An Epitope Platform for Safe and Effective HTLV-1-Immunization: Potential Applications for mRNA and Peptide-Based Vaccines
Abstract
1. Introduction
2. Methods
3. Results
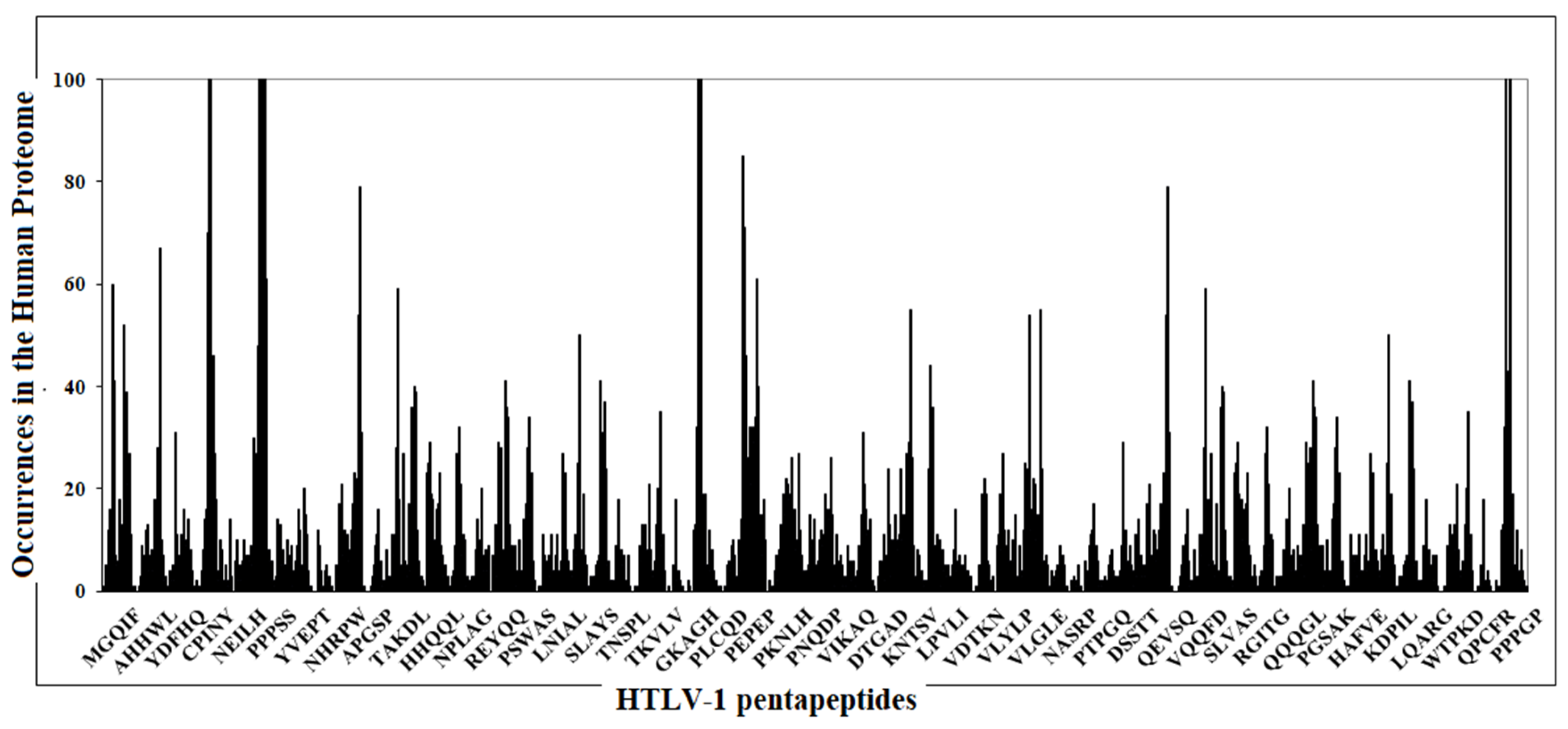
3.1. From Sequence Similarity Analyses to Anti-HTLV-1 Vaccination: Formulating a Specific and Effective Vaccine
3.2. From Sequence Similarity Analyses to Autoantibodies Generation: Potential Contribution to the HTLV-1 Pathological Burden
- A-kinase anchor protein 1, mitochondrial precursor (PSLALP) has a pivotal role in mitochondrial physiology, and its degradation leads to an increase in reactive oxygen species production, mitochondrial dysfunction, and ultimately cell death [16].
- A-kinase anchor protein 7 isoform gamma (HLTLPF) modulates L-type Ca2+ channels [17]. Alterations of L-type Ca2+ channels may affect cardiac contraction [18], associate with diabetes [19] and can cause autism [20] and relate to Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, neuropsychiatric diseases, and other CNS disorders [21].
- B-cell CLL/lymphoma 6 member B protein and B-cell CLL/lymphoma 9-like protein, both of which share the peptide PPTAPP with HTLV-1. When altered, these two proteins relate to tumorigenesis [26].
- Cadherin-2 (APQVLP) may be associated with reduction in bone mineral density or vertebral fracture prevalence in survivors of childhood acute lymphoblastic leukemia [27].
- Constitutive coactivator of PPAR-gamma-like protein 2 (QLPPTA) deletion is associated with autism [28].
- Dedicator of cytokinesis protein 9 (PPLLPH, PLLPHS, SLALPA) contributes to both risk and increased illness severity in bipolar disorder [29].
- DNA methyltransferase 1-associated protein 1 (APPLLP) is an essential regulator of activity and function of Ataxia Telangiectasia Mutated (ATM) kinase [30]. Alterations of the function and activity of ATM cause severe disability, poor coordination and telangiectasia, i.e., small dilated blood vessels [31].
- DNA polymerase nu (PPPGPC) plays a role in translesion DNA synthesis during interstrand cross-link repair in human cells [32].
- E3 ubiquitin-protein ligase HECTD1 (APGYDP) is an important factor in promoting base excision repair (BER), which is the major cellular DNA repair pathway that recognizes and excises damaged DNA bases to help maintain genome stability [33].
- Fertilization-influencing membrane protein (PSQLPP) plays a role in sperm–oocyte fusion during fertilization [34].
- FLYWCH-type zinc finger-containing protein 1 (PSLALP) has a possible tumor suppressive role in preventing colorectal cancer metastasis [35].
- Heterogeneous nuclear ribonucleoprotein C-like 1 (DLQAIK, LQAIKQ, QAKQE), alterations of which are found in sporadic and suspected Lynch syndrome endometrial cancer [36].
- Histone acetyltransferases KAT2A and KAT2B (DGRVIG), when altered, are associated with cardiovascular pathology [37].
- Islet cell autoantigen 1 (MKDLQA) Islet autoantibodies are typically associated with type 1 diabetes, but have been found in patients diagnosed with type 2 diabetes in whom they are associated with lower adiposity [38].
- La-related protein 1B (AIKQEV) is dysregulated in hepatocellular carcinoma [39].
- Mitochondrial dynamics protein MID49 (QPRPPP) is related to myopathies [40].
- Mucin-5AC precursor (SLSPVP, LSPVPT) is a major macromolecular component that determines the rheological properties of mucus; otherwise abnormal not-flowing mucus results in defective lung protection and leads to infection and inflammation [41].
- Neurofascin precursor (TGAVSS), which is a cell adhesion, ankyrin-binding protein, may be involved in neurite extension, axonal guidance, synaptogenesis, myelination, and neuron–glial cell interactions. Anti-neurofascin antibodies relate to optic, trigeminal, and facial neuropathy [42].
- Palmitoyltransferase ZDHHC1 (LALPAP) is involved in innate viral immune response [43].
- Protein piccolo (PPNHRP) is required for the development and function of neuronal networks formed between the brainstem and cerebellum. Alterations of protein piccolo may result in impaired motor coordination, cerebellar network dysfunction and pontocerebellar hypoplasia [44].
- Serine/threonine-protein kinase WNK1 (APQVLP) can cause hereditary sensory and autonomic neuropathy [45].
- Suppressor of tumorigenicity 7 protein (SLILPP) deficiency promotes laryngeal squamous cell carcinoma [46].
- Finally, to conclude this survey of pathologies that may be associated with HTLV-1 infection following cross-reactivity and autoantibody generation, we highlight the HTLV-1 epitope-derived VNFTQE peptide that is present in seventeen zinc finger proteins (ZFP) (numbered in Table 3 as: 57 homolog, 69, 101, 124, 136, 334, 439, 440, 442, 563, 669, 700, 709, 763, 823, 844, and 878). Zinc finger proteins comprise transcription factor families that are known for their ability to bind Zn2+ and are associated with numerous disorders. In the present context, the following are noteworthy: ZFP823 family, when mutated or dysregulated, is involved in acute leukemias [47]. Altered/dysregulated ZFPs may also associate with neurodevelopmental disorders such as intellectual disability, autistic features, psychiatric problems, and motor dysfunction [48]. Moreover, zinc finger proteins are implicated in the development and progression of several types of cancer [49]. Simply put, antibodies against the epitope-derived peptide VNFTQE alone might be sufficient to determine the wide range of pathologies that, in more or less severe forms, afflict HTLV-1-infected patients.
4. Discussion
4.1. Translational Potential and Advantages for mRNA Vaccines
4.2. Translational Potential and Advantages for Peptide-Based Vaccines
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schierhout, G.; McGregor, S.; Gessain, A.; Einsiedel, L.; Martinello, M.; Kaldor, J. Association between HTLV-1 infection and adverse health outcomes: A systematic review and meta-analysis of epidemiological studies. Lancet Infect. Dis. 2020, 20, 133–143. [Google Scholar] [CrossRef]
- Khan, M.Y.; Khan, I.N.; Farman, M.; Al Karim, S.; Qadri, I.; Kamal, M.A.; Al Ghamdi, K.; Harakeh, S. HTLV-1 Associated Neurological Disorders. Curr. Top. Med. Chem. 2017, 17, 1320–1330. [Google Scholar] [CrossRef] [PubMed]
- Romanelli, L.C.F.; Caramelli, P.; Martins, M.L.; Gonçalves, D.U.; Proietti, F.A.; Ribas, J.G.; Araújo, M.G.; Carneiro-Proietti, A.B.D.F. Incidence of Human T Cell Lymphotropic Virus Type 1-Associated Myelopathy/Tropical Spastic Paraparesis in a Long-Term Prospective Cohort Study of Initially Asymptomatic Individuals in Brazil. AIDS Res. Hum. Retrovir. 2013, 29, 1199–1202. [Google Scholar] [CrossRef] [PubMed]
- Quaresma, J.A.S.; Yoshikawa, G.T.; Koyama, R.V.L.; Dias, G.A.S.; Fujihara, S.; Fuzii, H.T. HTLV-1, Immune Response and Autoimmunity. Viruses 2015, 8, 5. [Google Scholar] [CrossRef]
- Levin, M.; Lee, S.M.; Kalume, F.; Morcos, Y.; Curtis Dohan, F., Jr.; Hasty, K.A.; Callaway, J.C.; Zunt, J.; Desiderio, D.M.; Stuart, J.M. Autoimmunity due to molecular mimicry as a cause of neurological disease. Nat. Med. 2002, 8, 509–513. [Google Scholar] [CrossRef]
- Kabiri, M.; Sankian, M.; Hosseinpour, M.; Tafaghodi, M. The novel immunogenic chimeric peptide vaccine to elicit potent cellular and mucosal immune responses against HTLV-1. Int. J. Pharm. 2018, 549, 404–414. [Google Scholar] [CrossRef]
- Kabiri, M.; Sankian, M.; Sadri, K.; Tafaghodi, M. Robust mucosal and systemic responses against HTLV-1 by delivery of multi-epitope vaccine in PLGA nanoparticles. Eur. J. Pharm. Biopharm. 2018, 133, 321–330. [Google Scholar] [CrossRef]
- Kannagi, M.; Hasegawa, A.; Nagano, Y.; Iino, T.; Okamura, J.; Suehiro, Y. Maintenance of long remission in adult T-cell leukemia by Tax-targeted vaccine: A hope for disease-preventive therapy. Cancer Sci. 2019, 110, 849–857. [Google Scholar] [CrossRef]
- Kanduc, D. “Self-Nonself” Peptides in the Design of Vaccines. Curr. Pharm. Des. 2009, 15, 3283–3289. [Google Scholar] [CrossRef] [PubMed]
- Kanduc, D. Peptide cross-reactivity the original sin of vaccines. Front. Biosci. 2012, S4, 1393–1401. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Chen, C.; Li, Z.; Huang, H.; Suzek, B.E.; Wu, C.H.; UniProt Consortium. A fast PeptideMatch service for UniProt knowledgebase. Bioinformatics 2013, 29, 2808–2809. [Google Scholar] [CrossRef]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef]
- Andreatta, M.; Nielsen, M. Gapped sequence alignment using artificial neural networks: Application to the MHC class I system. Bioinformatics 2016, 32, 511–517. [Google Scholar] [CrossRef]
- Alam, S.; Hasan, K.; Bin Manjur, O.H.; Khan, A.; Sharmin, Z.; Pavel, M.A.; Hossain, F. Predicting and Designing Epitope Ensemble Vaccines against HTLV-1. J. Integr. Bioinform. 2020, 16. [Google Scholar] [CrossRef]
- Czachor, A.; Failla, A.; Lockey, R.; Kolliputi, N. Pivotal role of AKAP121 in mitochondrial physiology. Am. J. Physiol. Physiol. 2016, 310, C625–C628. [Google Scholar] [CrossRef] [PubMed]
- Fraser, I.; Scott, J.D. Modulation of Ion Channels: A “Current” View of AKAPs. Neuron 1999, 23, 423–426. [Google Scholar] [CrossRef]
- Lu, F.; Pu, W.T. The architecture and function of cardiac dyads. Biophys. Rev. 2020, 12, 1007–1017. [Google Scholar] [CrossRef]
- Nieves-Cintrón, M.; Tamez, V.A.; Le, T.; Baudel, M.M.-A.; Navedo, M.F. Cellular and molecular effects of hyperglycemia on ion channels in vascular smooth muscle. Cell. Mol. Life Sci. 2021, 78, 31–61. [Google Scholar] [CrossRef] [PubMed]
- Marcantoni, A.; Calorio, C.; Hidisoglu, E.; Chiantia, G.; Carbone, E. Cav1.2 channelopathies causing autism: New hallmarks on Timothy syndrome. Pflügers Arch. Eur. J. Physiol. 2020, 472, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hopp, S.C. Targeting microglia L-type voltage-dependent calcium channels for the treatment of central nervous system disorders. J. Neurosci. Res. 2021, 99, 141–162. [Google Scholar] [CrossRef] [PubMed]
- Kaehler, C.; Isensee, J.; Nonhoff, U.; Terrey, M.; Hucho, T.; Lehrach, H.; Krobitsch, S. Ataxin-2-Like Is a Regulator of Stress Granules and Processing Bodies. PLoS ONE 2012, 7, e50134. [Google Scholar] [CrossRef] [PubMed]
- Panagopoulos, I.; Gorunova, L.; Spetalen, S.; Bassarova, A.; Beiske, K.; Micci, F.; Heim, S. Fusion of the genes ataxin 2 like, ATXN2L, and Janus kinase 2, JAK2, in cutaneous CD4 positive T-cell lymphoma. Oncotarget 2017, 8, 103775–103784. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.A.; Vonk, J.M.; Baurecht, H.; Marenholz, I.; Tian, C.; Hoffman, J.D.; Helmer, Q.; Tillander, A.; Ullemar, V.; Lu, Y.; et al. Eleven loci with new reproducible genetic associations with allergic disease risk. J. Allergy Clin. Immunol. 2019, 143, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Key, J.; Harter, P.N.; Sen, N.-E.; Gradhand, E.; Auburger, G.; Gispert, S. Mid-Gestation lethality of Atxn2l-Ablated Mice. Int. J. Mol. Sci. 2020, 21, 5124. [Google Scholar] [CrossRef]
- Wang, J.; Dong, L.; Xu, L.; Chu, E.S.; Chen, Y.; Shen, J.; Li, X.; Wong, C.C.; Sung, J.J.Y.; Yu, J. B cell CLL/lymphoma 6 member B inhibits hepatocellular carcinoma metastases in vitro and in mice. Cancer Lett. 2014, 355, 192–200. [Google Scholar] [CrossRef]
- Aaron, M.; Nadeau, G.; Ouimet-Grennan, E.; Drouin, S.; Bertout, L.; Beaulieu, P.; St-Onge, P.; Shalmiev, A.; Veilleux, L.-N.; Rauch, F.; et al. Identification of a single-nucleotide polymorphism within CDH2 gene associated with bone morbidity in childhood acute lymphoblastic leukemia survivors. Pharmacogenomics 2019, 20, 409–420. [Google Scholar] [CrossRef]
- De Wolf, V.; Crepel, A.; Schuit, F.; van Lommel, L.; Ceulemans, B.; Steyaert, J.; Seuntjens, E.; Peeters, H.; Devriendt, K. A complex Xp11.22 deletion in a patient with syndromic autism: Exploration ofFAM120Cas a positional candidate gene for autism. Am. J. Med. Genet. Part A 2014, 164, 3035–3041. [Google Scholar] [CrossRef]
- Detera-Wadleigh, S.D.; Liu, C.; Maheshwari, M.; Cardona, I.; Corona, W.; Akula, N.; Steele, C.; Badner, J.A.; Kundu, M.; Kassem, L.; et al. Sequence variation in DOCK9 and heterogeneity in bipolar disorder. Psychiatr. Genet. 2007, 17, 274–286. [Google Scholar] [CrossRef]
- Penicud, K.; Behrens, A. DMAP1 is an essential regulator of ATM activity and function. Oncogene 2014, 33, 525–531. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, B.S.P.; Putti, S.; Naro, F.; Pellegrini, M. Bone Marrow Transplantation as Therapy for Ataxia-Telangiectasia: A Systematic Review. Cancers 2020, 12, 3207. [Google Scholar] [CrossRef]
- Zietlow, L.; Smith, L.A.; Bessho, M.; Bessho, T. Evidence for the Involvement of Human DNA Polymerase N in the Repair of DNA Interstrand Cross-Links. Biochemistry 2009, 48, 11817–11824. [Google Scholar] [CrossRef] [PubMed]
- Bennett, L.; Madders, E.C.E.T.; Parsons, J.L. HECTD1 promotes base excision repair in nucleosomes through chromatin remodelling. Nucleic Acids Res. 2019, 48, 1301–1313. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, Y.; Lu, Y.; Noda, T.; Oji, A.; Larasati, T.; Kojima-Kita, K.; Yu, Z.; Matzuk, R.M.; Matzuk, M.M.; Ikawa, M. Spermatozoa lacking Fertilization Influencing Membrane Protein (FIMP) fail to fuse with oocytes in mice. Proc. Natl. Acad. Sci. USA 2020, 117, 9393–9400. [Google Scholar] [CrossRef]
- Muhammad, B.A.; Almozyan, S.; Babaei-Jadidi, R.; Onyido, E.; Saadeddin, A.; Kashfi, S.H.; Spencer-Dene, B.; Ilyas, M.; Lourdusamy, A.; Behrens, A.; et al. FLYWCH1, a Novel Suppressor of Nuclear β-Catenin, Regulates Migration and Morphology in Colorectal Cancer. Mol. Cancer Res. 2018, 16, 1977–1990. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, X.; Wang, T.; Zhang, Y.; Wang, Q.; Hu, Y. HNRNPCL1, PRAMEF1, CFAP74, and DFFB: Common Potential Biomarkers for Sporadic and Suspected Lynch Syndrome Endometrial Cancer. Cancer Manag. Res. 2020, 12, 11231–11241. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Tanios, F.; Reeps, C.; Zhang, J.; Schwamborn, K.; Eckstein, H.-H.; Zernecke, A.; Pelisek, J. Histone acetylation and histone acetyltransferases show significant alterations in human abdominal aortic aneurysm. Clin. Epigenetics 2016, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pilla, S.J.; Balasubramanyam, A.; Knowler, W.C.; Lazo, M.; Nathan, D.M.; Pi-Sunyer, X.; Clark, J.M.; Maruthur, N.M.; The Look AHEAD Research Group. Islet autoantibody positivity in overweight and obese adults with type 2 diabetes. Autoimmunity 2018, 51, 408–416. [Google Scholar] [CrossRef]
- Wang, M.; Huang, S.; Chen, Z.; Han, Z.; Li, K.; Chen, C.; Wu, G.; Zhao, Y. Development and validation of an RNA binding protein-associated prognostic model for hepatocellular carcinoma. BMC Cancer 2020, 20, 1–14. [Google Scholar] [CrossRef]
- Bartsakoulia, M.; Pyle, A.; Troncoso-Chandía, D.; Vial-Brizzi, J.; Paz-Fiblas, M.V.; Duff, J.; Griffin, H.; Boczonadi, V.; Lochmüller, H.; Kleinle, S.; et al. A novel mechanism causing imbalance of mitochondrial fusion and fission in human myopathies. Hum. Mol. Genet. 2018, 27, 1186–1195. [Google Scholar] [CrossRef]
- Ridley, C.; Thornton, D.J. Mucins: The frontline defence of the lung. Biochem. Soc. Trans. 2018, 46, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Zhang, X.; Inamizu, S.; Yamashita, K.; Yamasaki, R.; Matsushita, T.; Isobe, N.; Hiwatashi, A.; Tobimatsu, S.; Kira, J. Optic, trigeminal, and facial neuropathy related to anti-neurofascin 155 antibody. Ann. Clin. Transl. Neurol. 2020, 7, 2297–2309. [Google Scholar] [CrossRef]
- Sowers, M.L.; Tang, H.; Tian, B.; Goldblum, R.; Midoro-Horiuti, T.; Zhang, K. Bisphenol A Activates an Innate Viral Immune Response Pathway. J. Proteome Res. 2019, 19, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Falck, J.; Bruns, C.; Hoffmann-Conaway, S.; Straub, I.; Plautz, E.J.; Orlando, M.; Munawar, H.; Rivalan, M.; Winter, Y.; Izsvák, Z.; et al. Loss of Piccolo Function in Rats Induces Cerebellar Network Dysfunction and Pontocerebellar Hypoplasia Type 3-like Phenotypes. J. Neurosci. 2020, 40, 2943–2959. [Google Scholar] [CrossRef] [PubMed]
- Lafrenière, R.G.; Macdonald, M.L.E.; Dubé, M.-P.; Macfarlane, J.; O’Driscoll, M.; Brais, B.; Meilleur, S.; Brinkman, R.R.; Dadivas, O.; Pape, T.; et al. Identification of a Novel Gene (HSN2) Causing Hereditary Sensory and Autonomic Neuropathy Type II through the Study of Canadian Genetic Isolates. Expand. Spectr. BAF Relat. Disord. De Novo Var. SMARCC2 Cause Syndr. Intellect. Disabil. Dev. Delay 2004, 74, 1064–1073. [Google Scholar] [CrossRef]
- Qin, H.; Xu, J.; Gong, L.; Jiang, B.; Zhao, W. The long noncoding RNA ST7-AS1 promotes laryngeal squamous cell carcinoma by stabilizing CARM1. Biochem. Biophys. Res. Commun. 2019, 512, 34–40. [Google Scholar] [CrossRef]
- Schuschel, K.; Helwig, M.; Hüttelmaier, S.; Heckl, D.; Klusmann, J.-H.; Hoell, J.I. RNA-Binding Proteins in Acute Leukemias. Int. J. Mol. Sci. 2020, 21, 3409. [Google Scholar] [CrossRef]
- Al-Naama, N.; Mackeh, R.; Kino, T. C2H2-Type Zinc Finger Proteins in Brain Development, Neurodevelopmental, and Other Neuropsychiatric Disorders: Systematic Literature-Based Analysis. Front. Neurol. 2020, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Liu, J.; Xie, K. Zinc finger proteins and regulation of the hallmarks of cancer. Histol. Histopathol. 2019, 34, 18121. [Google Scholar]
- Greinacher, A.; Thiele, T.; Warkentin, T.E.; Weisser, K.; Kyrle, P.A.; Eichinger, S. Thrombotic Thrombocytopenia after ChAdOx1 nCov-19 Vaccination. N. Engl. J. Med. 2021, 384, 2092–2101. [Google Scholar] [CrossRef]
- Greinacher, A.; Selleng, K.; Mayerle, J. Anti-SARS-CoV-2 Spike Protein and Anti-Platelet Factor 4 Antibody Responses Induced by COVID-19 Disease and ChAdOx1 nCov-19 vaccination. Res. Sq. 2021, preprint. [Google Scholar] [CrossRef]
- Lucchese, G. Epitopes for a 2019-nCoV vaccine. Cell. Mol. Immunol. 2020, 17, 539–540. [Google Scholar] [CrossRef]
- Patel, A.; Dong, J.C.; Trost, B.; Richardson, J.S.; Tohme, S.; Babiuk, S.; Kusalik, A.; Kung, S.K.P.; Kobinger, G.P. Pentamers Not Found in the Universal Proteome Can Enhance Antigen Specific Immune Responses and Adjuvant Vaccines. PLoS ONE 2012, 7, e43802. [Google Scholar] [CrossRef]
- Mallery, D.L.; McEwan, W.; Bidgood, S.; Towers, G.; Johnson, C.M.; James, L.C. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21). Proc. Natl. Acad. Sci. USA 2010, 107, 19985–19990. [Google Scholar] [CrossRef] [PubMed]
- Greenlee, J.E.; Clawson, S.A.; Hill, K.E.; Wood, B.; Clardy, S.L.; Tsunoda, I.; Carlson, N.G. Anti-Yo Antibody Uptake and Interaction with Its Intracellular Target Antigen Causes Purkinje Cell Death in Rat Cerebellar Slice Cultures: A Possible Mechanism for Paraneoplastic Cerebellar Degeneration in Humans with Gynecological or Breast Cancers. PLoS ONE 2015, 10, e0123446. [Google Scholar] [CrossRef] [PubMed]
- Enose-Akahata, Y.; Abrams, A.; Massoud, R.; Bialuk, I.; Johnson, K.R.; Green, P.L.; Maloney, E.M.; Jacobson, S. Humoral immune response to HTLV-1 basic leucine zipper factor (HBZ) in HTLV-1-infected individuals. Retrovirology 2013, 10, 19. [Google Scholar] [CrossRef]
- Zeng, C.; Zhang, C.; Walker, P.G.; Dong, Y. Formulation and Delivery Technologies for mRNA Vaccines. In Current Topics in Microbiology and Immunology; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Igyártó, B.Z.; Jacobsen, S.; Ndeupen, S. Future considerations for the mRNA-lipid nanoparticle vaccine platform. Curr. Opin. Virol. 2021, 48, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Crommelin, D.J.; Anchordoquy, T.J.; Volkin, D.B.; Jiskoot, W.; Mastrobattista, E. Addressing the Cold Reality of mRNA Vaccine Stability. J. Pharm. Sci. 2021, 110, 997–1001. [Google Scholar] [CrossRef]
- Tinari, S. The EMA covid-19 data leak, and what it tells us about mRNA instability. BMJ 2021, 372, n627. [Google Scholar] [CrossRef]
- Brader, M.L.; Williams, S.J.; Banks, J.M.; Hui, W.H.; Zhou, Z.H.; Jin, L. Encapsulation state of messenger RNA inside lipid nanoparticles. Biophys. J. 2021, 25, S0006-3495(21)00241-1. [Google Scholar] [CrossRef]
- Valleriani, A.; Zhang, G.; Nagar, A.; Ignatova, Z.; Lipowsky, R. Length-dependent translation of messenger RNA by ribosomes. Phys. Rev. E 2011, 83, 042903. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. Comment on “Length-dependent translation of messenger RNA by ribosomes”. Phys. Rev. E 2012, 85, 023901. [Google Scholar] [CrossRef]
- Liao, H.X.; Spremulli, L.L. Effects of length and mRNA secondary structure on the interaction of bovine mitochondrial ribo-somes with messenger RNA. J. Biol. Chem. 1990, 265, 11761–11765. [Google Scholar] [CrossRef]
- Malonis, R.J.; Lai, J.R.; Vergnolle, O. Peptide-Based Vaccines: Current Progress and Future Challenges. Chem. Rev. 2020, 120, 3210–3229. [Google Scholar] [CrossRef] [PubMed]
- Ryzhikov, A.B.; Ryzhikov, E.A.; Bogryantseva, M.P.; Usova, S.V.; Danilenko, E.D.; Nechaeva, E.A.; Pyankov, O.V.; Pyankova, O.G.; Gudymo, A.S. A single blind, placebo-controlled randomized study of the safety, reactogenicity and immunogenicity of the “EpiVacCorona” Vaccine for the prevention of COVID-19, in volunteers aged 18–60 years (phase I–II). Russ. J. Infect. Immun. 2021, 11, 283–296. [Google Scholar] [CrossRef]
- Ryzhikov, A.B. Peptide Immunogens and Vaccine Composition against Coronavirus Infection COVID-19 Using Peptide Immunogens. Russian Federration. Patent No. 2738081, 7 December 2020. [Google Scholar]
- Lucchese, G.; Stufano, A.; Kanduc, D. Searching for an effective, safe and universal anti-HIV vaccine: Finding the answer in just one short peptide. Self Nonself 2011, 2, 49–54. [Google Scholar] [CrossRef][Green Version]
| HTLV-1 Protein | AaPosition | Peptide 1 | MHC Binding 2 |
|---|---|---|---|
| Gag polyprotein | 132–138 | VMHPHGA | HLA-B*07:02 |
| Gag polyprotein | 165–171 | PQFMQTI | HLA-B*07:02 |
| Gag polyprotein | 331–337 | ACQTWTP | n.a. |
| Gag polyprotein | 364–370 | GHWSRDC | n.a. |
| Gag-Pro-Pol polyprotein | 915–921 | SKEQWPL | n.a. |
| Gag-Pro-Pol polyprotein | 1086–1092 | RSWRCLN | n.a. |
| Gag-Pro-Pol polyprotein | 1351–1358 | IALWTINH | n.a. |
| Gag-Pro-Pol polyprotein | 1366–1374 | HKTRWQLHH | n.a. |
| Gag-Pro-Pol polyprotein | 1390–1398 | KQTHWYYFK | HLA-A*03:01 HLA-B*27:05 |
| Gag-Pro-Pol polyprotein | 1429–1435 | SAQWIPW | HLA-A*24:02 HLA-A*02:01 HLA-B*35:01 HLA-B*58:01 |
| Protein Tax-1 | 49–56 | CPEHQITW | n.a. |
| Envelope glycoprotein gp62 | 116–122 | GCQSWTC | n.a. |
| Envelope glycoprotein gp62 | 131–137 | PYWKFQH | n.a. |
| Envelope glycoprotein gp62 | 170–176 | YDPIWFL | n.a. |
| Envelope glycoprotein gp62 | 272–278 | NWTHCFD | n.a. |
| IEDB ID 1 | EPITOPE SEQUENCE 2 | VIRAL PROTEIN | ASSAY |
|---|---|---|---|
| 7612 | dapgYDPIWFLntepsqlpptappllphsnldhile | Envelope glycoprotein | B cell |
| 17248 | fNWTHCFDpqiqaivsspchnslilppfslspvpt | Envelope glycoprotein | B cell |
| 18895 | GCQSWTCpytgavssPYWKFQHdvn | Envelope glycoprotein | B cell |
| 20199 | GHWSRDCtqprpppgpcplcqdp | Pr gag-pro-pol.1 | B cell |
| 34858 | lalpaphltlpfNWTHCFDpqiq | Envelope glycoprotein | B cell |
| 38482 | lpfNWTHCFDpq | Envelope glycoprotein | B cell |
| 47047 | pcslkcpylGCQSWTCpytgavs | Envelope glycoprotein | B cell |
| 49915 | pVMHPHGAppnhrpwqmkdlqaikqevsqa | Pr gag-pro-pol.1 | B cell |
| 59328 | sllvdapgYDPIWFLntepsqlpptappllphsnldhilepsipwks | Envelope glycoprotein | B cell |
| 61300 | ssPYWKFQHdvnftqevsrln | Envelope glycoprotein | B cell |
| 62700 | sysdpcslkcpylGCQSWTCpyt | Envelope glycoprotein | B cell |
| 65003 | tlpfNWTHCFDpqiqaivs | Envelope glycoprotein | B cell |
| 65645 | tpllypslalpaphltlpfNWTHCFDpqiq | Envelope glycoprotein | B cell |
| 78192 | lalpaphltlpfNWTHCFDpqiqaivsspchnsli | Envelope glycoprotein | B cell |
| 78715 | latCPEHQITWdpidgrvig | Transcriptional activator Tax | B cell |
| 94435 | apgYDPIWFL | Envelope glycoprotein | T cell |
| 96592 | lpfNWTHCFDpqiqaivsspc | Envelope glycoprotein | T cell |
| 97892 | apqvlpVMHPHGAppn | Pr gag | B cell |
| Peptides | Human Proteins |
|---|---|
| SLILPP | (Pyruvate dehydrogenase (acetyl-transferring)) kinase isozyme 2, mitochondrial, Cilia- and flagella-associated protein 44, F-BAR domain only protein 1, Suppressor of tumorigenicity 7 protein, Suppressor of tumorigenicity 7 protein-like |
| PSLALP | 3-oxo-5-alpha-steroid 4-dehydrogenase 1, A-kinase anchor protein 1 mitochondrial precursor |
| QPRPPP | Acrosin precursor, Adipocyte enhancer-binding protein 1 precursor, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, Copine-9, Spectrin beta chain non-erythrocytic 2, Mitochondrial dynamics protein MID49, Synapsin-3, TANK-binding kinase 1-binding protein 1, Tensin-1 |
| PCSLKC | ADAMTS-like protein 1 precursor |
| HLTLPF | A-kinase anchor protein 7 isoform gamma |
| LALPAP | Alpha-mannosidase 2C1, Membrane-associated phosphatidylinositol transfer protein 2, Palmitoyltransferase ZDHHC1, Phosphatidylinositol 3-kinase regulatory subunit beta, Probable threonine protease PRSS50 precursor, Serine protease 56 precursor, Uncharacterized protein C11orf24 precursor |
| QAIVSS | Ataxin-2-like protein |
| PPTAPP | B-cell CLL/lymphoma 6 member B protein, B-cell CLL/lymphoma 9-like protein |
| SLSPVP | BCLAF1 and THRAP3 family member 3, Mucin-5AC precursor, Zinc finger and SCAN domain-containing protein 22 |
| APQVLP | Cadherin-2 precursor |
| QEVSRL | Calpain-10, Caseinolytic peptidase B protein homolog precursor, Dehydrogenase/reductase SDR family member 7C precursor, Kinesin-like protein KIFC3, Leucine-rich repeat-containing protein 69, Nucleobindin-1 precursor, Transcription factor MafB |
| RPPPGP | Carbohydrate-responsive element-binding protein, Cyclic GMP-AMP synthase, Helicase SRCAP, Homeobox protein HMX1, Membrane protein FAM174B precursor, Monocarboxylate transporter 3, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2, Proline-rich protein Y-linked, Sprouty-related, EVH1 domain-containing protein 3 |
| APGYDP | Carboxypeptidase N catalytic chain precursor |
| TQPRPP | CCR4-NOT transcription complex subunit 3 |
| KDLQAI, DLQAIK | Cilia- and flagella-associated protein 44 |
| VLPVMH | Coiled-coil domain-containing protein 77 |
| QLPPTA | Constitutive coactivator of PPAR-gamma-like protein 2 |
| AIVSSP | Cullin-9 |
| CTQPRP | Cyclin-G-associated kinase |
| PPLLPH | Dedicator of cytokinesis protein 9 |
| PLLPHS | Dedicator of cytokinesis protein 9, LIM/homeobox protein Lhx8, Probable palmitoyltransferase ZDHHC11B |
| SLALPA | Dedicator of cytokinesis protein 9, Transmembrane protein 130 precursor |
| APPLLP | DNA methyltransferase 1-associated protein 1, Protein AF-17, Synaptojanin-2, Transcription factor HES-4 |
| PPPGPC | DNA polymerase nu, Protein FAM214B, Tastin |
| NTEPSQ | DNA topoisomerase 2-binding protein 1 |
| IDGRVI | DnaJ homolog subfamily B member 5 |
| PLCQDP | Dynein heavy chain 14, axonemal |
| QEVSQA | Dynein heavy chain 3, axonemal |
| APGYDP | E3 ubiquitin-protein ligase HECTD1 |
| LPPFSL | E3 ubiquitin-protein ligase ZNF598 |
| PYTGAV | Endoplasmic reticulum resident protein 29 precursor |
| PSQLPP | Fertilization-influencing membrane protein, Mediator of RNA polymerase II transcription subunit 15, Mitochondrial import inner membrane translocase subunit Tim17-B, Protein KRBA1, Zinc finger and BTB domain-containing protein 32 |
| PSLALP | FLYWCH-type zinc finger-containing protein 1 |
| TAPPLL | Forkhead box protein Q1 |
| PPTAPP | Forkhead box protein Q1, Immediate early response gene 2 protein |
| LQAIKQ, QAKQE | Heterogeneous nuclear ribonucleoprotein C-like 1 |
| DLQAIK | Heterogeneous nuclear ribonucleoprotein C-like 1, Heterogeneous nuclear ribonucleoproteins C1/C2 |
| DGRVIG | Histone acetyltransferase KAT2A, Histone acetyltransferase KAT2B |
| LYPSLA | Host cell factor 2 |
| YPSLAL | Integrin beta-5 precursor |
| MKDLQA | Islet cell autoantigen 1 |
| GAVSSP | Kallikrein-9 precursor |
| PRPPPG | Carbohydrate-responsive element-binding protein, Cyclic GMP-AMP synthase, Homeobox protein HMX1, Kelch domain-containing protein 7A, Microtubule organization protein AKNA, Proline-rich protein 33, RING finger protein 225, Splicing factor 3B subunit 4, Tau-tubulin kinase 2, Trinucleotide repeat-containing gene 6B protein, Proline-rich protein, Y-linked |
| NLDHIL | Kinocilin |
| AIKQEV | La-related protein 1B |
| DPQIQA | Mediator of RNA polymerase II transcription subunit 17 |
| EPSQLP | Membrane primary amine oxidase |
| LSPVPT | Mucin-5AC precursor |
| TQPRPP | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase 1 |
| TGAVSS | Neurofascin precursor |
| LLYPSL | Olfactory receptor 6P1 |
| PLLYPS | Olfactory receptor 6P1, Trace amine-associated receptor 5 |
| ALPAPH | PR domain zinc finger protein 12 |
| PPGPCP | Proline-rich protein 13 |
| AIKQEV | Protein C-ets-1 |
| SQLPPT | Protein KRBA1 |
| PPNHRP | Protein piccolo |
| PPTAPP | Protein sidekick-2 precursor, Stanniocalcin-2 precursor |
| PFSLSP | Protein-glucosylgalactosylhydroxylysine glucosidase |
| LVDAPG | Receptor-type tyrosine-protein kinase FLT3 precursor |
| APQVLP | Regulating synaptic membrane exocytosis protein 1, Serine/threonine-protein kinase WNK1 |
| GAVSSP | RELT-like protein 1 precursor |
| LPPTAP | SH3 and multiple ankyrin repeat domains protein 1 |
| TGAVSS | Sorbin and SH3 domain-containing protein 1 |
| TQPRPP | Spectrin beta chain, non-erythrocytic 2 |
| KFQHDV | Sterile alpha motif domain-containing protein 9-like |
| LATCPE | Testis-expressed protein 38 |
| KDLQAI | Thrombospondin-1 precursor |
| ILEPSI | WW domain-binding protein 4 |
| CPLCQD | Zinc finger homeobox proteins 3 and 4 |
| LEPSIP | Zinc finger homeobox protein 4 |
| VNFTQE | Zinc finger protein 57 homolog, Zinc finger proteins 69, 101, 124, 136, 334, 439, 440, 442, 563, 669, 700, 709, 763, 823, 844, 878 |
| DVNFTQ | Zinc finger protein 136 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lucchese, G.; Jahantigh, H.R.; De Benedictis, L.; Lovreglio, P.; Stufano, A. An Epitope Platform for Safe and Effective HTLV-1-Immunization: Potential Applications for mRNA and Peptide-Based Vaccines. Viruses 2021, 13, 1461. https://doi.org/10.3390/v13081461
Lucchese G, Jahantigh HR, De Benedictis L, Lovreglio P, Stufano A. An Epitope Platform for Safe and Effective HTLV-1-Immunization: Potential Applications for mRNA and Peptide-Based Vaccines. Viruses. 2021; 13(8):1461. https://doi.org/10.3390/v13081461
Chicago/Turabian StyleLucchese, Guglielmo, Hamid Reza Jahantigh, Leonarda De Benedictis, Piero Lovreglio, and Angela Stufano. 2021. "An Epitope Platform for Safe and Effective HTLV-1-Immunization: Potential Applications for mRNA and Peptide-Based Vaccines" Viruses 13, no. 8: 1461. https://doi.org/10.3390/v13081461
APA StyleLucchese, G., Jahantigh, H. R., De Benedictis, L., Lovreglio, P., & Stufano, A. (2021). An Epitope Platform for Safe and Effective HTLV-1-Immunization: Potential Applications for mRNA and Peptide-Based Vaccines. Viruses, 13(8), 1461. https://doi.org/10.3390/v13081461









