and colony formation in Mycobacterium. DNA Repair 2015, 30, 21–27. [Google Scholar] [CrossRef] [Green Version]
- Hampton, S.E.; Baragana, B.; Schipani, A.; Bosch-Navarrete, C.; Musso-Buendia, J.A.; Recio, E.; Kaiser, M.; Whittingham, J.L.; Roberts, S.M.; Shevtsov, M. Design, synthesis, and evaluation of 5’-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase. ChemMedChem 2011, 6, 1816–1831. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A.M.; Ariza, M.E.; Williams, M.; Jason, L.; Beqaj, S.; Fitzgerald, J.T.; Lemeshow, S.; Glaser, R. Antibody to Ep-stein-Barr virus deoxyuridine triphosphate nucleotidohydrolase and deoxyribonucleotide polymerase in a chronic fatigue syndrome subset. PLoS ONE 2012, 7, e47891. [Google Scholar] [CrossRef] [Green Version]
- Waldman, W.J.; Williams, M.V., Jr.; Lemeshow, S.; Binkley, P.; Guttridge, D.; Kiecolt-Glaser, J.K.; Knight, D.A.; Ladner, K.J.; Glaser, R. Epstein-Barr virus-encoded dUTPase enhances proinflammatory cytokine production by macrophages in contact with endothelial cells: Evidence for depression-induced atherosclerotic risk. Brain Behav. Immun. 2008, 22, 215–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freeman, L.; Buisson, M.; Tarbouriech, N.; Van der Heyden, A.; Labbe, P.; Burmeister, W.P. The flexible motif V of Ep-stein-Barr virus deoxyuridine 5’-triphosphate pyrophosphatase is essential for catalysis. J. Biol. Chem. 2009, 284, 25280–25289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pecsi, I.; Szabo, J.E.; Adams, S.D.; Simon, I.; Sellers, J.R.; Vertessy, B.G.; Toth, J. Nucleotide pyrophosphatase employs a P-loop-like motif to enhance catalytic power and NDP/NTP discrimination. Proc. Natl. Acad. Sci. USA 2011, 108, 14437–14442. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Anti-E165R mAb | Western Blotting | IFA | Epitope Region aa | |||||
|---|---|---|---|---|---|---|---|---|
| 1–165 aa | 1–86 aa | 1–100 aa | 1–120 aa | 1–140 aa | 37–165 aa | |||
| 5C11 | + | − | − | − | − | + | + | 140–165 |
| 4C3 | + | − | − | − | + | + | − | 120–140 |
| 10E9 | + | − | − | + | + | + | − | 100–120 |
| 8F9 | + | − | − | − | − | + | + | 140–165 |
| 5D1 | + | − | − | − | − | + | + | 140–165 |
| 8E6 | + | − | − | + | + | + | − | 100–120 |
| 11B11 | + | − | − | − | − | + | + | 140–165 |
| 8A4 | + | − | − | − | − | + | + | 140–165 |
| 10C4 | + | − | − | − | − | + | − | 140–165 |
| 6A11 | + | + | + | + | + | + | − | 37–86 |
| 11D12 | + | − | − | − | − | + | + | 140–165 |
| 2C2 | + | − | − | + | + | + | + | 100–120 |
| 4F8 | + | − | − | − | + | + | − | 120–140 |
| 2G7 | + | − | − | − | + | + | − | 120–140 |
| 6D6 | + | − | − | − | + | + | + | 120–140 |
| 10A4 | + | − | − | − | + | + | − | 100–120 |
| 8A7 | + | − | − | − | + | + | − | 120–140 |
| 6A3 | + | − | − | − | − | + | + | 140–165 |
| 6G3 | + | − | − | − | + | + | + | 120–140 |
| NC | − | − | − | − | − | − | − | N/A |
| Antibody Volume | Buffer | Mouse Negative Serum + E165R | 8F9 + E165R | 5D1 + E165R | 6A3 + E165R | 6G3 + E165R |
|---|---|---|---|---|---|---|
| 0 μL | 10−52 | −1.6 × 10−3 | −1.6 × 10−3 | −1.6 × 10−3 | −1.6 × 10−3 | −1.6 × 10−3 |
| 20 μL | N/A | −1.6 × 10−3 | −1.2 × 10−3 | −1.3 × 10−3 | −1.3 × 10−3 | −1.2 × 10−3 |
| 40 μL | N/A | −1.6 × 10−3 | −1.1 × 10−3 | −1.1 × 10−3 | −1.1 × 10−3 | −1.2 × 10−3 |
| 100 μL | N/A | −1.6 × 10−3 | −6.0 × 10−4 | −8.0 × 10−4 | −9.0 × 10−4 | −8.0 × 10−4 |
| Max inhibition enzyme activity ratio (%) | N/A | N/A | 62.50 | 50.00 | 43.75 | 50.00 |
| Anti-E165R-mAbs | Antibody Subtype | Inhibitory Epitope |
|---|---|---|
| 8F9 | IgG2a | 149–RGEGRFGSTG–158 |
| 5D1 | IgG2a | 149–RGEGRFGSTG–158 |
| 6A3 | IgG1 | Undefined |
| 6G3 | IgG1 | N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Wang, R.; Zhu, X.; Jin, J.; Lu, W.; Zhao, X.; Wan, B.; Liao, Y.; Zhao, Q.; Netherton, C.L.; et al. Identification and Characterization of a Novel Epitope of ASFV-Encoded dUTPase by Monoclonal Antibodies. Viruses 2021, 13, 2175. https://doi.org/10.3390/v13112175
Zhang S, Wang R, Zhu X, Jin J, Lu W, Zhao X, Wan B, Liao Y, Zhao Q, Netherton CL, et al. Identification and Characterization of a Novel Epitope of ASFV-Encoded dUTPase by Monoclonal Antibodies. Viruses. 2021; 13(11):2175. https://doi.org/10.3390/v13112175
Chicago/Turabian StyleZhang, Shuai, Rui Wang, Xiaojing Zhu, Jiaxin Jin, Wenlong Lu, Xuyang Zhao, Bo Wan, Yifei Liao, Qin Zhao, Christopher L. Netherton, and et al. 2021. "Identification and Characterization of a Novel Epitope of ASFV-Encoded dUTPase by Monoclonal Antibodies" Viruses 13, no. 11: 2175. https://doi.org/10.3390/v13112175