Proteomic Analysis of Vero Cells Infected with Pseudorabies Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines, Viruses, Chemicals, and Antibodies
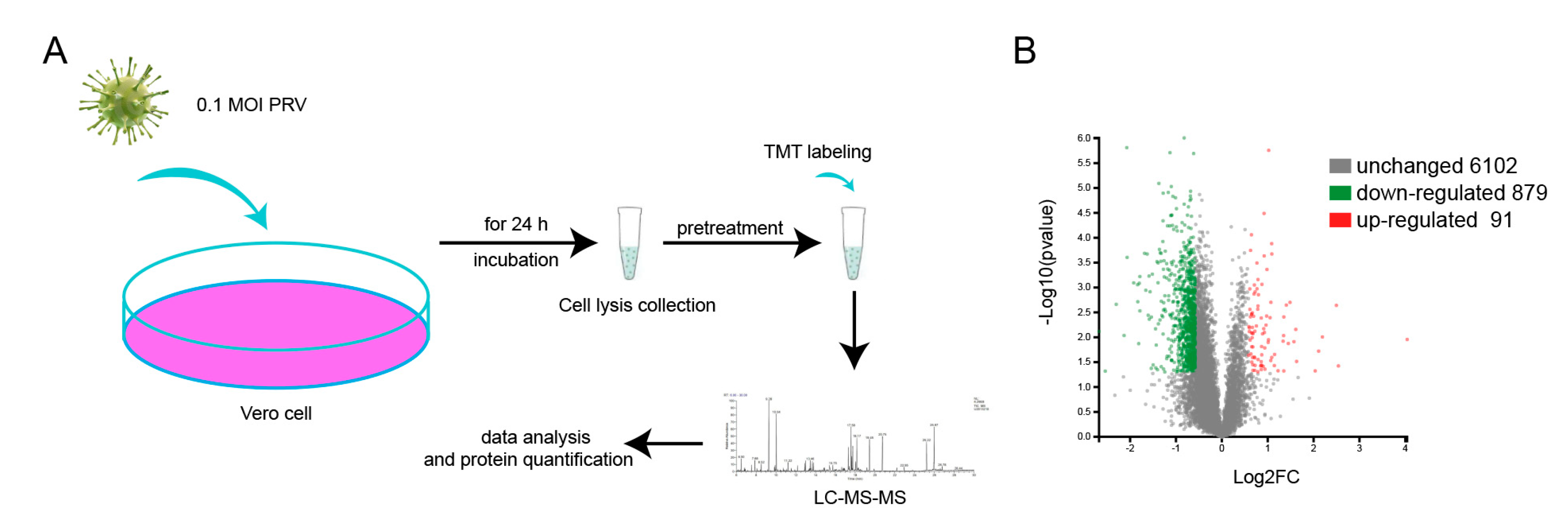
2.2. Virus Inoculation and Protein Preparation
2.3. Reductive Alkylation and TMT Labeling
2.4. Immunofluorescence Assay (IFA)
2.5. RNA Extraction and Real-Time PCR Analysis
2.6. Western Blot Analysis
2.7. Virus Titration
2.8. Data Analysis
3. Results
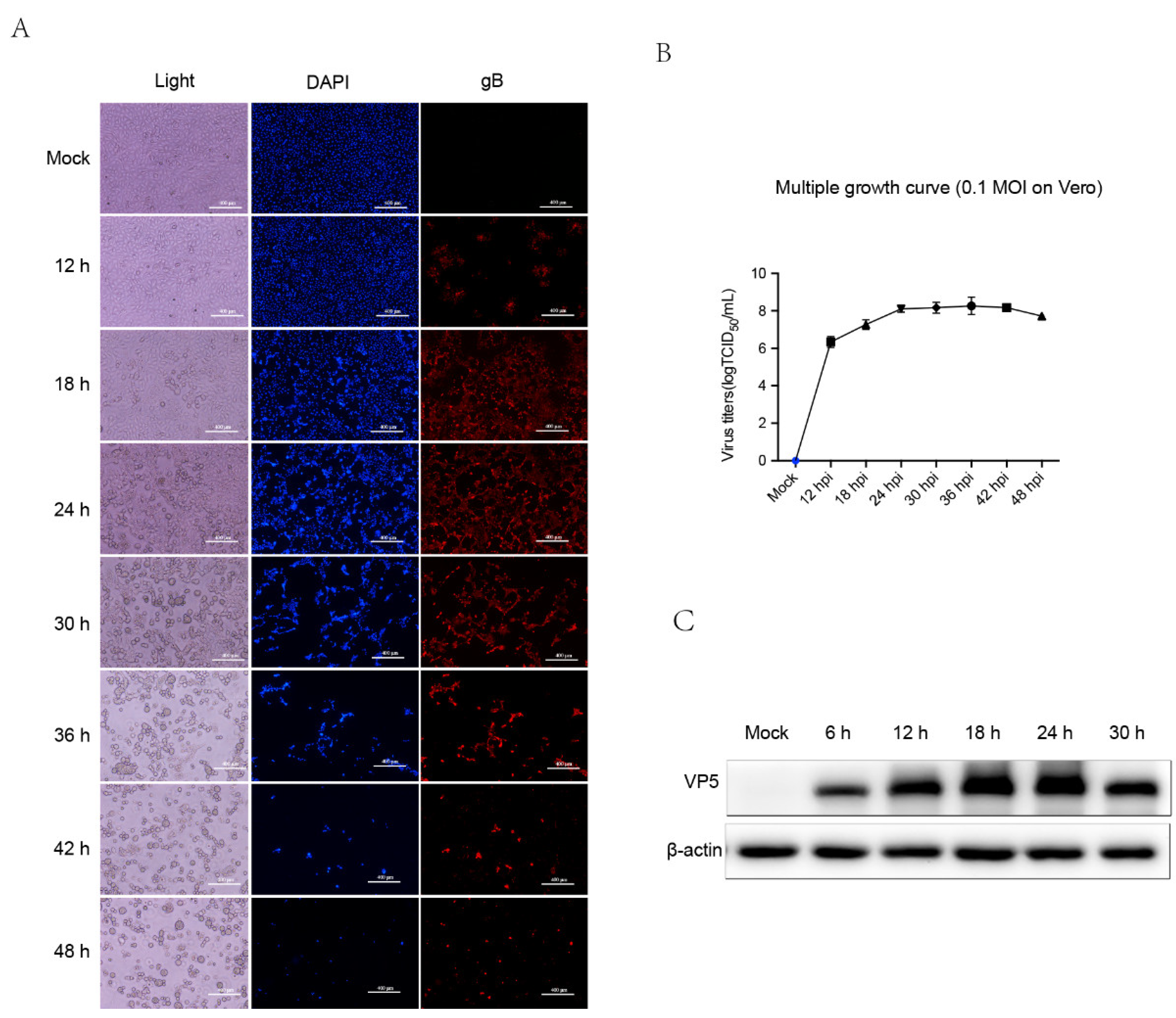
3.1. Kinetics of PRV HB1201 Replication in Vero Cells
3.2. Protein Profiles Determined by TMT/MS Analysis
3.3. Validation of TMT/MS Data by Western Blot and RT-qPCR
3.4. GO Analysis of The DEPs
3.5. KEGG Functional Annotation of DEPs
3.6. COG Annotation of DEPs
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Freuling, C.M.; Muller, T.F.; Mettenleiter, T.C. Vaccines against pseudorabies virus (PrV). Vet. Microbiol. 2017, 206, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Fons, F.; Vidal, D.; Hofle, U.; Vicente, J.; Gortazar, C. Aujeszky’s disease virus infection patterns in European wild boar. Vet. Microbiol. 2007, 120, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Muller, T.; Hahn, E.C.; Tottewitz, F.; Kramer, M.; Klupp, B.G.; Mettenleiter, T.C.; Freuling, C. Pseudorabies virus in wild swine: A global perspective. Arch. Virol. 2011, 156, 1691–1705. [Google Scholar] [CrossRef] [PubMed]
- Pomeranz, L.E.; Reynolds, A.E.; Hengartner, C.J. Molecular Biology of Pseudorabies Virus: Impact on Neurovirology and Veterinary Medicine. Microbiol. Mol. Biol. Rev. 2005, 69, 462–500. [Google Scholar] [CrossRef] [Green Version]
- Yan, K.; Liu, J.; Guan, X.; Yin, Y.X.; Peng, H.; Chen, H.C.; Liu, Z.F. The Carboxyl Terminus of Tegument Protein pUL21 Contributes to Pseudorabies Virus Neuroinvasion. J. Virol. 2019, 93, e02052-18. [Google Scholar] [CrossRef] [Green Version]
- An, T.Q.; Peng, J.M.; Tian, Z.J.; Zhao, H.Y.; Li, N.; Liu, Y.M.; Chen, J.Z.; Leng, C.L.; Sun, Y.; Chang, D.; et al. Pseudorabies virus variant in Bartha-K61-vaccinated pigs, China, 2012. Emerg. Infect. Dis. 2013, 19, 1749–1755. [Google Scholar] [CrossRef]
- Yu, X.; Zhou, Z.; Hu, D.; Zhang, Q.; Han, T.; Li, X.; Gu, X.; Yuan, L.; Zhang, S.; Wang, B.; et al. Pathogenic pseudorabies virus, China, 2012. Emerg. Infect. Dis. 2014, 20, 102–104. [Google Scholar] [CrossRef] [Green Version]
- Tong, W.; Liu, F.; Zheng, H.; Liang, C.; Zhou, Y.J.; Jiang, Y.F.; Shan, T.L.; Gao, F.; Li, G.X.; Tong, G.Z. Emergence of a Pseudorabies virus variant with increased virulence to piglets. Vet. Microbiol. 2015, 181, 236–240. [Google Scholar] [CrossRef]
- Ren, J.; Wang, H.; Zhou, L.; Ge, X.; Guo, X.; Han, J.; Yang, H. Glycoproteins C and D of PRV Strain HB1201 Contribute Individually to the Escape From Bartha-K61 Vaccine-Induced Immunity. Front. Microbiol. 2020, 11, 323. [Google Scholar] [CrossRef]
- He, W.; Auclert, L.Z.; Zhai, X.; Wong, G.; Zhang, C.; Zhu, H.; Xing, G.; Wang, S.; He, W.; Li, K.; et al. Interspecies Transmission, Genetic Diversity, and Evolutionary Dynamics of Pseudorabies Virus. J. Infect. Dis. 2019, 219, 1705–1715. [Google Scholar] [CrossRef]
- Yang, H.; Han, H.; Wang, H.; Cui, Y.; Liu, H.; Ding, S. A Case of Human Viral Encephalitis Caused by Pseudorabies Virus Infection in China. Front. Neurol. 2019, 10, 534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Nian, H.; Li, Z.; Wang, W.; Wang, X.; Cui, Y. Human encephalitis complicated with bilateral acute retinal necrosis associated with pseudorabies virus infection: A case report. Int. J. Infect. Dis. 2019, 89, 51–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Wang, X.; Xie, C.; Ding, S.; Yang, H.; Guo, S.; Li, J.; Qin, L.; Ban, F.; Wang, D.; et al. A novel human acute encephalitis caused by pseudorabies virus variant strain. Clin. Infect. Dis. 2021, 73, e3690–e3700. [Google Scholar] [CrossRef]
- Cheng, Z.; Kong, Z.; Liu, P.; Fu, Z.; Zhang, J.; Liu, M.; Shang, Y. Natural infection of a variant pseudorabies virus leads to bovine death in China. Transbound Emerg Dis 2020, 67, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Lian, K.; Zhang, M.; Zhou, L.; Song, Y.; Wang, G.; Wang, S. First report of a pseudorabies-virus-infected wolf (Canis lupus) in China. Arch. Virol. 2020, 165, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Nelemans, T.; Kikkert, M. Viral Innate Immune Evasion and the Pathogenesis of Emerging RNA Virus Infections. Viruses 2019, 11, 961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, C.; Zhang, R.; Lang, Y.; Shao, A.; Xu, A.; Feng, W.; Han, J.; Wang, M.; He, W.; Yu, C.; et al. Bclaf1 critically regulates the type I interferon response and is degraded by alphaherpesvirus US3. PLoS Pathog. 2019, 15, e1007559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Xu, A.; Qin, C.; Zhang, Q.; Chen, S.; Lang, Y.; Wang, M.; Li, C.; Feng, W.; Zhang, R.; et al. Pseudorabies Virus dUTPase UL50 Induces Lysosomal Degradation of Type I Interferon Receptor 1 and Antagonizes the Alpha Interferon Response. J. Virol. 2017, 91, e01148-17. [Google Scholar] [CrossRef] [Green Version]
- Romero, N.; Van Waesberghe, C.; Favoreel, H.W. Pseudorabies Virus Infection of Epithelial Cells Leads to Persistent but Aberrant Activation of the NF-κB Pathway, Inhibiting Hallmark NF-κB-Induced Proinflammatory Gene Expression. J. Virol. 2020, 94, e00196-20. [Google Scholar] [CrossRef]
- Wang, T.Y.; Yang, Y.L.; Feng, C.; Sun, M.X.; Peng, J.M.; Tian, Z.J.; Tang, Y.D.; Cai, X.H. Pseudorabies Virus UL24 Abrogates Tumor Necrosis Factor Alpha-Induced NF-κB Activation by Degrading P65. Viruses 2020, 12, 51. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Chen, D.; Chen, D.; Hu, Q.; Zhou, L.; Ge, X.; Han, J.; Guo, X.; Yang, H. Pseudorabies virus infection inhibits stress granules formation via dephosphorylating eIF2α. Vet. Microbiol. 2020, 247, 108786. [Google Scholar] [CrossRef] [PubMed]
- Munday, D.C.; Surtees, R.; Emmott, E.; Dove, B.K.; Digard, P.; Barr, J.N.; Whitehouse, A.; Matthews, D.; Hiscox, J.A. Using SILAC and quantitative proteomics to investigate the interactions between viral and host proteomes. Proteomics 2012, 12, 666–672. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Zhang, H.; Ding, Z.; Luo, R.; An, K.; Liu, L.; Bi, J.; Chen, H.; Xiao, S.; Fang, L. Proteome analysis of porcine epidemic diarrhea virus (PEDV)-infected Vero cells. Proteomics 2015, 15, 1819–1828. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Fang, L.; Wang, D.; Song, T.; Wang, T.; Xin, Y.; Chen, H.; Xiao, S. SILAC-based quantitative proteomic analysis of secretome of Marc-145 cells infected with porcine reproductive and respiratory syndrome virus. Proteomics 2016, 16, 2678–2687. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhou, L.; Ge, X.; Guo, X.; Han, J.; Zhang, Y.; Yang, H. Quantitative Proteomic Analysis of Porcine Intestinal Epithelial Cells Infected with Porcine Deltacoronavirus Using iTRAQ-Coupled LC-MS/MS. J. Proteome Res. 2020, 19, 4470–4485. [Google Scholar] [CrossRef] [PubMed]
- Volcy, K.; Fraser, N.W. DNA damage promotes herpes simplex virus-1 protein expression in a neuroblastoma cell line. J. Neurovirol. 2013, 19, 57–64. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Full, F.; Ensser, A. Early Nuclear Events after Herpesviral Infection. J. Clin. Med. 2019, 8, 1408. [Google Scholar] [CrossRef] [Green Version]
- Smith, S.; Weller, S.K. HSV-I and the cellular DNA damage response. Future Virol. 2015, 10, 383–397. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Pearlman, A.H.; Hsieh, P. DNA mismatch repair and the DNA damage response. DNA Repair 2016, 38, 94–101. [Google Scholar] [CrossRef] [Green Version]
- Dempsey, A.; Bowie, A.G. Innate immune recognition of DNA: A recent history. Virology 2015, 479–480, 146–152. [Google Scholar] [CrossRef]
- Ferguson, B.J.; Mansur, D.S.; Peters, N.E.; Ren, H.; Smith, G.L. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. eLife 2012, 1, e00047. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kang, L.; Song, D.; Liu, L.; Yang, S.; Ma, L.; Guo, Z.; Ding, H.; Wang, H.; Yang, B. Ku70 Senses HTLV-1 DNA and Modulates HTLV-1 Replication. J. Immunol. 2017, 199, 2475–2482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sui, H.; Zhou, M.; Imamichi, H.; Jiao, X.; Sherman, B.T.; Lane, H.C.; Imamichi, T. STING is an essential mediator of the Ku70-mediated production of IFN-λ1 in response to exogenous DNA. Sci. Signal. 2017, 10, eaah5054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Yang, S.; Liu, L.; Wang, H.; Yang, B. HTLV-1 Tax impairs K63-linked ubiquitination of STING to evade host innate immunity. Virus Res. 2017, 232, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Yuen, C.K.; Chan, C.P.; Fung, S.Y.; Wang, P.H.; Wong, W.M.; Tang, H.V.; Yuen, K.S.; Chan, C.P.; Jin, D.Y.; Kok, K.H. Suppression of Type I Interferon Production by Human T-Cell Leukemia Virus Type 1 Oncoprotein Tax through Inhibition of IRF3 Phosphorylation. J. Virol. 2016, 90, 3902–3912. [Google Scholar] [CrossRef] [Green Version]
- Johnston, B.P.; Pringle, E.S.; McCormick, C. KSHV activates unfolded protein response sensors but suppresses downstream transcriptional responses to support lytic replication. PLoS Pathog. 2019, 15, e1008185. [Google Scholar] [CrossRef] [Green Version]
- Isaac, R.; Goldstein, I.; Furth, N.; Zilber, N.; Streim, S.; Boura-Halfon, S.; Elhanany, E.; Rotter, V.; Oren, M.; Zick, Y. TM7SF3, a novel p53-regulated homeostatic factor, attenuates cellular stress and the subsequent induction of the unfolded protein response. Cell Death Differ. 2017, 24, 132–143. [Google Scholar] [CrossRef] [Green Version]
- Aloni-Grinstein, R.; Charni-Natan, M.; Solomon, H.; Rotter, V. p53 and the Viral Connection: Back into the Future. Cancers 2018, 10, 178. [Google Scholar] [CrossRef] [Green Version]
- Mehrbod, P.; Ande, S.R.; Alizadeh, J.; Rahimizadeh, S.; Shariati, A.; Malek, H.; Hashemi, M.; Glover, K.K.M.; Sher, A.A.; Coombs, K.M.; et al. The roles of apoptosis, autophagy and unfolded protein response in arbovirus, influenza virus, and HIV infections. Virulence 2019, 10, 376–413. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhang, W.; Liu, Y.; Xie, J.; Hu, C.; Wang, X. Role of p53 in pseudorabies virus replication, pathogenicity, and host immune responses. Vet. Res. 2019, 50, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laval, K.; Vernejoul, J.B.; Van Cleemput, J.; Koyuncu, O.O.; Enquist, L.W. Virulent Pseudorabies Virus Infection Induces a Specific and Lethal Systemic Inflammatory Response in Mice. J. Virol. 2018, 92, e01614-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laval, K.; Van Cleemput, J.; Vernejoul, J.B.; Enquist, L.W. Alphaherpesvirus infection of mice primes PNS neurons to an inflammatory state regulated by TLR2 and type I IFN signaling. PLoS Pathog. 2019, 15, e1008087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kammerl, I.E.; Meiners, S. Proteasome function shapes innate and adaptive immune responses. Am. J. Physiol. Lung Cell. Mol. Physiol. 2016, 311, L328–L336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, M.; Qiu, S.; Zhang, L.; Sun, Y.; Bao, E.; Lv, Y. Pseudorabies virus glycoprotein gE suppresses interferon-β production via CREB-binding protein degradation. Virus Res. 2021, 291, 198220. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Liu, T.; Zhao, J.; Xia, H.; Xie, J.; Guo, Y.; Zhong, L.; Li, M.; Yang, Q.; Peng, C.; et al. DNA-PK deficiency potentiates cGAS-mediated antiviral innate immunity. Nat. Commun. 2020, 11, 6182. [Google Scholar] [CrossRef]
- Jakobsson, A.; Westerberg, R.; Jacobsson, A. Fatty acid elongases in mammals: Their regulation and roles in metabolism. Prog. Lipid Res. 2006, 45, 237–249. [Google Scholar] [CrossRef]
- Lodes, D.E.; Zhu, J.; Tsai, N.P. E3 ubiquitin ligase Nedd4-2 exerts neuroprotective effects during endoplasmic reticulum stress. J. Neurochem. 2022, 160, 613–624. [Google Scholar] [CrossRef]




| Primer Name | Primer Sequence |
|---|---|
| XRCC6-F | GCTCCTTGGTGGATGAGTTT |
| XRCC6-R | CTTGCTGATGTGGGTCTTCA |
| XRCC5-F | TGACTTCCTGGATGCACTAATCGT |
| XRCC5-R | TTGGAGCCAATGGTCAGTCG |
| GAPDH-F | CCTTCCGTGTCCCTACTGCCAAC |
| GAPDH-R | GACGCCTGCTTCACCACCTTCT |
| Accession | Description | FC (P_24h/M_24h) | p Value (P_24h/M_24h) | Significant |
|---|---|---|---|---|
| XP_007997295.1 | 4-hydroxybenzoate polyprenyltransferase, mitochondrial | 16.440879 | 0.0111 | Yes |
| XP_008000412.1 | ATP synthase subunit gamma, mitochondrial isoform X2 | 5.82243 | 0.03774 | Yes |
| XP_007964526.1 | non-homologous end-joining factor 1 | 5.638689 | 0.002289 | Yes |
| XP_007966188.1 | transmembrane 7 superfamily member 3 isoform X1 | 4.570457 | 0.009848 | Yes |
| XP_007985885.1 | DNA-directed RNA polymerase III subunit RPC5 isoform X1 | 4.318305 | 0.01897 | Yes |
| XP_008001057.1 | proton myo-inositol cotransporter | 4.10077 | 0.04733 | Yes |
| XP_007978034.1 | hemoglobin subunit alpha | 3.092117 | 0.03053 | Yes |
| XP_007959284.1 | bromodomain-containing protein 9 isoform X1 | 3.054434 | 0.006955 | Yes |
| XP_008010294.1 | testis-expressed sequence 2 protein isoform X1 | 2.974047 | 0.01225 | Yes |
| XP_007995408.1 | relA-associated inhibitor isoform X1 | 2.802074 | 0.00199 | Yes |
| XP_008014790.1 | tropomodulin-2 isoform X1 | 2.732538 | 0.01336 | Yes |
| XP_007965594.1 | myeloid leukemia factor 2 | 2.647029 | 0.002251 | Yes |
| XP_007958764.1 | kinesin-like protein KIF16B isoform X1 | 2.550698 | 0.003898 | Yes |
| XP_007958522.1 | conserved oligomeric Golgi complex subunit 3 | 2.549788 | 0.04741 | Yes |
| XP_007997053.1 | serum albumin | 2.544889 | 0.009266 | Yes |
| XP_007977282.1 | elongation of very long chain fatty acids protein 1 | 2.521127 | 0.01201 | Yes |
| XP_008008965.1 | vitronectin | 2.339382 | 0.04719 | Yes |
| XP_008012665.1 | calcium signal-modulating cyclophilin ligand | 2.218158 | 0.02212 | Yes |
| XP_007995769.1 | splicing factor, arginine/serine-rich 19 isoform X1 | 2.208965 | 0.037 | Yes |
| XP_008014703.1 | E3 ubiquitin-protein ligase NEDD4 isoform X3 | 2.12792 | 0.000212 | Yes |
| Accession | Description | FC (P_24h/M_24h) | p Value (P_24h/M_24h) | Significant |
|---|---|---|---|---|
| XP_007995562.1 | glioma tumor suppressor candidate region gene 2 protein | 0.156304 | 0.007612 | Yes |
| XP_008007884.1 | UAP56-interacting factor isoform X1 | 0.171554 | 0.04785 | Yes |
| XP_007975472.1 | thioredoxin-interacting protein | 0.202567 | 0.002175 | Yes |
| XP_007971461.1 | tripartite motif-containing protein 40 | 0.2282 | 0.009207 | Yes |
| XP_008016505.1 | general transcription factor II-I | 0.237577 | 0.000002 | Yes |
| XP_008016152.1 | wolframin | 0.239067 | 0.000248 | Yes |
| XP_007972702.1 | structural maintenance of chromosomes flexible hinge domain-containing protein 1 isoform X1 | 0.263824 | 0.000806 | Yes |
| XP_007980315.1 | epsilon-sarcoglycan isoform X1 | 0.280037 | 0.000878 | Yes |
| XP_007966031.1 | zinc finger protein AEBP2 isoform X1 | 0.280576 | 0.001929 | Yes |
| XP_008013602.1 | histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific isoform X1 | 0.284503 | 0.01345 | Yes |
| XP_007962440.1 | regulator of G-protein signaling 10 isoform X1 | 0.287038 | 0.000822 | Yes |
| XP_007982422.1 | vasorin | 0.289883 | 0.000601 | Yes |
| XP_007977511.1 | probable U3 small nucleolar RNA-associated protein 11 isoform X1 | 0.289894 | 0.002949 | Yes |
| XP_008001366.1 | bax inhibitor 1 | 0.311135 | 0.000205 | Yes |
| XP_007962616.1 | antigen KI-67 isoform X1 | 0.316703 | 0.001139 | Yes |
| XP_007960624.1 | ribosome biogenesis protein BMS1 homolog | 0.323469 | 0.000212 | Yes |
| XP_008016787.1 | zinc finger and SCAN domain-containing protein 21 isoform X2 | 0.328314 | 0.003606 | Yes |
| XP_008012144.1 | alpha-protein kinase 2 isoform X1 | 0.332672 | 0.00059 | Yes |
| XP_008013174.1 | treacle protein isoform X1 | 0.334286 | 0.004324 | Yes |
| XP_008007268.1 | solute carrier organic anion transporter family member 2A1 | 0.338268 | 0.001218 | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Xu, S.; Chen, D.; Jiang, R.; Kang, H.; Ge, X.; Zhou, L.; Han, J.; Zhang, Y.; Guo, X.; et al. Proteomic Analysis of Vero Cells Infected with Pseudorabies Virus. Viruses 2022, 14, 755. https://doi.org/10.3390/v14040755
Yang X, Xu S, Chen D, Jiang R, Kang H, Ge X, Zhou L, Han J, Zhang Y, Guo X, et al. Proteomic Analysis of Vero Cells Infected with Pseudorabies Virus. Viruses. 2022; 14(4):755. https://doi.org/10.3390/v14040755
Chicago/Turabian StyleYang, Xintan, Shengkui Xu, Dengjin Chen, Ruijiao Jiang, Haoran Kang, Xinna Ge, Lei Zhou, Jun Han, Yongning Zhang, Xin Guo, and et al. 2022. "Proteomic Analysis of Vero Cells Infected with Pseudorabies Virus" Viruses 14, no. 4: 755. https://doi.org/10.3390/v14040755






