Abstract
Tomato-infecting viruses have been considered as a serious threat to tomato crops in Poland. Therefore, during 2014–2021, 234 tomato samples delivered directly by greenhouse tomato growers to Plant Disease Clinic of IPP-NRI were tested. Eight virus species: pepino mosaic virus (PepMV), tomato yellow ring orthotospovirus (TYRV), tomato spotted wilt orthotospovirus (TSWV), potato virus Y (PVY), cucumber mosaic virus (CMV), tomato black ring virus (TBRV) and tomato mosaic virus (ToMV) were detected in single or mixed infection in 89 samples. The presence of TYRV was established for the first time in Poland in 2014. Since then, its presence has been observed in single and mixed infection with TSWV and CMV. Here, we analysed the genetic variability of TYRV population based on complete nucleocapsid (N) protein gene sequence of 55 TYRV isolates. Maximum-likelihood reconstruction revealed the presence of three distinct, well-supported phylogroups. Moreover, the effect of host species on virus diversity was confirmed. Therefore, RT-LAMP assay was developed for the rapid and efficient detection of TYRV isolates that can be implemented in field and greenhouse conditions.
1. Introduction
Tomatoes are one of the world’s most consumed vegetable crop. On a global scale, the annual production of tomatoes is estimated to be over 180 million tons and their harvesting area is still increasing [1]. After Italy, Spain, Romania, Portugal and Greece, Poland is a significant producer and exporter of tomato in UE [2,3]. The tomato yield quantity and quality depend on many factors from which diseases caused by bacteria, fungi and viruses are one of the major limiting factors influencing the production. Among the tomato pathogens, viruses are particularly difficult to control due to their high level of genetic variability, rapid evolution and adaptation as a consequence of high mutation rates, large population sizes and very short generation times that can favour the evolution of new viral variants [4]. Furthermore, plants are frequently infected by more than one virus (mixed infection), often leading to synergism, resulting in the development of very severe symptoms on infected plants and their fruits. Therefore, the detailed characterisation of viral populations provides relevant information on the processes involved in virus evolution and epidemiology which is crucial for designing reliable diagnostic tools and developing efficient and durable disease control strategies [5]. During the surveys of tomato crops conducted in the past by the Department of Virology and Bacteriology IPP-NRI in Poznań the occurrence of nine virus species: cucumber mosaic virus (CMV, genus Cucumovirus), pepino mosaic virus (PepMV, genus Potexvirus), potato virus M (PVM, genus Carlavirus), potato virus Y (PVY, genus Potyvirus), tobacco mosaic virus (TMV, genus Tobamovirus), tomato black ring virus (TBRV, genus Nepovirus), tomato mosaic virus (ToMV, genus Tobamovirus), tomato torrado virus (ToTV, genus Torradovirus) and tomato spotted wilt orthotospovirus (TSWV, genus Orthotospovirus) was confirmed [6,7,8,9,10,11,12]. The seasonal and spatial variation in the prevalence of viral diseases was observed, but the most frequently found viruses were PepMV and PVY in greenhouse and field tomatoes, respectively [13,14]. The reported viruses occurred in single and mixed infection and caused variable symptoms on tomato plants from very mild to even severe necrosis leading to yield and quality losses. The severity of symptoms might also be correlated with the presence of additional subviral RNAs which can be associated with the genomes of CMV and TBRV [15,16,17,18,19,20]. These particles called satellite RNAs (satRNAs) are relatively short, non-infectious and their replication, encapsidation and spread depend on the helper virus. SatRNAs share little sequence similarity with the viral genomic RNAs and their presence might have a great impact on the virus replication, accumulation and symptoms observed on infected plants [21,22,23].
In 2014, new orthotospovirus species named tomato yellow ring orthotospovirus (TYRV), was found in greenhouse tomatoes for the first time [24] and since then its presence in Poland has been observed in a single or mixed infection. The virus was first reported in Iran as tomato yellow fruit ring virus (TYFRV) in 2002 [25] and later was detected on subsequent economic plants such as potato, soya, peanut pepper, and ornamental plants such as chrysanthemum, gazania, cineraria, anemone and alstroemeria [26,27,28,29,30]. In addition, in 2012 it was detected in tomato plants in Kenya [31]. TYRV has quasi-spherical particles measuring 80–120 nm in diameter and it is transmitted naturally in a persistent propagative manner by several thrips’ species [32,33]. Similar to other orthotospoviruses, the TYRV genome contains three single-stranded (ss) RNA segments designated as small (S), medium (M) and large (L) RNA. The L RNA encodes an RNA-dependent RNA polymerase (RdRp) in a negative-sense orientation, and it is involved in transcription and virus replication. In contrast, each of the M and S RNAs consists of two genes—one in the positive- and the other in the negative-sense orientation. The M RNA encodes a non-structural protein (NSM) involved in cell-to-cell movement and glycoprotein precursor that is post-translationally cleaved into GN and GC glycoproteins associated with thrips transmission. S RNA encodes the gene silencing suppressor (NSs) and nucleocapsid (N) protein [26]. TYRV represents a new threat to the Polish tomato crops and other cultivated plant species. Typically, infected tomato plants have fruit with severe symptoms which do not possess market value (Figure 1).

Figure 1.
Tomato fruits grown in commercial greenhouse infected with TYRV.
It is very likely that, due to the similarity of symptoms induced by TYRV with those induced by TSWV, its presence in Poland remained unnoticed for some time. Therefore, disease management strongly relies on a fast and accurate identification of the causal agent [10]. In recent years, considerable progress has been made in developing high specificity and low detection limit tools for virus identification. Loop-mediated Isothermal Amplification (LAMP) is a frequently used nucleotide amplification technique developed by Notomi et al. [34]. It is a rapid and cost-effective diagnostic tool that enables the isothermal amplification of the target gene, using a DNA polymerase from Bacillus stearothermophilus that has polymerase and reverse transcriptase activity [35]. Due to this activity, LAMP can be easily combined with the reverse transcription reaction, directly detecting target RNA without a separate RT step (RT-LAMP). The LAMP reaction uses pairs of inner and outer primers that can recognize a total of six regions in the target nucleic acid. Two extra loop primers can also be employed (Loop F and Loop B) to accelerate amplification and improve detection performance [35]. The use of six primers ensures high specificity for target amplification; moreover, the loop primers ultimately accelerate the reaction and enable carrying it out within a period of half an hour in comparison when the original LAMP method is used [36]. LAMP amplifies target DNA at isothermal conditions (usually 60–65 °C) and obviates the need for thermal cyclers and postamplification procedures for signal detection. The significant advantages of LAMP assay make it very popular and widely used by many researchers to detect plant and animal viruses [37,38,39,40,41,42].
The aim of this study was the analysis of the incidence of viral diseases in commercial greenhouses in Poland with particular emphasis on orthotospoviruses. Moreover, we analysed the genetic variability and evolutionary processes involved in TYRV diversification. Analyses were performed on 55 TYRV sequences, representing the largest data set that has been analysed for this virus to date. The contribution of the host plant in TYRV population structure was investigated using Bayesian approaches. Finally, we developed an RT-LAMP assay for the rapid and efficient detection of TYRV isolates.
2. Materials and Methods
2.1. Virus Source
During 2014–2021, 234 tomato (Solanum lycopersicum) samples, delivered directly by greenhouse tomato growers to Plant Disease Clinic of IPP-NRI, were tested. Collected tomatoes (var. Tomimaru Muchoo, Torero) originated from twelve different Polish voivodeships. The majority of them came from Greater Poland, Kuyavian-Pomeranian and Masovian. The tested tomato plants showed variable symptoms: chlorotic or necrotic mosaic on leaves and/or fruits, necrotic plots on fruits, bright yellow ring patterns on fruits, chlorotic rings on leaves and/or fruits, leaf malformation and plant stunting suggesting virus infection. From each sample, 100 mg of apical tomato leaves were used for total RNA isolation using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) in accordance with the manufacturer’s instructions.
2.2. Viruses Infecting Tomatoes in Poland
In order to identify the virus infection, the wide range of diagnostic primers detecting CMV, PepMV, PVY, TBRV, tomato brown rugose fruit virus (ToBRFV), tomato chlorosis virus (ToCV), ToMV, ToTV, TMV, TSWV and TYRV were used (Table 1). The selection of primers depended on the observed symptoms and the available information of the cultivation process (presence of vectors, symptoms, temperature conditions, origin of seedlings). Moreover, in case of samples suspected and/or confirmed to be infected with TBRV and CMV, the presence of satellite RNAs was also checked using an appropriate primers pair (Table 1). Isolated total RNAs were used in RT-PCR performed in a 50 μL mixture containing 25 μL of DreamTaq Green PCR Master Mix (ThermoFisher, Watham, MA, USA), 1 μL of RevertAid Reverse Transcriptase (ThermoFisher, Watham, MA, USA), 2 μL of specific primers pair (Table 1) and 22 μL of sterile water. The reaction conditions were as follows: reverse transcription at 42 °C for 20 min, 94 °C for 3 min followed by 30 cycles of 94 °C for 30 s, 45–66 °C (depending on the primers used) for 30 s, 72 °C for 30 s−1 min (depending on the primers used) and a final elongation at 72 °C for 5 min.

Table 1.
Primers used in tomato virus diagnostics.
In a negative control reaction, total RNA isolated from healthy tomatoes was used as a template. RT-PCR products were verified by electrophoresis in 1.5% agarose gel, purified from agarose gels (Zymoclean Gel DNA Recovery Kits, Zymo Research, Irvine, CA, USA) and sequenced by an outsourced company (Genomed S.A., Warsaw, Poland). Obtained sequences were analysed using Standard Nucleotide BLAST online tool (blastn, http://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 30 September 2021), assembled, edited and aligned using BioEdit [52] and SnapGene® software (from Insightful Science; available at snapgene.com).
2.3. Analysis of the TYRV Polish Population
In order to analyse the virus genetic variability, full length of nucleocapsid (N) protein gene sequences of 15 TYRV Polish isolates (previously reported and three described in this study) were compared with the other 40 complete nucleocapsid (N) protein gene sequences of TYRV isolates retrieved from the GenBank database (Supplementary Table S1). The sequence of Kenyan TYRV-Loitoktok was incomplete and therefore was not included in the analysis. The multiple sequence alignments were conducted using MUSCLE [53], as implemented in MEGA X [54]. Single-nucleotide polymorphisms (SNPs) were analysed using SnapGene Viewer software. Sequence identity matrices were displayed using BioEdit and Sequence Demarcation Tool Version 1.2 (SDTv1.2) [55]. Prior to analysis of phylogenetic relationships and selection pressure, the occurrence of recombination and location of recombination breakpoints were investigated using the Datamonkey Adaptive Evolution Server with the Genetic Algorithm for Recombination Detection (GARD) method [56]. Phylogenetic analyses were carried out using the maximum-likelihood method embedded in MEGA X and appropriate nucleotide substitution model (Tamura 3-parameter (T92) with gamma distribution (+G)). Confidence in branch points in the phylogenetic trees was assessed by the bootstrap method, with 1000 pseudorandom replicates. The visualisation of the phylogenetic tree was performed using Evolview v3 webserver [57]. The pervasive and episodic selection were estimated using different methods based on the ratio of nonsynonymous and synonymous substitutions (dN/dS). Diversifying/purifying selection was estimated for individual sites using: Fixed Effects Likelihood (FEL), Fast, Unconstrained Bayesian AppRoximation (FUBAR), Single Likelihood Ancestor Counting (SLAC) and Mixed Effects Model of Evolution (MEME) statistical methods in the Datamonkey Adaptive Evolution Server [58]. The significance value was set to p < 0.05 for the FEL, SLAC, and MEME. The results of FUBAR, based on a Bayesian approach, were accepted when the posterior probability was greater than 0.9.
2.4. Phylogenetic-Trait Association
The posterior distribution of TYRV phylogenies was obtained using the Bayesian Markov chain Monte Carlo (MCMC) approach as available in the BEAST version 2.6.6 package [59]. Analyses were performed based on 50 full-length nucleocapsid (N) protein gene sequences of TYRV isolates. The minimal number of isolates representing one host plant species was two; therefore, the sequences of TYRV-Tri1M, TYRV-Tob2M, TYRV-t/TR-Gaz8, TYRV-A/TR-Ane4 and TYRV-CI/TR-Cin5 isolates were deleted from the analyses. Firstly, the multiple sequence alignment was performed using the MUSCLE algorithm as implemented in MEGA X. Then, to find the best-fitting substitution model for our data the jModelTest was applied [60]. Based on Bayesian information criterion (BIC), Hasegawa-Kishino-Yano (HKY) model with gamma distribution (+G) was chosen as the most proper one. Subsequently, the input files for BEAST analyses were created in BEAST-associated program BEAUTi. Both strict and relaxed molecular clock models and different tree priors were tested and then the obtained results were compared to choose the most adequate combination for our data. The final analysis was performed with the relaxed clock exponential model and coalescent constant population tree prior. The chain length was run for 107 generations with first 104 samples discarded as a burn-in. Convergence of the particular parameters and their effective sample size (ESS > 200) was checked with Tracer v1.7.2 [61]. Finally, maximum clade credibility tree (MCC) was annotated and summarised from the posterior distribution of trees generated by BEAST (after ignoring 10% of trees) using TreeAnnotator v 2.6.3 (as a part of BEAST package). An MCC tree was visualised with FigTree v 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/, accessed on 25 November 2018). Subsequently, phylogeny-trait association was evaluated with BaTS 2.0 [62]. As a trait, the host origin of TYRV isolates was defined. The analysis was performed based on 9001 sample trees generated by BEAST (after removing first 10% of trees). To investigate the host origin distribution along the phylogenetic tree, three different statistics were obtained: parsimony score (PS), association index (AI) and the maximum monophyletic clade size (MC). Both AI and PS enabled us to investigate the overall degree of the host plant structure among the TYRV population, whereas particular MC values allowed us to assess the level of clustering in individual host plants. The values of PS, AI and MC were assigned as statistically significant when the p-value < 0.05.
2.5. RT-LAMP
Eight TYRV isolates were selected for RT-LAMP development including three described in this study. Total RNAs extracted from healthy and infected with TSWV tomato plants were used as negative controls. In the first step, the specific primer pairs were designed based on the complete genome alignment of TYRV isolates nucleocapsid (N) protein gene sequences retrieved from the GenBank database (Supplementary Table S1). The set of diagnostic primers (Table 2) was designed using LAMP Designer (OptiGene, Horsham, UK). The RT-LAMP was performed in a total volume of 25 μL. The reaction mixture contained 15 μL of Isothermal Mastermix (ISO-001nd) (Novazym, Poznań, Poland), 0.5 μL of reverse transcriptase (Roche, Basel, Switzerland), 2 μM of FIP and BIP, 0.5 μM of F3 and B3 primers, 1 μL of tested RNAs and 3.5 μM of sterile water. In order to optimize the reaction conditions, the mixture was incubated at 60 °C, 63 °C, 65 °C and 70 °C for 30 min and 1 h in Biometra T3000 thermocycler (Biometra, Göttingen, Germany). The RT-LAMP products were analysed by electrophoresis in 2% agarose gel and directly by the visual inspection of the reaction tube under UV light after adding 2 μL of EvaGreen®Dye (Biotium, Fremont, CA, USA). In addition, to establish primer specificity, the RT-LAMP was performed using LightCycler ®96 Instrument (Roche, Basel, Switzerland). The reaction consisted of 7.5 μL of Isothermal Mastermix Fluorescent Dye (ISO-001) (Novazym, Poznań, Poland), 1 μM of FIP and BIP, 0.25 μM of F3 and B3 primers, 0.25 μL reverse transcriptase (Roche, Basel, Switzerland) and 0.5 μL of target RNA in a final volume of 12.5 μL. The fluorescence signals were recorded in FAM channel (excitation at 470 nm, detection at 510 nm) for 30 min. The sensitivity of RT-LAMP assay and conventional RT-PCR were estimated and compared. For this purpose, tenfold dilutions of TYRV-H total RNA were used. The RNA concentration was measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), adjusted to 100 ng/μL, and serially diluted to serve as a template for both RT-LAMP and conventional RT-PCR methods. The RT-PCR was performed using 2 μL of TY2-F/TY2-R primers (Table 1), 1 μL of diluted RNA, 25 μL of DreamTaq Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA), 1 μL RevertAid Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) and 22 μL of sterile water. The reaction conditions were as described hereinabove. The detection limit of compared techniques was determined by electrophoresis in 1.5% agarose gel.

Table 2.
Primers used in RT-LAMP reaction.
3. Results
3.1. Viruses Infecting Tomato in Poland
Out of 234 collected tomato samples, 89 were infected by viruses in single or mixed infection (Table 3). The results of RT-PCR were confirmed by sequencing. Predominantly detected virus was PepMV (66.2%), found in tomatoes every year. The results also indicated a high incidence of orthotospoviruses, followed by TYRV (16.9%) and TSWV (13.4%). CMV and PVY were detected at a lower frequency of 7.8% and 2.2%, respectively, whereas TBRV and ToMV were indicated only once. Mixed infection with two of the above- mentioned viruses were observed in 9% of the analysed samples. In case of mixed infection, the symptoms observed on the tested plants were usually more severe. Samples originated from tomato plants which exhibited necrotic mosaic on leaves and considered to be infected with CMV or TBRV were also tested for the presence of satRNAs. The presence of satRNAs was not confirmed in CMV neither TBRV-infected samples.

Table 3.
Tomato samples tested by RT-PCR for 11 virus species grouped by year.
3.2. Analysis of TYRV Population
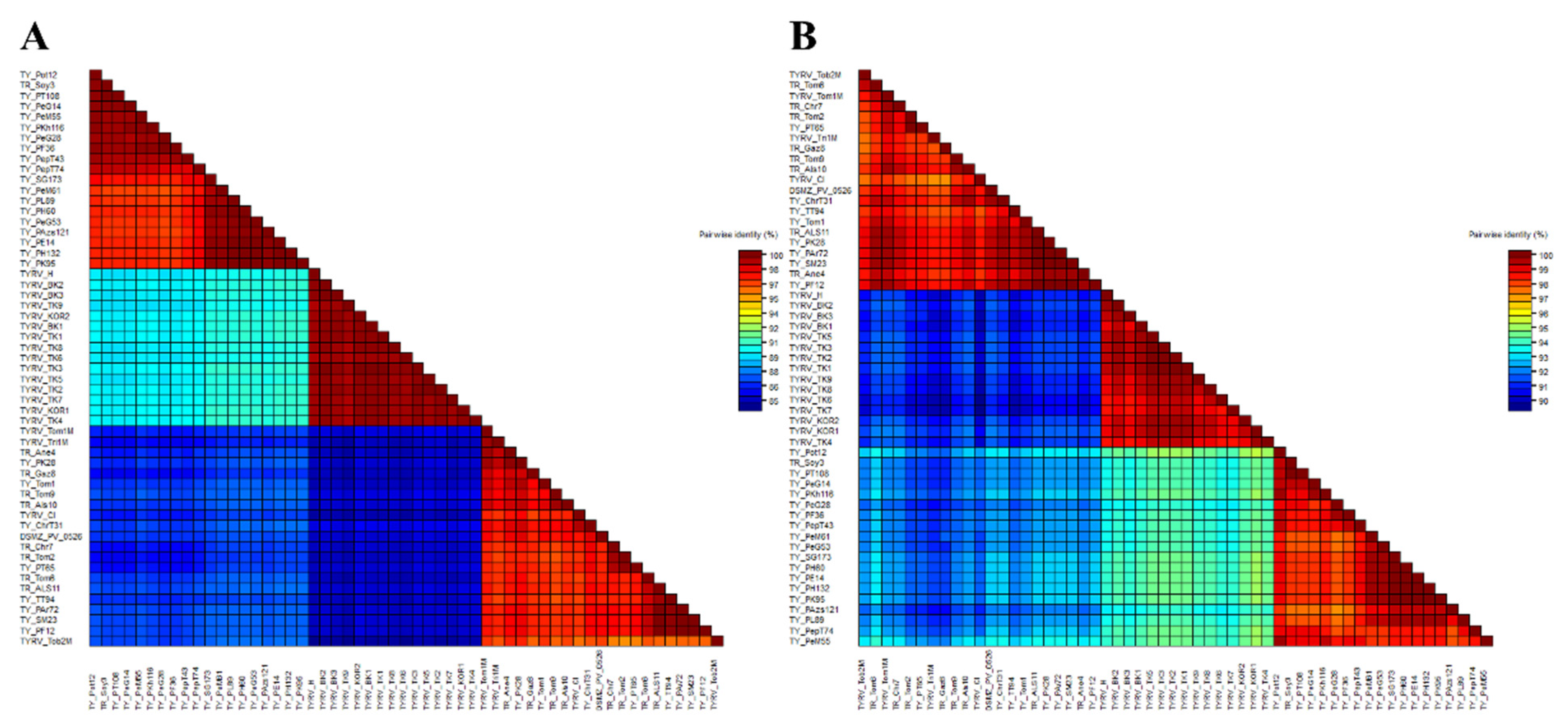
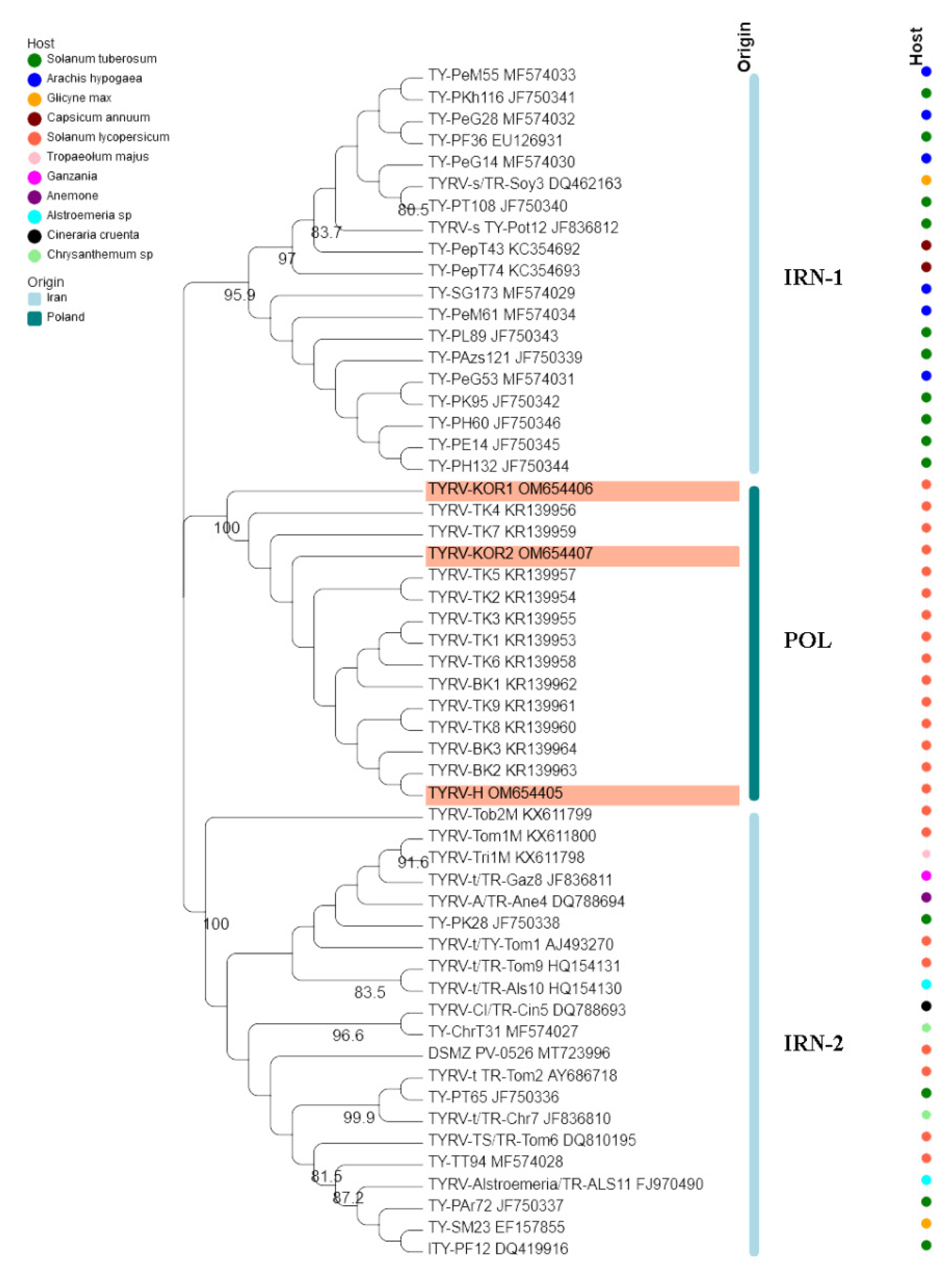
The nucleocapsid (N) protein gene sequences of three new Polish TYRV isolates (TYRV-H, TYRV-KOR1, TYRV-KOR2) of 825 nucleotides in length were deposited in GenBank under accession numbers: OM654405-OM654407, respectively. The nucleotide and amino acid sequence identity of the 15 Polish TYRV nucleocapsid (N) protein gene sequences were relatively high and ranged from 99.2% to 99.8% and from 99.2% to 100%, respectively (Supplementary Table S2). A comparative study of nucleocapsid (N) protein gene sequences of the Polish isolates (compared to TYRV-TK1 sequence) revealed the presence of several point mutations from which 15 were nonsynonymous ones (Supplementary Table S3). The level of genetic diversity within the Polish TYRV population was rather low, but the Polish isolates differ significantly from those collected in Iran. The nucleotide and amino acid nucleocapsid (N) protein gene sequence identity of all 55 TYRV isolates ranged from 83.9% to 100% and from 89.4% to 100%, respectively (Figure 2 and Supplementary Table S4). The sequences of TY-PH60, TY-PE14, TY-PH132 and TY-PK95 were identical, whereas those belonging to the isolates TYRV Tob2-M, TYRV-Cl and TYRV-Tri1M were characterised as the most divergent ones. Recombination analysis showed no evidence of recombination events within the analysed population. Phylogenetic analysis performed on the nucleotide sequences revealed defined clustering by geographical origin (Figure 3). Three geographical subpopulations can be distinguished: cluster of the Polish isolates (POL) and two separate clusters gathering Iranian isolates (IRN-1 and IRN-2). Newly identified TYRV isolates were closely related to the ones previously reported in Poland. Interestingly, no clear clustering according to the host plant was observed. Isolates originated from S. lycopersicum grouped in POL and IRN-1 clusters, whereas isolates from S. tuberculosum and Glycine max were located in both Iranian clusters. Nevertheless, isolates from Arachis hypogaea, Capsicum annum, Alstroemeria and Chrysanthemum isolates grouped together within appropriative Iranian clusters. All isolates from ornamental plants were found in IRN-1 cluster. Selective pressure analysis indicated that TYRV population is driven by purifying selection. The evidence of potential negative selection was obtained for 87 codons, from which 52 were detected by three of the used algorithms (FEL, SLAC, FUBAR) (Supplementary Table S5). Only three codons were identified by two algorithms to be under positive selection: codon 2, 57 and 185.
Figure 2.
Two-dimensional visualization of nucleotide (A) and amino acid (B) nucleocapsid (N) protein gene sequence identity of 55 tomato yellow ring virus (TYRV) isolates used in this study. The matrices were performed using SDTv1.2.
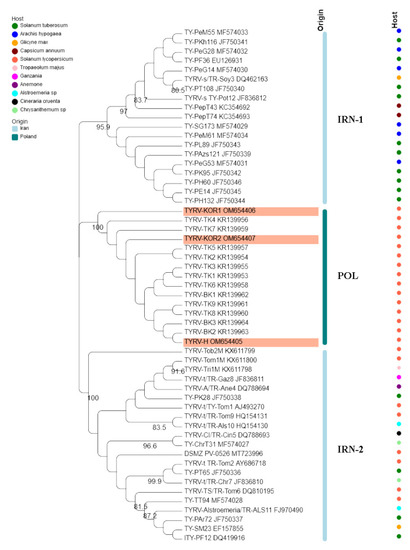
Figure 3.
Phylogenetic tree constructed based on nucleocapsid (N) protein gene nucleotide sequences of 55 tomato yellow ring virus (TBRV) isolates. Double names of some isolates correspond to names of isolates in GenBank and those used by Golnaraghi et al. in their work [30]. Host species and country of origin were represented by dots and lines, respectively. Newly described Polish TYRV isolates are highlighted.
3.3. Host Driven Structure of the TYRV Population
In order to identify the host-driven structure in the TYRV population, firstly, the Bayesian phylogeny for 50 nucleocapsid (N) protein gene sequences was inferred (Supplementary Figure S1). Secondly, three summary statistics were calculated: AS, PS and MC. The values of AS, PS and MC and their statistical support were presented in (Table 4). Analyses included TYRV isolates originated from: S. lycopersicum (22 isolates), Alstroemeria sp. (2 isolates), Chrysanthemum sp. (2 isolates), G. max (2 isolates), S. tuberosum (14 isolates), A. hypoagea (6 isolates) and C. annuum (2 isolates) (Suplementary Table S1). The sample size required for analyses was at least two and, therefore, the isolates from Tropaeolum majus (1 sample), N. tabacum (1 sample), Gazania sp. (1 sample), Anemone (1 sample) and Cineraria cruenta (1 sample) were excluded due to an insufficient sample size. Despite the reduced number of isolates used in analyses, the topology of obtained MCC tree was consistent with this presented by a maximum likelihood tree (Supplementary Figure S1). A phylogenetic trait association test allowed us to confirm that there is an overall effect of host species in the distribution of TYRV variability, as shown by significant AI and PS values (p-value < 0.00). Nonetheless, only two subpopulations of TYRV isolates retrieved from S. lycopersicum and S. tuberosum showed differentiation, whereas no statistically significant results were obtained for Alstroemeria sp., Chrysanthemum sp., G. max, A. hypoagea, and C. annuum (Table 4).

Table 4.
Bayesian statistic measuring the association between host plant and clustering observed in the MCC tree. Statistical supported values are bolded.
3.4. Optimisation of the RT-LAMP Conditions
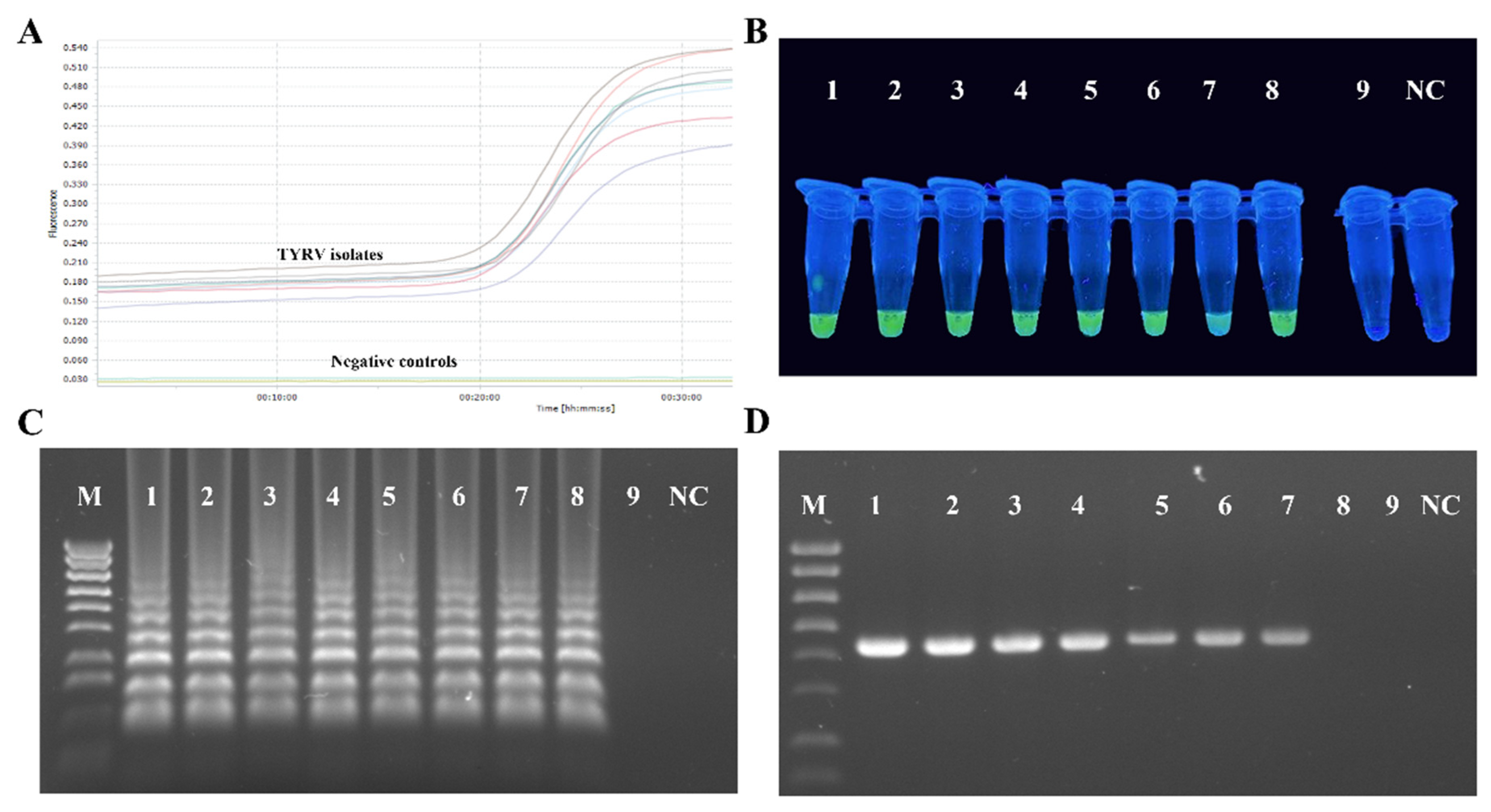
RT-LAMP primers designed in this study efficiently amplified all the tested TYRV isolates (Figure 4A). The amplification products, visible as ladder-like DNA fragments, were obtained in all the tested temperatures and time periods, but the most optimal conditions were 63 °C and 60 min of reaction time. Samples from TSWV-infected and healthy plants were negative. Green colour, indicating a positive reaction, was observed in UV light after adding the dye only for the TYRV-infected samples and no colour changes were seen for the negative controls (Figure 4B). A sensitivity test for the RT-LAMP method revealed it was ten times more sensitive than conventional RT-PCR (Figure 4C,D).
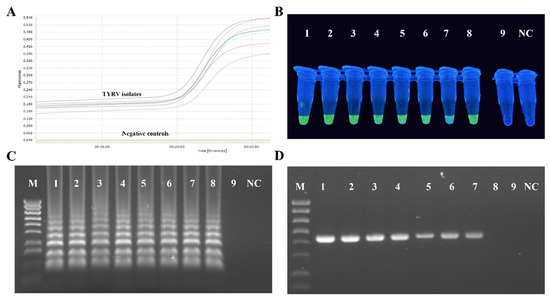
Figure 4.
Results of RT-LAMP optimalization. (A) Amplification plots (showed with different colours) of eight tested TYRV samples monitored in the LightCycler® 96 (Roche, Basel, Switzerland). Total RNAs isolated from healthy and TSWV-infected plants were used as negative controls. (B) Visual detection of RT-LAMP products using EvaGreen®Dye (Biotium, Fremont, CA, USA) of eight TYRV isolates (TYRV-TK1, TYRV-TK2, TYRV-TK4, TYRV-BK1, TYRV-BK2, TYRV-H, TYRV-KOR1, TYRV-KOR2, probes 1–8, respectively), TSWV isolate (probe 9) and healthy plant (NC). (C) Electrophoretic separation of RT-LAMP products in a 2% agarose gel. M- Gene Ruler 1 Kb Plus Ladder (ThermoFisher Scientific, Watham, MA, USA), lines 1–9—ten-fold dilutions of total RNA of TYRV-H isolates, NC-negative control. (D) Electrophoretic separation of RT-PCR products in 1.5% agarose gel. M-Gene Ruler 1 Kb Plus Ladder (ThermoFisher Scientific, Watham, MA, USA), lines 1–9 ten-fold dilutions of total RNA of TYRV-H isolates, NC-negative control.
4. Discussion
Plant viral diseases cause epidemics in crops that are often too complex to control and threaten food security. This difficulty is mainly due to the lack of early detection and ineffective countermeasures, especially for emerging viruses. Furthermore, mixed infection affecting a single plant or crop are recognised to be common in plant disease epidemics. Mixed infection can generate a range of ecological interactions that have an impact on viral fitness, and consequently have far-reaching epidemiological implications. From over 130 viruses knowing to infect tomatoes, about 50 species are considered as serious threat causing several problems in field-grown and greenhouse crops [63,64]. During 2014–2021, in order to analyse the occurrence of viruses infecting greenhouse tomato in Poland 234 plant samples were tested. The majority of the collected samples with virus-like symptoms were not infected with any of tested viruses. It is likely that visible symptoms were caused by physiological plant issues, inadequate dose of fertilisers or inappropriate use of plant protection products. We also cannot exclude the possibility that samples were infected with other virus species, designed and used primer pairs were not complementary to present viral isolates or the virus concentration in tissue was too low to be detected using conventional PCR. These problems can be solved by using high-throughput sequencing (HTS), which was proved to be very successful for virus discovery to resolve disease etiology in many agricultural crops. The greatest advantage of HTS over other diagnostic approaches is that it gives a complete view of the viral phytosanitary status of a plant [65]. As a diagnostic tool, HTS is perhaps more broad-spectrum than any previously used assay. Nevertheless, using HTS for routine virus detection is limited due to high costs, equipment requirement and computing equipment capable of storing and processing large datasets, as well as expertise on genome assembly analysis pipelines and packages [66].
Among infected samples, PepMV was the most frequently detected. It is a rapidly emerging virus, which has been established as one of the most important viral diseases in tomato production worldwide [63]. In Poland, several isolates belonging to the European and Chilean 2 genotypes have been found since 2003 [8]. Since then, the virus has been constantly detected in tomato crops in single and mixed infection [8,67]. In 2017 and 2018, plant protection products PMV®-01 of Belgian DCM company and V10 of Dutch Valto company were registered in Poland, respectively. The vaccination strategy is based on the principle of cross-protection, where a mild isolate of the virus is used to protect plants against more severe ones. Since then, the number of PepMV-infected samples has been decreasing in comparison to previous years. Nevertheless, PepMV still remains one of the most important threats to tomato cultivation due to its high genetic variability, quasispecies nature and creation of new variants by mutations and recombinations [8,68]. TYRV in single and mixed infection with TSWV was found in tomato plants for the first time in 2014 [24]. Orthotospoviruses are one of the major threats to vegetable crops worldwide. Research conducted in Plant Disease Clinic of IPP-NRI, Poznań, revealed that TSWV has been mostly identified in ornamental plants such as chrysanthemum or gerbera. On the other hand, in the following years, its presence was also confirmed in tomato, and the infected fruits did not have any commercial value. Apart from TSWV, TYRV was also detected in a mixed infection with other orthotospoviruses such as iris yellow spot orthotospovirus (IYSV) and impatiens necrotic spot orthotospovirus (INSV) [69]. In 2021, otherwise than earlier, TYRV was detected in a mixed infection with CMV, which is the first evidence of a TYRV infection with the virus representing a different genus. CMV (genus Cucumovirus) has one of the broadest host range among plant viruses and is responsible for important agronomic losses in many crops worldwide [70]. During tomato surveys in Poland, CMV has been occasionally detected, in single infection in four samples and in mixed infection with PepMV and TYRV. Co-infection with CMV and viruses from another genus (such as Crinivirus, Potexvirus, Potyvirus, Tobamovirus) led to synergists interactions affecting the symptoms development and increasing the CMV accumulation [70]. In Poland, CMV is definitely more often identified on cucumber and zucchini than tomato [71,72]. What is more, the CMV genome might be associated with satRNAs which are responsible for the development of a lethal disease in tomato. Epidemics of satellite-RNA-containing isolates have been reported in France, Spain, Italy and Greece [73,74,75,76]. The D-satRNA, B-satRNA, and WL1-satRNA induce necrosis, chlorosis, and attenuation, respectively [23]. Therefore, tomato samples exhibited necrotic mosaic on leaves and identified to be infected with CMV were additionally tested for the presence of satRNAs but no additional subviral particles were detected. Another virus species identified in tomato population was PVY. It was first reported in tomato greenhouse production in 2002 [10] and since then the presence of the virus in Poland has been occasionally detected. The situation changed in 2008 when the increased distribution of the PVYNWi-P strain was observed [77]. The surveys conducted in greenhouse and fields during 2012–2013 revealed that PVY population is much more diverse and the isolates representing different strains were identified: PVYNTN, PVY0, PVYNWi-P, PVYNN242 and new recombinant variant between PVYNTN and PVYNWi-P. The predominant strain in greenhouse population was PVYNWi-P and this strain was also identified in 2020 suggesting its prevalence in Poland. The remaining viruses: TBRV and ToMV were detected only once, and it seems that they are not a threat to tomato production. Tobamoviruses on tomato have been rarely detected but lately they were identified in irrigation ditches and drainage canals surrounding fields [78] which can be potential virus source for tomato field infection. In case of TBRV, the presence of satRNAs was also verified as they might have an impact on virus replication and accumulation. None of the subviral RNAs were detected in the tested sample.
Recently, new emerging virus species have been identified in the EU. One of them was ToBRV, firstly reported in a greenhouse tomato crop in 2014 in Jordan [79]. To date, the incidence of ToBRV infection has been reported in 34 countries, including Poland, where the virus was found for the first time in tomato plant and seeds in 2020 [80]. The second one is ToCV, which is considered as a serious production problem for field and greenhouse tomato growers [81]. Therefore, tomato samples, which exhibited symptoms such as mild foliar symptoms with clearly brown rugose symptoms on fruits, interveinal chlorosis, leaf brittleness and limited necrotic flecking or leaf bronzing on leaves, were tested for the occurrence of those viruses. No samples were confirmed to be infected with ToBRFV and ToCV.
Pathogen diagnostics plays an important role in crop protection and disease management. The detailed analysis of genetic structure of virus population is essential to develop effective, sensitive, low-time and cost detection method to identify viruses especially when new pathogen species emerged. Therefore, in this work we decided to analyse the evolutionary dynamics of TYRV which presence has been observed in Poland since 2014. The phylogenetic analyses divided TYRV populations into three clusters corresponding to geographical regions of their origin. The phylogenetic division into two Iranian clusters was reported previously [30,82]. Based on the results obtained by Golnaraghi et al. the isolate from Kenya grouped in Iranian cluster 2 [30]. The Polish isolates came from two neighbouring provinces known for tomato greenhouse production (Kuyavian-Pomeranian and Greater Poland) [50], whereas Iranian ones were collected from distinct provinces such as Fars, Teheran, Markazi, Mazandaran [25,26,28,58]. These distant locations suggest virus transmission with plant propagative materials [83]. Additional analyses revealed that the TYRV population exhibits some host-driven structure, especially in reference to isolates originated from tomato and potato. These results generally correspond to grouping on the phylogenetic tree, but they should be interpreted with caution due to the overabundance of tomato and potato isolates in comparison to the representants of other host plants.
The genetic structure and diversity of viral populations can be affected by several factors, such as recombination and selective pressure. Recombination clearly plays a significant role in the evolution of RNA viruses by generating genetic variation, and is beneficial in the case of genes that are under diversifying selection [84]. The recombination analysis performed based on full-length nucleocapsid (N) protein gene sequences of 55 TYRV isolates showed no evidence of recombination. In addition, within this population, purifying selection was found to be predominant. It is assumed that purifying selection acts on codons, in which change may adversely affect the properties of the virus and, therefore, genetic variants with these mutations will be eliminated from the virus population. Similar results were obtained by Golnaraghi group [30] who used a smaller number of isolates and different methods to analyse the TYRV population structure. In the same work, the authors indicated that the TYRV Polish population recently increases in size preceded by a genetic bottleneck, i.e., a founder effect [30].
Disease management strongly relies on a fast and accurate identification of the causal agent [85]. Several diagnostic techniques have been developed for TYRV detection including both serological and molecular methods [31,51,86]. Double antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA) is widely used in large-scale screening for TYRV infection in various types of plants; however, the molecular techniques are more sensitive than the serological ones. Low virus titre can limit sensitivity, producing false negatives when the virus concentration is under the technique detection threshold [85]. Therefore, we aimed to develop an RT-LAMP assay for the reliable and rapid diagnostics of TYRV. LAMP exhibits a higher sensitivity in comparison to PCR and is less affected by inhibitors present in tomato plants (e.g., phenols and complex polysaccharides), which are often a cause of false negative results. LAMP assay does not require a highly purified nucleic acid template and it also may be performed effectively with crude extract from an infected plant. Therefore, it can be carried out using lateral flow devices, making it suitable for onsite detection or testing in the field. The products of LAMP can be detected much faster than in standard techniques, sometimes only requiring analysis with the naked eye [87]. It has been reported that LAMP assay can be even 1000 times more sensitive than conventional PCR with equivalent specificity [88]. The primers for RT-LAMP were designed based on the alignment of the complete nucleocapsid (N) protein gene of TYRV in order to amplify the wide range of TYRV isolates. RT-LAMP assay developed in this study is capable of detecting TYRV isolates within 1 h. The result of RT-LAMP was direct visualized under UV light by adding fluorescent dye and in real-time conditions. The sensitivity was tenfold higher than that of RT-PCR. Our results indicate that the TYRV RT-LAMP assay is sensitive and specific, and has the potential to be developed into a field or greenhouse diagnostic test.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v14071405/s1, Table S1. List of TYRV isolates of which nucleocapsid (N) protein gene sequences were used in this study. Table S2. Percentage of nucleotide (bottom part of table, nt) and amino acid (upper part of table, aa) sequences identity of nucleocapsid (N) protein gene sequences of Polish TYRV isolates. Table S3. Number of mutations observed in the nucleocapsid (N) protein gene sequences of the TYRV Polish isolates, in comparison to TYRV-TK1 sequence. Table S4. Percentage of nucleotide (bottom part of table, nt) and amino acid (upper part of table, aa) sequences identity of N gene sequences of 55 TYRV isolates. Table S5. Codon positions detected under selective pressure using FEL, SLAC, FUBAR and MEME algorithms. Figure S1. MCC tree constructed based on 50 TYRV full-length nucleocapsid (N) protein gene nucleotide sequences. Numbers above the brunches represent Bayesian posterior probabilities (BPP).
Author Contributions
Conceptualization, A.Z.-N. and B.H.-J.; Methodology, A.Z.-N. and B.H.-J.; Software, A.Z.-N., D.B. and A.T.; Formal Analysis, A.Z.-N., J.M., D.B., A.T. and N.J.; Resources, N.B.-F., B.H.-J., A.Z.-N., J.M. and D.B.; Writing—Original Draft Preparation, A.Z.-N.; Writing—Review and Editing, A.Z.-N., B.H.-J., D.B., A.T., J.M. and N.B.-F.; Visualization, A.Z.-N., D.B. and A.T.; Supervision, B.H.-J.; Project Administration, B.H.-J.; Funding Acquisition, B.H.-J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially supported by the Polish National Science Centre project number 2017/25/B/NZ9/01715.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to kindly thank our former co-worker Natalia Rymelska for excellent technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/home/en (accessed on 17 March 2022).
- Statistics|Eurostat. Available online: https://ec.europa.eu/eurostat/databrowser/view/tag00115/default/table?lang=en (accessed on 25 April 2022).
- Statistics Poland—Production and Foreign Trade of Agricultural Products in 2019. Available online: https://stat.gov.pl/obszary-tematyczne/rolnictwo-lesnictwo/uprawy-rolne-i-ogrodnicze/produkcja-i-handel-zagraniczny-produktami-rolnymi-w-2019-roku,1,16.html (accessed on 25 April 2022).
- Elena, S.F.; Fraile, A.; García-Arenal, F. Evolution and Emergence of Plant Viruses. Adv. Virus Res. 2014, 88, 161–191. [Google Scholar] [CrossRef] [PubMed]
- Rubio, L.; Guerri, J.; Moreno, P. Genetic Variability and Evolutionary Dynamics of Viruses of the Family Closteroviridae. Front. Microbiol. 2013, 4, 151. [Google Scholar] [CrossRef] [PubMed]
- Jończyk, M.; Le Gall, O.; Pałucha, A.; Borodynko, N.; Pospieszny, H. Cloning and Sequencing of Full-Length CDNAs of RNA1 and RNA2 of a Tomato Black Ring Virus Isolate from Poland. Arch. Virol. 2004, 149, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Pospieszny, H.; Borodynko, N.; Obrępalska-Stęplowska, A.; Hasiów, B. The First Report of Tomato Torrado Virus in Poland. Plant Dis. 2007, 91, 1364. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Borodynko, N.; Jackowiak, P.; Figlerowicz, M.; Pospieszny, H. Pepino Mosaic Virus—A Pathogen of Tomato Crops in Poland: Biology, Evolution and Diagnostics. J. Plant Prot. Res. 2010, 50, 470–476. [Google Scholar] [CrossRef]
- Laskowska, D. Characterization of Tomato Spotted Wilt Disease on Tomato and Vector Role in Transmission. Stud. Rep. Inst. Soil Sci. Plant Cultiv. 2008, 13, 43–50. [Google Scholar]
- Pospieszny, H. Nasilone wystepowanie chorob wirusowych pomidora szklarniowego w Wielkopolsce, w roku 2002. Prog. Plant Prot. 2003, 43, 321–324. [Google Scholar]
- Moycho, W.; Gubanski, M.; Fomaidis, B.; Lemanska, M.; Wajsbard, E. Występowanie wirusa mozaiki tytoniowej na plantacjach pomidorów miasta Łodzi i okolicy. Postępy Nauk Rolniczych 1960, 7, 79–82. [Google Scholar]
- Minicka, J.; Zarzyńska-Nowak, A.; Budzyńska, D.; Borodynko-Filas, N.; Hasiów-Jaroszewska, B. High-Throughput Sequencing Facilitates Discovery of New Plant Viruses in Poland. Plants 2020, 9, 820. [Google Scholar] [CrossRef]
- Minicka, J.; Hasiów-Jaroszewska, B.; Borodynko-Filas, N.; Pospieszny, H.; Hanssen, I.M. Rapid Evolutionary Dynamics of the Pepino Mosaic Virus—Status and Future Perspectives. J. Plant Prot. Res. 2016, 56, 337–345. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Stachecka, J.; Minicka, J.; Sowiński, M.; Borodynko, N. Variability of Potato Virus Y in Tomato Crops in Poland and Development of a Reverse-Transcription Loop-Mediated Isothermal Amplification Method for Virus Detection. Phytopathology 2015, 105, 1270–1276. [Google Scholar] [CrossRef] [PubMed]
- Gould, A.R.; Palukaitis, P.; Symons, R.H.; Mossop, D.W. Characterization of a Satellite RNA Associated with Cucumber Mosaic Virus. Virology 1978, 84, 443–455. [Google Scholar] [CrossRef]
- Aranda, M.A.; Fraile, A.; Garcia-Arenal, F. Genetic Variability and Evolution of the Satellite RNA of Cucumber Mosaic Virus during Natural Epidemics. J. Virol. 1993, 67, 5896–5901. [Google Scholar] [CrossRef] [PubMed]
- Murant, A.F.; Mayo, M.A.; Harrison, B.D.; Goold, R.A.Y. Evidence for Two Functional RNA Species and a ‘Satellite’ RNA in Tomato Black Ring Virus. J. Gen. Virol. 1973, 19, 275–278. [Google Scholar] [CrossRef]
- Oncino, C.; Hemmer, O.; Fritsch, C. Specificity in the Association of Tomato Black Ring Virus Satellite RNA with Helper Virus. Virology 1995, 213, 87–96. [Google Scholar] [CrossRef]
- Hemmer, O.; Meyer, M.; Greif, C.; Fritsch, C.Y. Comparison of the Nucleotide Sequences of Five Tomato Black Ring Virus Satellite RNAs. J. Gen. Virol. 1987, 68, 1823–1833. [Google Scholar] [CrossRef]
- Shimura, H.; Pantaleo, V.; Ishihara, T.; Myojo, N.; Inaba, J.; Sueda, K.; Burgyán, J.; Masuta, C. A Viral Satellite RNA Induces Yellow Symptoms on Tobacco by Targeting a Gene Involved in Chlorophyll Biosynthesis Using the RNA Silencing Machinery. PLoS Pathog. 2011, 7, e1002021. [Google Scholar] [CrossRef]
- Escriu, F.; Fraile, A.; García-Arenal, F. Evolution of Virulence in Natural Populations of the Satellite RNA of Cucumber Mosaic Virus. Phytopathology 2000, 90, 480–485. [Google Scholar] [CrossRef]
- Takanami, Y. A Striking Change in Symptoms on Cucumber Mosaic Virus-Infected Tobacco Plants Induced by a Satellite RNA. Virology 1981, 109, 120–126. [Google Scholar] [CrossRef]
- Xu, P.; Roossinck, M.J. Cucumber Mosaic Virus D Satellite RNA–Induced Programmed Cell Death in Tomato. Plant Cell 2000, 12, 1079–1092. [Google Scholar] [CrossRef]
- Zarzyńska-Nowak, A.; Rymelska, N.; Borodynko, N.; Hasiów-Jaroszewska, B. The Occurrence of Tomato Yellow Ring Virus on Tomato in Poland. Plant Dis. 2016, 100, 234. [Google Scholar] [CrossRef]
- Winter, S.; Koerbler, M.; Shahraeen, N.; Katul, L.; Lesemann, D.E. Characterization of a New Tospovirus Species Infecting Tomato in Iran. In Proceedings of the First Joint Conference of International Working Groups on Legume Viruses and Vegetable Viruses, Bonn, Germany, 4–9 August 2002. [Google Scholar]
- Hassani-Mehraban, A.; Saaijer, J.; Peters, D.; Goldbach, R.W.; Kormelink, R. Molecular and Biological Comparison of Two Tomato Yellow Ring Virus (TYRV) Isolates: Challenging the Tospovirus Species Concept. Arch. Virol. 2007, 152, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Rasoulpour, R.; Izadpanah, K. Characterisation of Cineraria Strain of Tomato Yellow Ring Virus from Iran. Australas. Plant Pathol. 2007, 36, 286–294. [Google Scholar] [CrossRef]
- Golnaraghi, A.R.; Pourrahim, R.; Ahoonmanesh, A.; Zamani-Zadeh, H.R.; Farzadfar, S. Detection and Characterization of a Distinct Isolate of Tomato Yellow Fruit Ring Virus from Potato. Plant Dis. 2008, 92, 1280–1287. [Google Scholar] [CrossRef] [PubMed]
- Beikzadeh, N.; Bayat, H.; Jafarpour, B.; Rohani, H.; Peters, D.; Hassani-Mehraban, A. Infection of Alstroemeria Plants with Tomato Yellow Ring Virus in Iran. J. Phytopathol. 2012, 160, 45–47. [Google Scholar] [CrossRef]
- Golnaraghi, A.; Shahraeen, N.; Nguyen, H.D. Characterization and Genetic Structure of a Tospovirus Causing Chlorotic Ring Spots and Chlorosis Disease on Peanut; Comparison with Iranian and Polish Populations of Tomato Yellow Fruit Ring Virus. Plant Dis. 2018, 102, 1509–1519. [Google Scholar] [CrossRef]
- Birithia, R.; Subramanian, S.; Villinger, J.; Muthomi, J.W.; Narla, R.D.; Pappu, H.R. First Report of Tomato Yellow Ring Virus (Tospovirus, Bunyaviridae) Infecting Tomato in Kenya. Plant Dis. 2012, 96, 1384. [Google Scholar] [CrossRef]
- Mortazavi, N.; Aleosfoor, M.; Minaei, K. Transmission of Cineraria Isolate of Tomato Yellow Ring Virus by Frankliniella occidentalis and Thrips tabaci (Thysanoptera, Thripidae). Linz. Biol. Beiträge J. 2014, 45, 2011–2018. [Google Scholar]
- Mortazavi, N.; Aleosfoor, M. Efficiency of Thrips tabaci and Frankliniella occidentalis Populations in Transmission of Tomato Yellow Ring Virus. Zool. Ecol. 2015, 25, 241–246. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Chaouch, M. Loop-Mediated Isothermal Amplification (LAMP): An Effective Molecular Point-of-Care Technique for the Rapid Diagnosis of Coronavirus SARS-CoV-2. Rev. Med. Virol. 2021, 31, e2215. [Google Scholar] [CrossRef] [PubMed]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated Reaction by Loop-Mediated Isothermal Amplification Using Loop Primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Trzmiel, K.; Hasiów-Jaroszewska, B. Development of Loop-Mediated Isothermal Amplification Assay for Rapid Detection of Genetically Different Wheat Dwarf Virus Isolates. Mol. Biol. Rep. 2020, 47, 8325–8329. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Liang, L.; Ren, X.; Jia, Y.; Han, K.; Zhao, J.; Song, C.; Cui, S. Development of a TaqMan Loop-Mediated Isothermal Amplification Assay for the Rapid Detection of Pigeon Paramyxovirus Type 1. Arch. Virol. 2021, 166, 1599–1605. [Google Scholar] [CrossRef]
- Jang, W.S.; Lim, D.H.; Yoon, J.; Kim, A.; Lim, M.; Nam, J.; Yanagihara, R.; Ryu, S.-W.; Jung, B.K.; Ryoo, N.-H.; et al. Development of a Multiplex Loop-Mediated Isothermal Amplification (LAMP) Assay for on-Site Diagnosis of SARS CoV-2. PLoS ONE 2021, 16, e0248042. [Google Scholar] [CrossRef]
- Zarzyńska-Nowak, A.; Hasiów-Jaroszewska, B.; Jeżewska, M. Molecular Analysis of Barley Stripe Mosaic Virus Isolates Differing in Their Biological Properties and the Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays for Their Detection. Arch. Virol. 2018, 163, 1163–1170. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Borodynko, N. Detection of Pepino Mosaic Virus Isolates from Tomato by One-Step Reverse Transcription Loop-Mediated Isothermal Amplification. Arch. Virol. 2013, 158, 2153–2156. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Kasprowicz, M.; Borodynko-Filas, N. Rapid Detection of Cucumber Mosaic Virus Isolates Representing Distinct Phylogenetic Subgroups by Reverse Transcription, Loop-Mediated Isothermal Amplification. J. Plant Dis. Prot. 2018, 125, 227–230. [Google Scholar] [CrossRef]
- Bashir, N.S.; Kalhor, M.R.; Zarghani, S.N. Detection, Differentiation and Phylogenetic Analysis of Cucumber Mosaic Virus Isolates from Cucurbits in the Northwest Region of Iran. Virus Genes 2006, 32, 277–288. [Google Scholar] [CrossRef]
- Škorić, D.; Krajačić, M.; Perica, M.Ć.; Halupecki, E.; Topić, J.; Igrc-Barčić, J. Cucumber Mosaic Virus (Cucumovirus) and Associated SatRNA in Weed Species under the Natural Epidemic Conditions of Tomato Lethal Necrosis in Croatia/Gurken Mosaik Virus (Cucumovirus) Und Verwandte SatRNA, Die in Kroatien Unter Natürlichen Bedingungen in Unkrautarten Gefunden Wurden, Führten Zu Letaler Tomatennekrose. Z. Pflanzenkrankh. Pflanzenschutz/J. Plant Dis. Prot. 2000, 107, 304–309. [Google Scholar]
- Rigotti, S.; Gugerli, P. Rapid Identification of Potato Virus Y Strains by One-Step Triplex RT-PCR. J. Virol. Methods 2007, 140, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Zarzyńska-Nowak, A.; Hasiów-Jaroszewska, B.; Budzyńska, D.; Trzmiel, K. Genetic Variability of Polish Tomato Black Ring Virus Isolates and Their Satellite RNAs. Plant Pathol. 2020, 69, 1034–1041. [Google Scholar] [CrossRef]
- PM 7/146 (1); Tomato Brown Rugose Fruit Virus. EPPO: Luxembourg, 2021; Volume 51, pp. 178–197. [CrossRef]
- Segev, L.; Wintermantel, W.M.; Polston, J.E.; Lapidot, M. First Report of Tomato Chlorosis Virus in Israel. Plant Dis. 2004, 88, 1160. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Udaya Shankar, A.C.; Nayaka, S.C.; Lund, O.S.; Prakash, H.S. Detection of Tobacco Mosaic Virus and Tomato Mosaic Virus in Pepper and Tomato by Multiplex RT–PCR. Lett. Appl. Microbiol. 2011, 53, 359–363. [Google Scholar] [CrossRef]
- Budziszewska, M.; Obrepalska-Steplowska, A.; Wieczorek, P.; Pospieszny, H. The Nucleotide Sequence of a Polish Isolate of Tomato Torrado Virus. Virus Genes 2008, 37, 400–406. [Google Scholar] [CrossRef]
- Zarzyńska-Nowak, A.; Hasiów-Jaroszewska, B.; Korbecka-Glinka, G.; Przybyś, M.; Borodynko-Filas, N. A Multiplex RT-PCR Assay for Simultaneous Detection of Tomato Spotted Wilt Virus and Tomato Yellow Ring Virus in Tomato Plants. Can. J. Plant Pathol. 2018, 40, 580–586. [Google Scholar] [CrossRef]
- Hall, T. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Edgar, R.C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A Virus Classification Tool Based on Pairwise Sequence Alignment and Identity Calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef]
- Kosakovsky Pond, S.L.; Posada, D.; Gravenor, M.B.; Woelk, C.H.; Frost, S.D.W. Automated Phylogenetic Detection of Recombination Using a Genetic Algorithm. Mol. Biol. Evol. 2006, 23, 1891–1901. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A Webserver for Visualization, Annotation, and Management of Phylogenetic Trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef] [PubMed]
- Weaver, S.; Shank, S.D.; Spielman, S.J.; Li, M.; Muse, S.V.; Kosakovsky Pond, S.L. Datamonkey 2.0: A Modern Web Application for Characterizing Selective and Other Evolutionary Processes. Mol. Biol. Evol. 2018, 35, 773–777. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; Maio, N.D.; et al. BEAST 2.5: An Advanced Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [PubMed]
- Posada, D. JModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Parker, J.; Rambaut, A.; Pybus, O.G. Correlating Viral Phenotypes with Phylogeny: Accounting for Phylogenetic Uncertainty. Infect. Genet. Evol. 2008, 8, 239–246. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Lapidot, M.; Thomma, B.P.H.J. Emerging Viral Diseases of Tomato Crops. Mol. Plant Microbe Interact. 2010, 23, 539–548. [Google Scholar] [CrossRef]
- Oladokun, J.O.; Halabi, M.H.; Barua, P.; Nath, P.D. Tomato Brown Rugose Fruit Disease: Current Distribution, Knowledge and Future Prospects. Plant Pathol. 2019, 68, 1579–1586. [Google Scholar] [CrossRef]
- Maree, H.J.; Fox, A.; Al Rwahnih, M.; Boonham, N.; Candresse, T. Application of HTS for Routine Plant Virus Diagnostics: State of the Art and Challenges. Front. Plant Sci. 2018, 9, 1082. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Boezen, D.; Zwart, M.P. Metagenomic Studies of Viruses in Weeds and Wild Plants: A Powerful Approach to Characterise Variable Virus Communities. Viruses 2021, 13, 1939. [Google Scholar] [CrossRef] [PubMed]
- Hasiów-Jaroszewska, B.; Pospieszny, H.; Borodynko, N. New Necrotic Isolates of Pepino Mosaic Virus Representing the Ch2 Genotype. J. Phytopathol. 2009, 157, 494–496. [Google Scholar] [CrossRef]
- Minicka, J.; Rymelska, N.; Elena, S.F.; Czerwoniec, A.; Hasiów-Jaroszewska, B. Molecular Evolution of Pepino Mosaic Virus during Long-Term Passaging in Different Hosts and Its Impact on Virus Virulence. Ann. Appl. Biol. 2015, 166, 389–401. [Google Scholar] [CrossRef]
- Ghotbi, T.; Shahraeen, N.; Winter, S. Occurrence of Tospoviruses in Ornamental and Weed Species in Markazi and Tehran Provinces in Iran. Plant Dis. 2005, 89, 425–429. [Google Scholar] [CrossRef]
- Jacquemond, M. Cucumber Mosaic Virus. In Advances in Virus Research; Loebenstein, G., Lecoq, H., Eds.; Viruses and Virus Diseases of Vegetables in the Mediterranean Basin; Academic Press: London, UK, 2012; Chapter 13; Volume 84, pp. 439–504. [Google Scholar]
- Hasiów-Jaroszewska, B.; Budzyńska, D.; Rymelska, N.; Korpys, P.; Borodynko-Filas, N. Phylogenetic Evidence of Natural Reassortants in the Cucumber Mosaic Virus Population in Poland. Can. J. Plant Pathol. 2018, 40, 587–593. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Chrzanowski, M.; Budzyńska, D.; Rymelska, N.; Borodynko-Filas, N. Genetic Diversity, Distant Phylogenetic Relationships and the Occurrence of Recombination Events among Cucumber Mosaic Virus Isolates from Zucchini in Poland. Arch. Virol. 2017, 162, 1751–1756. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.M.; Waterworth, H.E. Cucumber Mosaic Virus Associated RNA 5: Causal Agent for Tomato Necrosis. Science 1977, 196, 429–431. [Google Scholar] [CrossRef] [PubMed]
- Gallitelli, D.; Di Franco, A.; Vovlas, C.; Kaper, J. Infezioni Miste del Virus del Mosaico del Cetriolo (CMV) e di Potyvirus in Colture Ortive di Puglia e Basilicata. Inf. Fitopatol. 1988, 38, 57–64. [Google Scholar]
- Jorda, C.; Alfaro, A.; Aranda, A.; Moriones, E.; Garcia, F. Epidemic of Cucumber Mosaic Virus plus Satellite RNA in Tomatoes of Eastern Spain. Plant Dis. 1992, 76, 363–366. [Google Scholar] [CrossRef]
- Giakountis, A.; Tsarmpopoulos, I.; Chatzivassiliou, E.K. Cucumber Mosaic Virus Isolates from Greek Legumes Are Associated with Satellite RNAs That Are Necrogenic for Tomato. Plant Dis. 2018, 102, 2268–2276. [Google Scholar] [CrossRef]
- Pospieszny, H.; Borodynko, N.; Hasiow-Jaroszewska, B. Wirus Y ziemniaka (Potato Virus Y, PVY) na pomidorze szklarniowym. Prog. Plant Prot. 2009, 49, 1327–1330. [Google Scholar]
- Jeżewska, M.; Trzmiel, K.; Zarzyńska-Nowak, A. Detection of Infectious Tobamoviruses in Irrigation and Drainage Canals in Greater Poland. J. Plant Prot. Res. 2018, 58, 202–205. [Google Scholar] [CrossRef]
- Salem, N.; Mansour, A.; Ciuffo, M.; Falk, B.W.; Turina, M. A New Tobamovirus Infecting Tomato Crops in Jordan. Arch. Virol. 2016, 161, 503–506. [Google Scholar] [CrossRef] [PubMed]
- EPPO Tomato Brown Rugose Fruit Virus (TOBRFV) [World Distribution]. EPPO Global Database. Available online: https://gd.eppo.int/taxon/TOBRFV/distribution (accessed on 15 March 2022).
- Wintermantel, W.M.; Wisler, G.C. Vector Specificity, Host Range, and Genetic Diversity of Tomato Chlorosis Virus. Plant Dis. 2006, 90, 814–819. [Google Scholar] [CrossRef]
- Pourrahim, R.; Golnaraghi, A.; Farzadfar, S.; Ohshima, K. Partial Biological and Molecular Characterization of Tomato Yellow Fruit Ring Virus Isolates from Potato. Plant Pathol. J. 2012, 28, 390–400. [Google Scholar] [CrossRef][Green Version]
- Golnaraghi, A.R.; Pourrahim, R.; Farzadfar, S.; Ahoonmanesh, A. Identification and Partial Characterization of a Tospovirus Causing Leaf and Stem Necrosis on Potato. Plant Pathol. J. 2007, 6, 227–234. [Google Scholar]
- Worobey, M.; Holmes, E.C. Evolutionary Aspects of Recombination in RNA Viruses. J. Gen. Virol. 1999, 80, 2535–2543. [Google Scholar] [CrossRef]
- Rubio, L.; Galipienso, L.; Ferriol, I. Detection of Plant Viruses and Disease Management: Relevance of Genetic Diversity and Evolution. Front. Plant Sci. 2020, 11, 1092. [Google Scholar] [CrossRef]
- Beikzadeh, N.; Jafarpour, B.; Rouhani, H.; Peters, D.; Hassani Mehraban, A. Molecular Diagnosis of Tomato Yellow Ring Virus (Tyrv) On Alstroemeria in Khorasan Razavi Province, Iran. Iran. Plant Prot. Res. J. Plant Prot. 2011, 25, 313–324. [Google Scholar]
- Bhat, A.I.; Aman, R.; Mahfouz, M. Onsite Detection of Plant Viruses Using Isothermal Amplification Assays. Plant Biotechnol. J. 2022. [Google Scholar] [CrossRef]
- Tomlinson, J.; Boonham, N. Potential of LAMP for Detection of Plant Pathogens. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 2008, 3, 1–7. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).