Abstract
Although placental small extracellular vesicles (sEVs) are extensively studied in the context of pregnancy, little is known about their role during viral congenital infection, especially at the beginning of pregnancy. In this study, we examined the consequences of human cytomegalovirus (hCMV) infection on sEVs production, composition, and function using an immortalized human cytotrophoblast cell line derived from first trimester placenta. By combining complementary approaches of biochemistry, electron microscopy, and quantitative proteomic analysis, we showed that hCMV infection increases the yield of sEVs produced by cytotrophoblasts and modifies their protein content towards a potential proviral phenotype. We further demonstrate that sEVs secreted by hCMV-infected cytotrophoblasts potentiate infection in naive recipient cells of fetal origin, including human neural stem cells. Importantly, these functional consequences are also observed with sEVs prepared from an ex vivo model of infected histocultures from early placenta. Based on these findings, we propose that placental sEVs could be important actors favoring viral dissemination to the fetal brain during hCMV congenital infection.
1. Introduction
Human cytomegalovirus (hCMV) belongs to the Herpesviridae family, and its prevalence is of 50 to 90% in global human population. Most of the hCMV infections occurring among immunocompetent adults induce an asymptomatic acute replication phase followed by a lifelong persistent latent state. However, hCMV primary infection, reinfection, and/or reactivation may severely compromise the health of immunocompromised people and is a major issue during pregnancy [1,2]. Congenital infection by hCMV affects 1% of live births in western countries, making hCMV the most frequently transmitted virus in utero [2,3], causing placental and fetal impairments of variable severity. The most severe consequences are observed when transmission occurs during the peri-conceptional period or first trimester [4]. Infection of the placenta itself allows the virus to actively replicate and enable its further access to the fetus [3,5,6]. The infected placenta can develop a pathology that may lead to miscarriage, premature delivery, intra uterine growth retardation, or even fetal death [2,4]. On the other side, the infection of the fetus causes various visceral and central nervous system damage, congenital hCMV infection being the most common cause of brain malformations and deafness of infectious origin [2,7,8]. Despite the extensive research conducted so far, the pathophysiology of hCMV infection remains unclear, especially concerning potential factors which may explain the wide variety of clinical manifestations and their severity [2].
In the course of hCMV congenital infection, the placenta is a central key organ, i.e., the target of viral replication allowing further vertical transmission towards the fetus. Amongst the numerous placental functions, a recently described and extensively studied mode of communication between both maternal and fetal sides consists in the production of placental extracellular vesicles (EVs) [9,10]. EVs are membranous vesicles secreted by cells in both physiological and pathological situations, whose main subtypes can be distinguished depending on their biogenesis and size into small EVs (sEVs) and large EVs. They are specifically composed of various molecules such as proteins, lipids, and coding and non-coding RNAs [11,12]. Once released into the extracellular space, EVs can be internalized by other recipient cells, in their immediate environment or within long distances, wherein they exert regulatory roles [13]. For example, placental EVs can be uptaken by natural killer cells [14,15] or by primary placental fibroblasts [16]. Although the understanding of the biological relevance of placental EVs in vivo remains limited, recent findings highlight their roles in cell-cell communication underlying the feto-placenta-maternal dialogue during pregnancy [17,18]. Interestingly, placental EV content is altered upon gestational diseases such as preeclampsia, preterm birth, or gestational diabetes mellitus, and recent literature points towards a putative role of dysregulated placental EVs during pathological pregnancies [19,20,21,22,23,24].
Although placental sEVs are extensively studied in the context of pregnancy diseases, little is currently known about their role during hCMV congenital infection, especially at the very beginning of pregnancy where the most severe sequelae take their origin. Our recent data described a dysregulation of the surface expression of placental sEV markers upon hCMV infection in an ex vivo model of first trimester placental histocultures, suggesting a putative role for viral dissemination [25]. In the present study, we used immortalized cytotrophoblasts derived from first trimester placenta, named HIPEC (human invasive proliferative extravillous cytotrophoblast) [26], to comprehensively examine the consequences of hCMV infection on sEV production, content, and function in recipient cells. We show that hCMV increases the sEV production by HIPECs and alters their protein content towards a potential proviral phenotype. Finally, we observe that sEVs secreted by hCMV-infected HIPECs potentiate infection in naive recipient cells, including human neural stem cells (NSCs). Importantly, this enhancement of hCMV infection is also observed with sEVs from ex vivo early placental histocultures. Hence, we propose that placental sEVs could be important actors favoring viral dissemination towards the fetal brain during hCMV congenital infection.
2. Materials and Methods
2.1. Human Ethic Approval
The use of NSCs from human embryonic stem cells was approved by the French authorities (Agence de la Biomédecine, authorization number SASB0920178S).
The biological resource center Germethèque (BB-0033-00081; declaration: DC-2014-2202; authorization: AC-2015-2350) obtained the written consent from each patient (CPP.2.15.27) for the use of human samples and their associated data. For first trimester placenta explants, the steering committee of Germethèque gave its approval for the realization of this study on 5 February 2019. The hosting request made to Germethèque bears the number 20,190,201 and its contract is referenced under the number 19 155C.
2.2. Cell Lines
HIPECs were obtained from Dr T. Fournier (Inserm, Paris; Transfer agreement n°170,448). The expression of the cytokeratin 7 specific cytotrophoblastic marker was verified (data not shown). They were cultured in DMEM/F12 medium (Gibco, Whaltam, MA, USA) at 50/50 ratio (v/v), in the presence of 10% fetal bovine serum (FBS, Sigma-Aldrich, Saint-Louis, MO, USA), 100 U/mL penicillin-100 µg/mL streptomycin (Gibco), and 100 µg/mL normocin (Invivogen, Carlsbad, CA, USA).
MRC5 cells (RD-Biotech, Besançon, France) were cultured in Dulbecco’s Modified Eagle Medium (DMEM with Glutamax, Gibco) with the same supplementation as HIPECs.
NSCs were obtained from Dr A. Benchoua (I-Stem, Evry, France); they were produced from ES human cells (SA001, I-STEM, UMR861 France) [27] and maintained in growth medium as described [7]. Stem character of NSCs was systematically assessed by immunofluorescence against Nestin and SOX2 proteins (data not shown).
Cell cultures were checked for the absence of mycoplasma (Plasmotest, Invivogen).
2.3. Virus Production, Titration and Infection
The endotheliotropic VHL/E strain of hCMV—a gift from Dr C. Sinzger, University of Ulm, Germany—was used in this study [28]. Viral stocks were obtained upon amplification of the virus on MRC5 cells (maximum three to four passages), concentrated by ultracentrifugation, and titrated by indirect immunofluorescence as described [7]. In some experiments, virus titration was also performed by qPCR from cell culture supernatants [29].
2.4. sEV Preparation
Culture medium was previously depleted from EVs to obtain “Exofree” medium: DMEM supplemented with 20% FBS was ultracentrifuged at 100,000 g for 16 h at 4 °C and filtered at 0.22 µm. Exofree medium was then obtained by a 1:1 dilution with F12 to reach 10% FBS.
A total of 4 million HIPECs were seeded in 150 cm2 flask, with 6 flasks per condition. Then, 24 h later, cells were infected or not by hCMV at multiplicity of infection (MOI) of 10 (Supplementary Figure S1a). Culture medium was replaced by Exofree medium 24 h post-infection, after having previously ensured that this did not affect the cell growth (Supplementary Figure S1e). Culture supernatants were collected at 48 h and 72 h post-infection. Medium were pooled for each condition (non-infected or infected) and submitted to sEV preparation protocol. Cell number at the end of the experiment was determined by counting upon trypsinization (Supplementary Figure S1d) and cell viability evaluated by trypan blue.
Procedures for sEV preparation and PKH67 staining were realized as described previously [25], according to ISEV guidelines [30]. All relevant data were submitted to the EV-TRACK knowledgebase (EV-TRACK ID: EV210154) and obtained an EV-METRIC score of 100% for HIPEC and placental explant sEVs [31].
2.5. Nanoparticle Tracking Analysis (NTA)
sEV preparations were tracked using a NanoSight LM10 (Malvern Panalytical, Malvern, UK) equipped with a 405 nm laser. Videos were recorded three times for each sample at constant temperature (22 °C) during 60 s and analyzed with NTA Software 2.0 (Malvern instruments Ltd.). Data were analyzed with Excel and GraphPad Prism (v8) software.
2.6. Transmission Electron Microscopy and Immunolabeling Electron Microscopy
Procedures were performed essentially as described [25,32,33]. Immunodetection was carried out with the following primary antibodies: mouse anti-human CD63 (Abcam ab23792), mouse anti-human CD9, or mouse anti-human CD81 (both from Dr E. Rubinstein, Université Paris-Sud, Institut André Lwoff, Villejuif, France). Secondary incubation was performed with a rabbit anti mouse Fc fragment (Dako Agilent Z0412), then grids were incubated with Protein A-Gold 10 nm (Cell Microscopy Center, Department of Cell Biology, Utrecht University). All samples were observed with a Tecnai Spirit electron microscope (FEI, Eindhoven, The Netherlands), and digital acquisitions were made with a numeric 4k CCD camera (Quemesa, Olympus, Münster, Germany). Images were analysed with iTEM software (EMSIS) and statistical studies were done with GraphPad Prism software (v8).
2.7. Western Blot
Western blots were realized as previously described [25], by using the following primary antibodies: mouse anti-CD81 (200 ng/mL, Santa-Cruz, Dallas, TX, USA), mouse anti-CD63 (500 ng/mL, BD Pharmingen, NJ, USA), mouse anti-CD9 (100 ng/mL, Millipore, Burlington, MA, USA), rabbit anti-Tsg101 (1 µg/mL, Abcam, Cambridge, UK), rabbit anti-Alix (1 µg/mL, Abcam), mouse anti-Thy1 (0.5 µg/mL, Biolegend, San Diego, CA, USA), rabbit anti-Tom20 (1/500, Sigma), or goat anti-Calnexin (2 µg/mL, Abcam). After incubation with the secondary antibody, membranes were visualized using the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE, USA).
2.8. Quantitative Proteomic Analysis
2.8.1. Sample Preparation
Protein samples in Laemmli buffer (3 biological replicates of sEVs preparation from non-infected and hCMV-infected cytotrophoblasts cells) were submitted to reduction and alkylation (30 mM DTT and 90 mM iodoacetamide, respectively). Protein samples were digested with trypsin on S-trap Micro devices (Protifi, Farmingdale, NY, USA) according to manufacturer’s protocol, with the following modifications: precipitation was performed using 545 µL S-Trap buffer and 1 µg Trypsin was added per sample for digestion.
2.8.2. NanoLC-MS/MS Analysis
Peptides were analyzed by nanoLC-MS/MS using an UltiMate 3000 RSLCnano system coupled to a Q-Exactive-Plus mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Five µL of each sample were loaded on a C-18 precolumn (300 µm ID × 5 mm, Dionex) in a solvent made of 5% acetonitrile and 0.05% TFA and at a flow rate of 20 µL/min. After 5 min of desalting, the precolumn was switched online with the analytical C-18 column (75 µm ID × 15 cm, Reprosil C18) equilibrated in 95% solvent A (5% acetonitrile, 0.2% formic acid) and 5% solvent B (80% acetonitrile, 0.2% formic acid). Peptides were eluted using a 5 to 50% gradient of solvent B over 105 min at a flow rate of 300 nL/min. The Q-Exactive-Plus was operated in a data-dependent acquisition mode with the XCalibur software. Survey scan MS were acquired in the Orbitrap on the 350–1500 m/z range with the resolution set to a value of 70,000. The 10 most intense ions per survey scan were selected for HCD fragmentation. Dynamic exclusion was employed within 30 s to prevent repetitive selection of the same peptide. At least 3 injections were performed for each sample.
2.8.3. Bioinformatics Data Analysis of Mass Spectrometry Raw Files
Raw MS files were processed with the Mascot software for database search and with Proline [34] for label-free quantitative analysis. Data were searched against human herpesvirus 5 and human entries of the UniProtKB protein database (Human betaherpesvirus 5 clone VHL-E-BAC19 and release Uniprot Swiss-Prot February 2018). Carbamidomethylation of cysteines was set as a fixed modification, whereas oxidation of methionine was set as variable modification. Specificity of trypsin/P digestion was set for cleavage after K or R, and two missed trypsin cleavage sites were allowed. The mass tolerance was set to 10 ppm for the precursor and to 20 mmu in tandem MS mode. Minimum peptide length was set to 7 amino acids, and identification results were further validated in Proline by the target decoy approach using a reverse database at both a PSM and protein false-discovery rate of 1%. After mean of replicate injections, the abundance values were log2 transformed and missing values were replaced by random numbers drawn from a normal distribution with a width of 0.3 and down shift of 1.8 using the Perseus toolbox (version 1.6.7.0). For statistical analysis, a Student t-test (two-tailed t-test, equal variances) was then performed on log2 transformed values to analyse differences in protein abundances in all biologic group comparisons. Significance level was set at p = 0.05, and log2 ratios were considered relevant if higher than 1 or lower than −1. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [35] partner repository with the dataset identifier PXD029146.
2.9. Functional Proteomic Data Analysis
Volcano plot was established for proteins whose mean abundance exhibited a log2 ratio higher than 1 or lower than −1 and when Student’s t-test p-values were ≤ 0.05 between the infected and the non-infected conditions. The list of human proteins exhibiting a normalized mean protein abundance log2 ratio > 1 or < −1 between sEVs from non-infected or hCMV-infected samples was used as an input for analysis with QIAGEN Ingenuity Pathway Analysis (IPA) [36]. Results from IPA biological functions and diseases analysis were filtered to retrieve annotations having an absolute activation z-score > 1 and defined by less than 150 molecules. The resulting annotations were manually curated to remove redundant annotations sharing identical genes, keeping annotations defined by the greater number of molecules.
2.10. Flow Cytometry Analysis
After incubation of cells with PKH67-stained sEVs, cells were washed twice with PBS and trypsinized, before proceeding to flow cytometry analysis. PKH67 positive cells were analyzed on a Macsquant VYB Flow Cytometer (Miltenyi Biotec, Paris, France), by using FCS and FITC fluorescence parameters, and by subtracting cell autofluorescence background. Data were analyzed with FlowJo (BD) and GraphPad Prism (v8) software.
2.11. Immunofluorescence
Cells were fixed using 4% PFA (Electron microscopy Sciences, Hatfield, PA, USA) at room temperature for 20 min. Permeabilization was performed with PBS 0.3% Triton-X100 (Thermofisher scientific) for 10 min, followed by 1 h incubation in blocking buffer (PBS with 5% FBS). Incubation with primary antibodies diluted in blocking buffer was carried out overnight at 4 °C, against hCMV immediate early protein 1 and 2 (1 µg/mL; Abcam IE1/IE2 CH160 ab53495), nestin (4 µg/mL; Abcam 10C2 ab 22035), or SOX2 (1/500 of stock; Cell Signaling D6D9 #3579). Secondary antibody incubation (Goat anti mouse or rabbit-Alexa-fluor 488 or 594 (2 µg/mL; Thermo Fischer Scientific, Whaltham, Ma, USA)) was performed at room temperature for 1 h. For actin staining, Alexa-fluor 568 phalloidin (5 µg/mL; Thermo Fischer Scientific A12380) was incubated on cells overnight at 4 °C. DAPI staining (1 µg/mL; Sigma) was performed for 10 min at room temperature. ProLong Gold without DAPI (Thermo Fischer Scientific) was used for coverslip mounting.
Widefield acquisitions were realized using Apotome microscope (Zeiss, Iena, Germany) and confocal acquisitions were made on SP8-STED microscope (Leica, Wetzlar, Germany). Image processing was performed using ImageJ. GraphPad Prism (v8) software was used to perform data statistical analysis.
2.12. Placental Histoculture
Placental histocultures and infection were carried out as described [25,37] on first trimester placentas (4 placentas; mean = 13.11 ± 0.49 (SEM) weeks of amenorrhea, i.e., 11.11 ± 0.49 weeks of pregnancy; age of the women: mean = 23 ± 1.5 (SEM) year-old). Briefly, trophoblastic villi were dissected in small explants and infected or not by hCMV overnight before extensive washing and deposition on gelatin sponges (Gelfoam, Pfizer, New York, NY, USA) in Exofree medium. Conditioned medium was collected and renewed every 3 to 4 days. At 14 days of culture, collected medium was pooled for each condition and used to perform sEV preparation. Placental explants were weighed for normalization of resuspension volume.
3. Results
3.1. HIPEC Infection by hCMV Leads to an Increase of sEV Secretion without Modifying Their General Features
To study the consequences of hCMV infection on placental sEV secretion, composition, and function in early pregnancy, conditioned media of HIPECs, cells described to be fully permissive for hCMV [38,39], were collected between 48 and 72 h post-infection before sEV preparation, times at which 50–80% of cells were infected (Supplementary Figure S1b). To exclude any viral contamination of sEVs, an infectivity test was systematically carried out for each preparation. No infection was detected in cells incubated with sEVs alone (Supplementary Figure S1c). Moreover, no structure evoking viral particles was observed by transmission electron microscopy (TEM) on sEVs from infected HIPECs (Supplementary Figure S2, and data not shown).
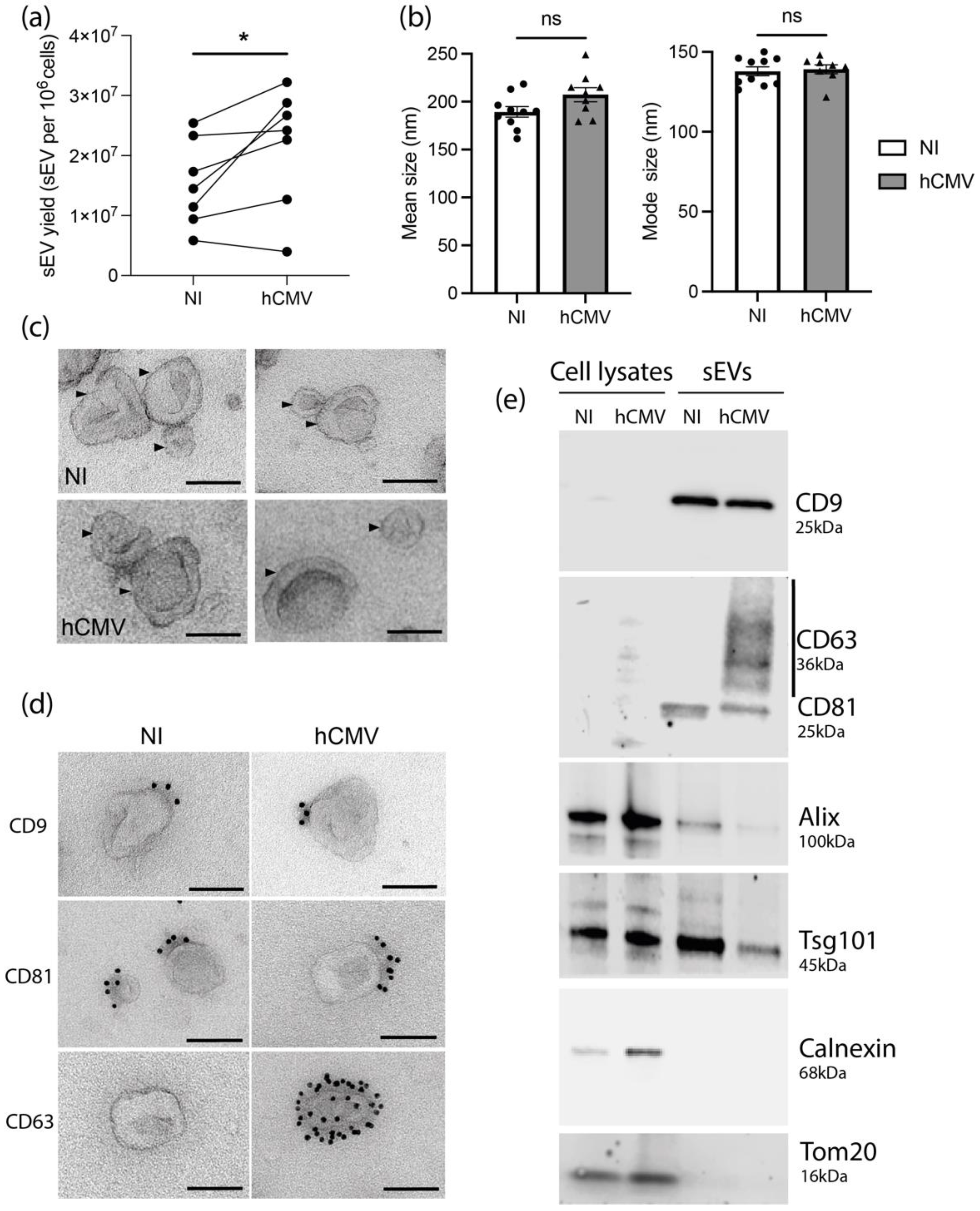
Despite a decrease of cell number upon hCMV infection compared to non-infected (NI) HIPECs (Supplementary Figure S1d), the quantification of sEVs isolated per cell showed a significant higher yield upon infection, with an increase of around 40% (Figure 1a). No difference in either the mean size or the mode size of the sEVs was observed upon infection when analyzed by NTA (Figure 1b) and sEVs from both NI or infected HIPECs exhibited the same typical structure and shape as evidenced by TEM (Figure 1c and Supplementary Figure S2).
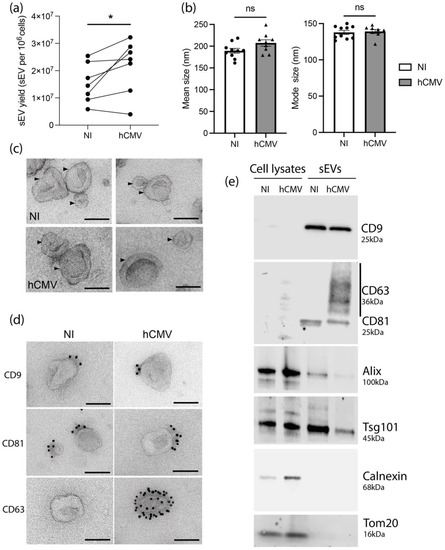
Figure 1.
Characterization of sEVs from NI and hCMV infected HIPECs. (a) Yield of sEV recovered from HIPECs either non-infected (NI) or infected (hCMV) at a multiplicity of infection of 10, between 48 and 72 h post-infection. *, p = 0.0464 by paired t-test for seven independent experiments. (b) Comparison of mean size (left histogram) and mode size (right histogram) between sEVs from non-infected (NI) or infected (hCMV) HIPECs. Histograms show the mean ± SEM of three independent experiments. ns: non-significant by Mann–Whitney test. (c) Electron microscopy images of sEVs (indicated by an arrow) prepared from non-infected (NI) or infected (hCMV) HIPECs. Magnification = 26,000×. Scale bar = 100 nm. Images are representative of at least three independent experiments. (d) TEM observation of sEV—isolated from non-infected (NI) or infected (hCMV) HIPECs—which were immunogold-labelled for CD9, CD81, or CD63, and revealed with Protein A-gold particle of 10 nm diameter. Scale bar = 100 nm. Magnification = 26,000×. In CD63 IEM, only one example of positive vesicle, representing around 1–5% of sEVs isolated upon infection, is shown, the other being negative (see Supplementary Figure S3 for wide field image). (e) Western blot realized on either whole cell lysates (left wells) or purified sEVs (right wells), from non-infected (NI) or infected (hCMV) HIPECs. Proteins of interest and their corresponding molecular weight are indicated on the right of the Figure, with a smear for CD63 due to the non-reducing conditions of the western blot, which preserve its rich glycosylated pattern.
We next assessed the impact of hCMV infection on the expression of sEV canonical markers. By combining western-blotting, immunolabeling electron microscopy (IEM), and multiplex bead-based flow cytometry, we observed that sEV preparations expressed specific vesicular markers including CD9, CD81, Alix, and Tsg101, but no endoplasmic reticulum or mitochondrial markers (Figure 1d,e and Supplementary Figure S3), attesting the purity of the sEV preparations [30]. No drastic differences were observed in their expression levels upon infection as compared to NI cells, except for CD63 protein, which was not detected in whole cell lysates, but enriched in sEVs from hCMV-infected HIPECs (Figure 1e). By IEM, at the level of individual vesicles, we noticed that this increase was correlated to the presence of a small proportion of sEVs highly positive for CD63 (between 1 and 5%), while the others remained negative (Figure 1d and Supplementary Figure S4). By multiplex bead-based flow cytometry assay, no significant increase in CD63 expression was detected between sEVs from NI or infected HIPECs upon infection, certainly due to the low proportion of positive vesicles (Supplementary Figure S3). Hence, hCMV infection increased HIPEC-sEV production but did not globally impact on their global features, except for CD63 that is detected in a subpopulation of vesicles upon infection.
3.2. sEVs Secreted by Infected HIPECs Harbor a Potential Proviral Protein Cargo
To extend the molecular characterization of sEVs secreted by hCMV-infected HIPECs, a comprehensive proteomic approach that allows for a deeper analysis of sEV composition was carried out. The analysis by mass spectrometry-based quantitative proteomics of equivalent amounts of sEVs from NI or hCMV-infected HIPECs led to the identification of 3079 proteins across all samples (3048 human and 31 viral; Supplementary Table S1). Among the 3048 human proteins identified, the gProfiler2 R package was able to interrogate 2936 proteins, for which the term “extracellular exosome” (Gene ontology GO:0070062) appeared as the most significantly enriched (false discovery rate (FDR) = 1.087859e-259, R package gProfiler2), with 962 of them (32.8%) associated with the “extracellular exosome” GO term. Conversely, these 962 proteins constituted 44.1% of the proteins which define the GO term, and the 94 of the top 100 most frequently identified exosomal proteins, as defined by the Exocarta database (http://exocarta.org/exosome_markers_new (accessed on 1 November 2021)), were detected in the sEV preparations.
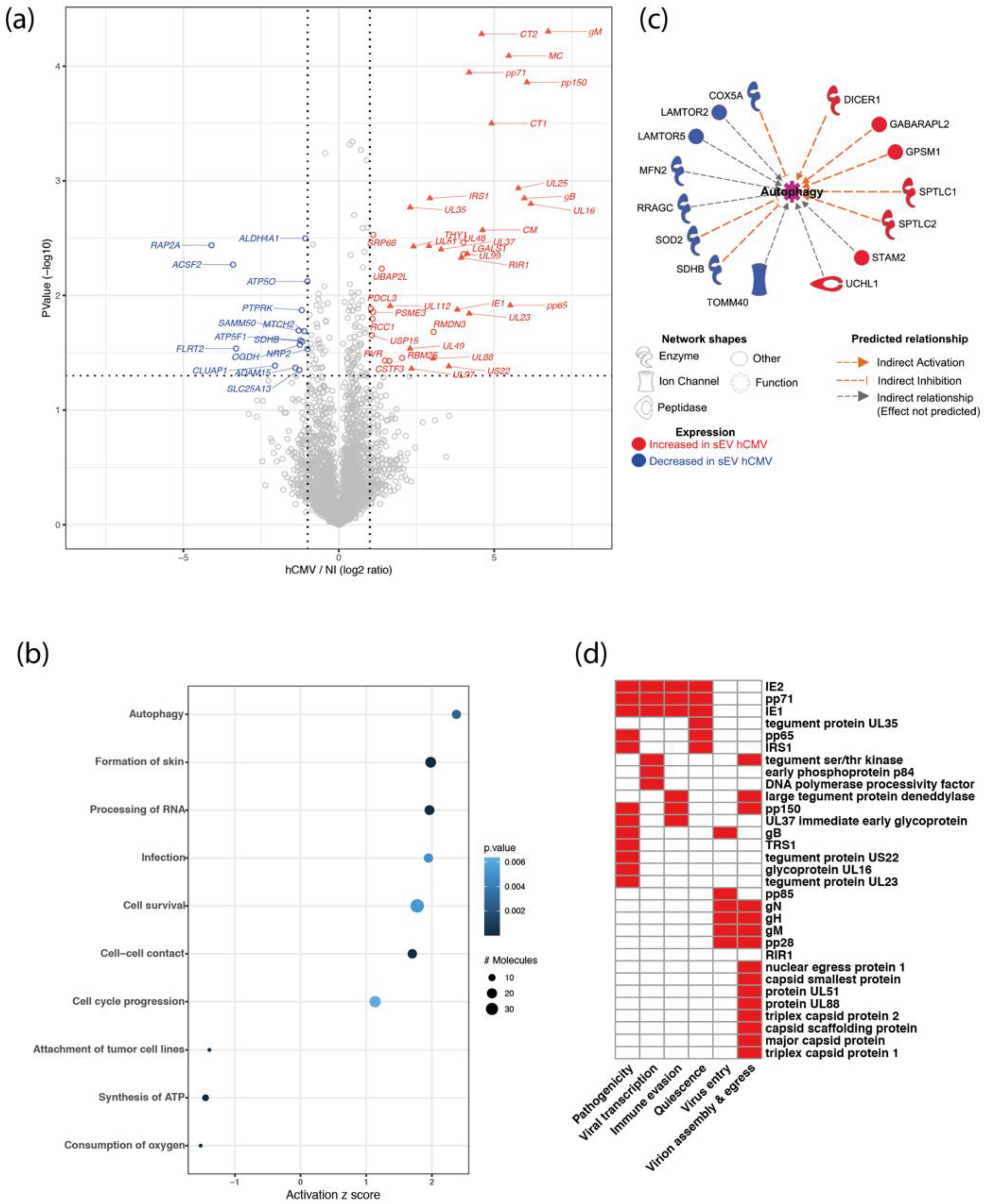
Proteomics data indicated that 31 viral proteins were loaded in sEVs from hCMV-infected HIPECs, as well as 6 human proteins that were significantly over-represented and 15 under-represented (Figure 2a). Interestingly, the Thy-1 cellular protein, known to play an important role to facilitate hCMV entry into cells via macropinocytosis [40,41], was significantly enriched in sEVs upon infection (Supplementary Figure S5). Moreover, by using the Ingenuity Pathway Analysis (IPA) tool, several biological functions were found to be significantly over- or under-represented in sEVs upon infection (Figure 2b). The most modulated pathway identified was “autophagy”, with several actors showing altered expression in sEVs from infected HIPECs, leading to an overall pattern of autophagy activation (Figure 2c). Since the EV content primarily reflects the composition of the cells they are derived from, this may also reflect the activation of the autophagy pathway induced during early hCMV infection of host cells [42,43,44]. On the other hand, two pathways related to mitochondrial functions (i.e., “consumption of oxygen” and “ATP synthesis”) were found to be lower in sEVs from hCMV-infected HIPECs than in NI HIPECs (Figure 2b). This may also be the consequence of hCMV-induced mitochondrial dysfunctions [45,46,47,48], likely via interference with the antiviral Viperin protein, which leads to decreased cellular ATP levels [49].
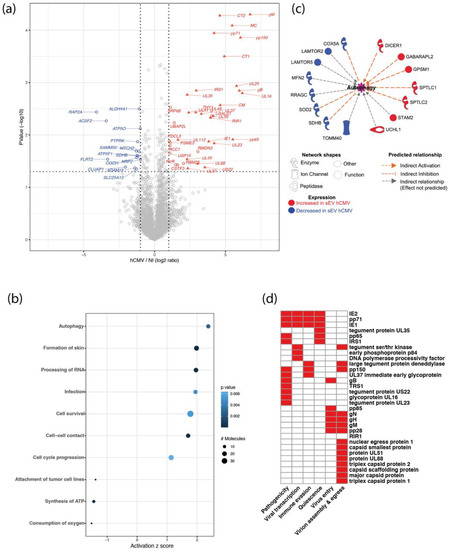
Figure 2.
Proteomic analysis of sEV composition upon infection of HIPECs by hCMV. (a) Volcano-plot representing differences in normalized mean protein abundance in sEVs hCMV versus sEVs NI. Human and viral proteins exhibiting significant differences between the two conditions are represented by circles and triangles, respectively (Student T-test p-value ≤ 0.05 and log2 ratio ≥ 1 or ≤ −1). Red: over-represented proteins; Blue: under-represented proteins. (b) Dot plot representation of the top diseases and biological functions associated with human proteins exhibiting an absolute normalized mean abundance log2 ratio greater than 1 or lower than −1, in sEVs hCMV versus sEVs NI. Top diseases and biological functions associated with changes in the protein content of sEVs upon hCMV-infection were identified using QIAGEN Ingenuity Pathway Analysis (IPA). Size of the circles depends on the number of the proteins identified in the corresponding pathway; level of blue intensity depends on the p-value. (c) Human proteins associated with the predicted increased activation state of autophagy pathway as determined by IPA. (d) Heatmap representation of the biological functions associated with hCMV viral proteins expressed in sEVs hCMV. In red are indicated the functions attributed to the viral proteins (See Supplementary Table S2 for bibliography references).
Importantly, the proteomic data also revealed that 31 proteins found in sEVs secreted by infected HIPECs were of viral origin. They are involved in different aspects of hCMV infection, from viral entry to egress, quiescence, as well as pathogenicity and immune evasion, and are mainly immediate, early, or late proteins (Figure 2d and Supplementary Table S2). Interestingly, although sEV preparations were devoid of viral particles as assessed by infectivity assays and TEM (Supplementary Figures S1c and S2), some of the viral proteins in sEV from hCMV-infected HIPECs are structural proteins (Figure 2d and Supplementary Table S2), i.e., the envelope proteins gB, gH, and gM, as it has already been described in other studies [50,51,52]. Most of the other proteins identified were capsid and tegument proteins that are delivered to host cells upon infection, or proteins immediately expressed after virus entry such as IE1 and IE2, which play a role in early transcriptions [53,54,55]. We also identified pp65, which participates to the transactivation of viral major immediate early genes [56,57], as well as pp71, which stimulates viral immediate early transcription and inhibits the host innate response by targeting STING [58,59]. Finally, IRS1 and TRS1, which inhibit the establishment of an antiviral state in infected cells, in particular by antagonizing the autophagy pathway induced upon hCMV infection [42,43], were also detected.
Altogether, analysis of the proteomic data suggests that sEVs secreted from infected HIPECs carry a protein cargo with potential proviral properties. The incorporation of these viral and cellular proteins into sEVs could enhance the viral spread by providing the recipient cells with elements that may facilitate the early steps of a further hCMV infection.
3.3. sEVs Are Efficiently Uptaken by Recipient MCR5 Cells
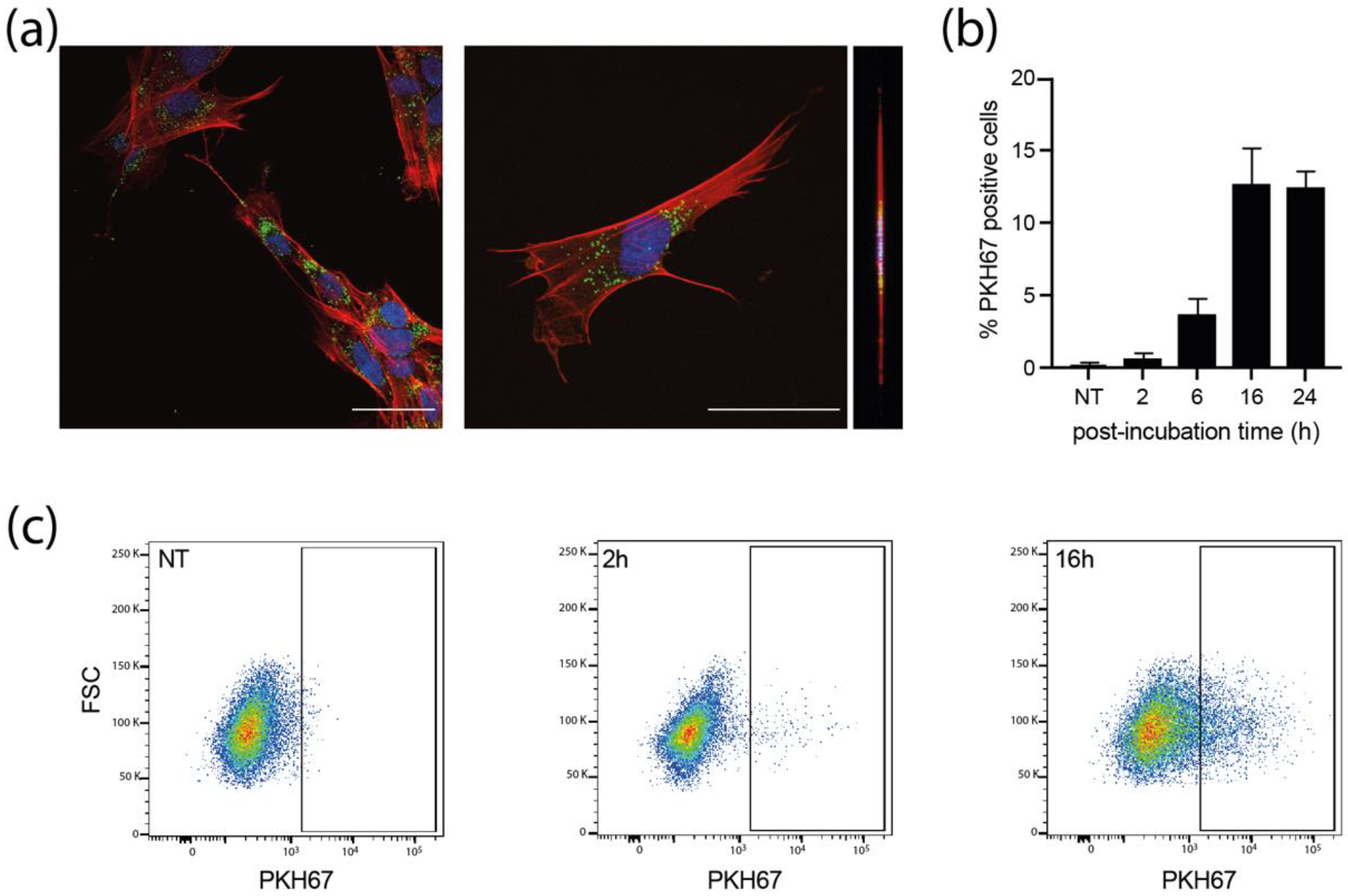
We next examined whether MRC5 cells could uptake sEVs. PKH67-stained sEVs were incubated with MRC5 cells for different times and the internalization of labeled-sEVs was evaluated both by confocal fluorescence (Figure 3a and Supplementary Figure S6) and flow cytometry (Figure 3b,c). As early as 2 h post-incubation, some MRC5 cells already showed numerous cytoplasmic puncta for both sEVs produced by NI and infected HIPECs (Supplementary Figure S6). At 16 h, many cells showed bright green puncta in their cytoplasm (Figure 3a), with a percentage of positive cells evaluated to 13% by flow cytometry, which was not altered up to 24 h (Figure 3b,c). Moreover, these puncta were visible in the same confocal plane as actin (revealed by phalloidin staining), corroborating the intracytoplasmic localization of PKH67 fluorescence, and excluding therefore a possible binding of sEVs to the cell surface (Figure 3a, Supplementary Figure S6b,c). However, in comparison to the fluorescence data (Figure 3a), this percentage may be somewhat underestimated, as subtraction of cell autofluorescence background may mask a significant amount of uptaken sEVs, which fluoresce poorly, given the small size of the vesicles, and thus the small number of PKH67 molecules incorporated. Altogether, these data indicate that the sEVs secreted by HIPECs are largely uptaken by recipient cells, and also suggest that the internalized sEVs may thereafter exert a biological function.
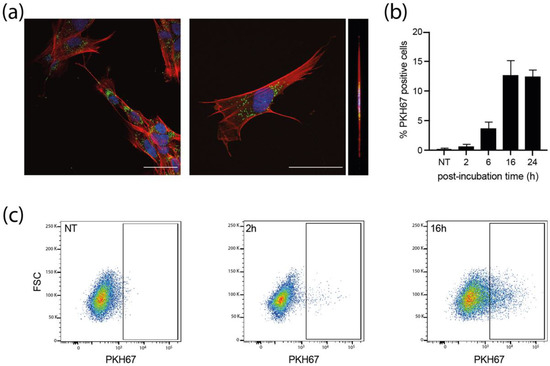
Figure 3.
Internalization of sEVs isolated from non-infected HIPECs in fetal MRC5 cells. (a) Confocal images of fluorescence microscopy carried out on MRC5 cells after 16 h incubation with PKH67-labelled sEVs. Blue: DAPI; Red: Phalloidin; Green: PKH67. Scale bar: 100 μm. Magnification = 63×. The right image corresponds to the orthogonal projection of the cell z-axis. (b) Histogram representing the percentage of PKH67 positive cells along time, upon incubation of MRC5 cells with sEVs. Bars represent the mean ± SEM of three independent experiments. (c) Monitoring of PKH67-labeled sEVs internalization by MRC5 cells by flow cytometry. Dot plots represent MRC5 cell fluorescence upon incubation with PKH67-stained sEVs (200 sEVs/cell) for cells that have not been incubated with sEVs (NT, non-treated), or upon 2 h or 16 h of incubation. X-axis: PKH-67 fluorescence intensity; Y-axis: FSC. Gate indicates cells positive for PKH67.
3.4. sEVs from hCMV-Infected HIPECs Potentialize Further Infection of Recipient MRC5 Cells
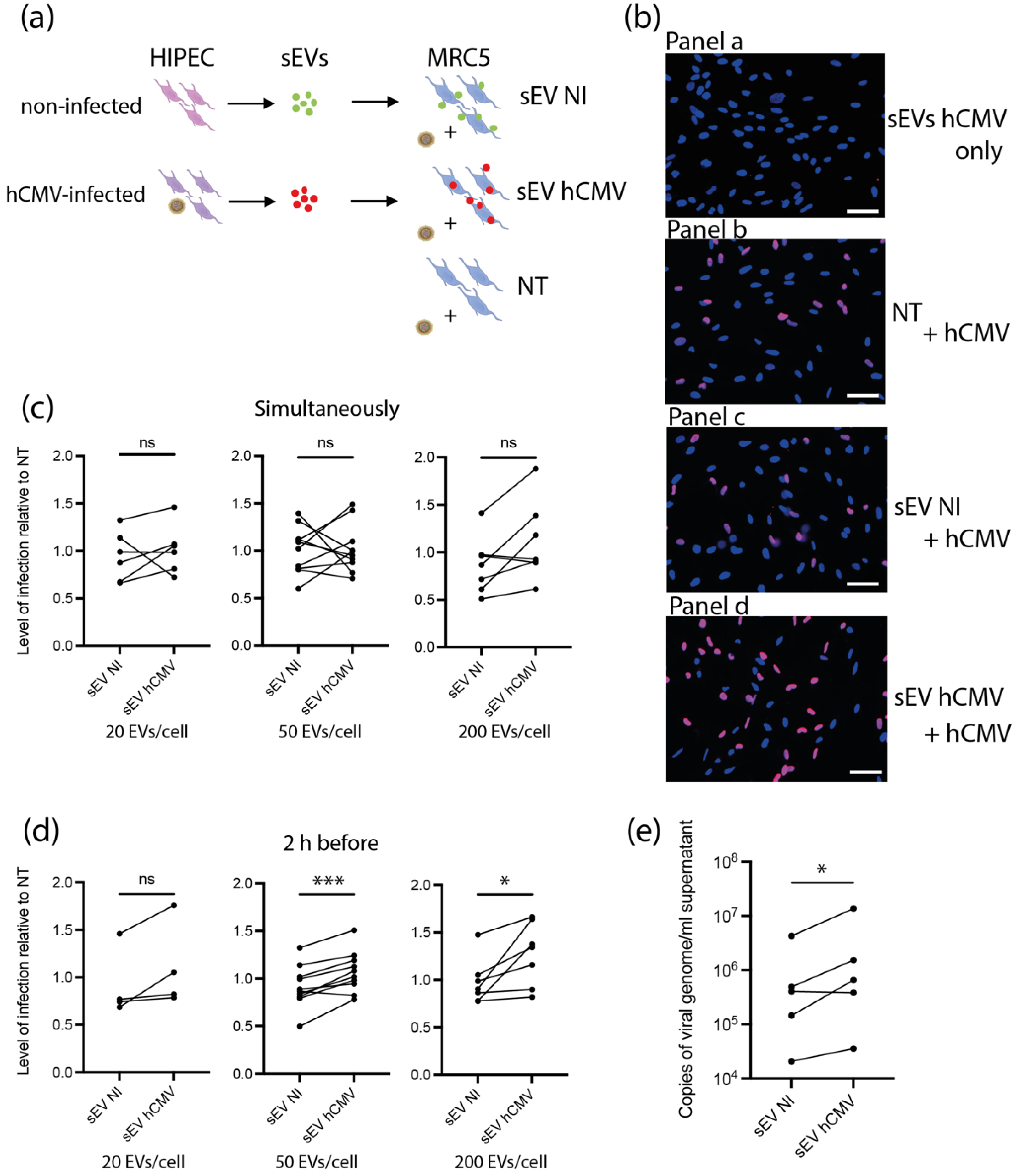
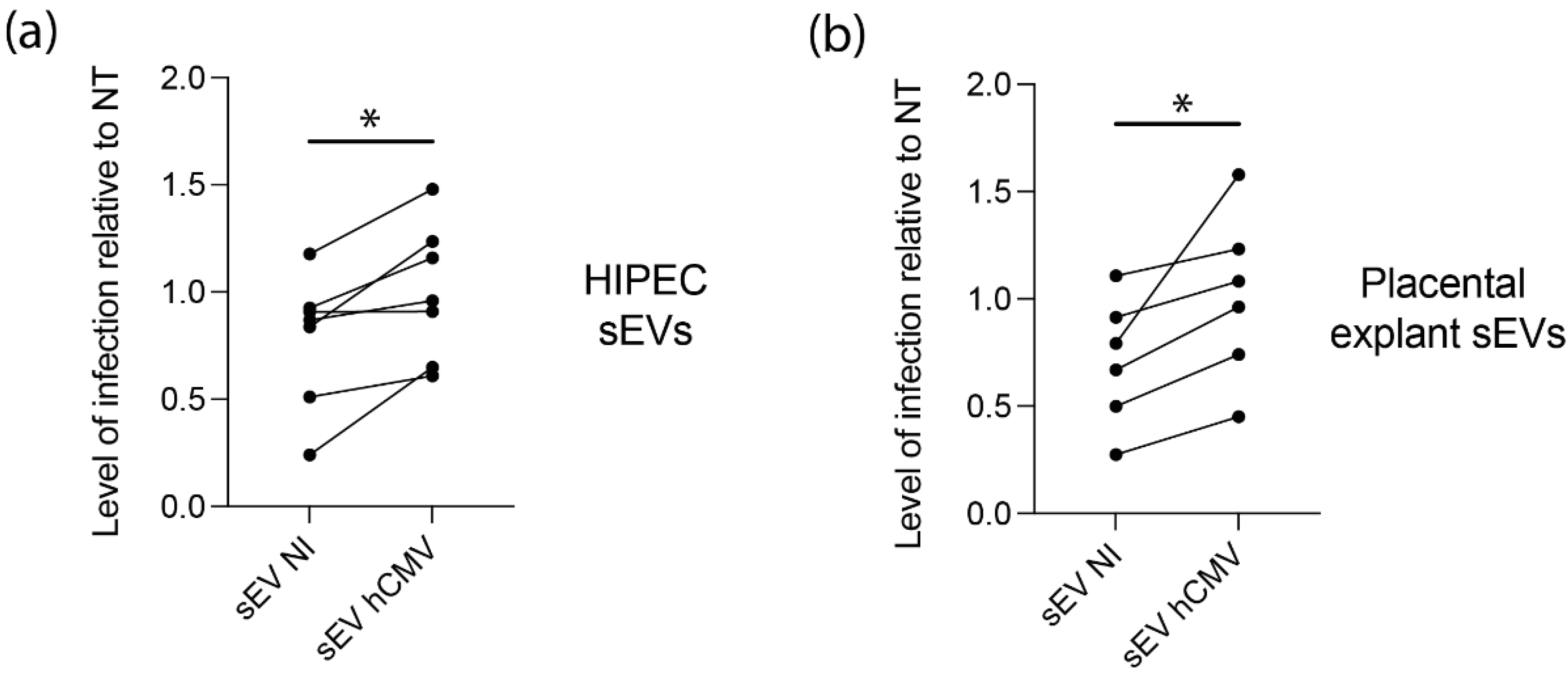
Based on the proteomic data suggesting a potential proviral activity of sEVs, we assessed their ability to modulate hCMV infection. As sEV content was composed of proteins potentially prone to act on early steps of infection-entry and immediate early transcriptions, we reasoned that any putative action of sEVs should take place immediately after delivery of their cargo into recipient cells. In this regard, different amounts of sEVs from NI or hCMV-infected HIPECs were incubated with MRC5 cells either concomitantly or 2 h before hCMV infection (Figure 4a). Then, 24 h later, cells were subjected to an anti-IE1/2 immunofluorescence (Figure 4b). When added alone, sEVs from infected HIPECs did not lead to any detectable expression of IE1/2 in MRC5 cells (Figure 4b panel a), confirming that they were not contaminated by residual infectious viral particle. This also indicates that the detection of IE proteins upon infection was due to viral gene expression and not to the presence of IE proteins carried by sEV from hCMV-infected HIPECs, even if IE proteins 1 and 2 were detected in proteomic analysis. As shown in Figure 4c, when added simultaneously with hCMV, sEVs did not influence the level of MRC5 cell infection, whatever their origin and quantity, as compared to non-treated cells. In contrast, the addition of increasing doses of sEV from infected HIPECs 2 h before infection led to a potentiation of the infection, when compared to cells treated with sEV from NI HIPECs. This increase was significant for the two highest doses of sEVs applied, with a stimulation of infection of around 17% and 30% when 50 and 200 sEVs were added per cell, respectively (Figure 4b,d). Finally, the proviral effect of sEVs from infected HIPECs was observed throughout the viral cycle, with an approximately 2.7-fold increase in the release of viral genome copies in the cell supernatant compared to cells incubated with sEVs of the NI HIPECs when 50 sEVs was added per cell (Figure 4e).
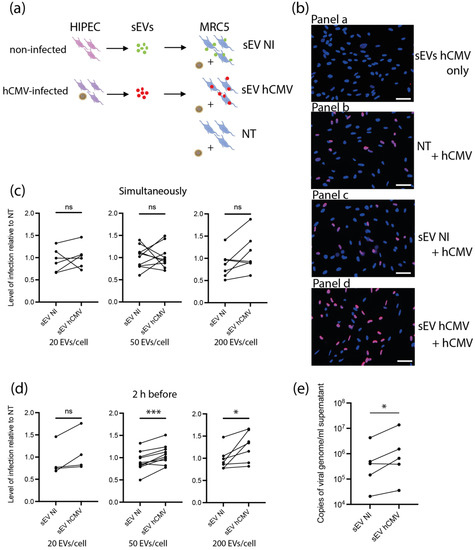
Figure 4.
Effect of HIPEC sEVs on MRC5 cell permissiveness for hCMV. (a) Experimental procedure (NT = non-treated). (b) Immunofluorescence performed on MRC5 cells against IE viral antigen (blue: DAPI; red: IE1/2). Panel a- Non-infected control MRC5 cells upon 24 h incubation with sEVs isolated from hCMV-infected HIPECs. Panels b,c,d- MRC5 cells were either non-treated (Panel b- NT) or incubated during 2 h with sEVs prepared from non-infected (Panel c- sEV NI) or infected HIPECs (Panel d- sEV hCMV) with 50 sEV per cell, then infected during 24 h with hCMV at a MOI of 0.5 before proceeding to immunofluorescence. Scale bar: 200 μm. (c,d) MRC5 cells were incubated with sEVs prepared from non-infected (sEV NI) or hCMV-infected (sEV hCMV) HIPECs and infected by hCMV at a MOI of 0.5, concomitantly (c) or 2 h after sEV incubation (d). Three increasing doses of sEVs were used in these experiments (20, 50, or 200 sEV per MRC5 cell, from left to right). Then, 24 h later, expression of IE antigen was assessed by immunofluorescence. Quantification of the percentage of IE positive cells was carried out and normalized by the percentage of cells infected by hCMV without any sEV (NT). Each dot is an independent experiment and corresponds to the mean of the counting of 10 fields, with around 70 cells counted, i.e., around 700 cells per dot. n = 4 to 10 independent experiments. Since sEVs used in infection assays were prepared each time in parallel between non-infected and hCMV-infected HIPECs from a given batch, statistical analysis was done by pairing the results between sEV NI and sEV hCMV for each independent experiment. ns, non-significant; *, p < 0.05; ***, p < 0.001 by paired t-test. (e) MRC5 cells were incubated with sEVs prepared from non-infected (sEV NI) or hCMV-infected (sEV hCMV) HIPECs and infected by hCMV at a MOI of 0.5, 2 h after sEV incubation, with 50 sEVs per cells. At 72 h post-infection, virus titration was performed by qPCR from cell culture supernatants. n = 5 independent experiments. *, p < 0.05 by ratio paired t-test.
3.5. sEVs from hCMV-Infected HIPECs and Placental Tissues Enhance Infection of Human Neural Stem Cells
Since placental sEVs are found in high amount in fetal blood, representing 45% of total fetal plasma EVs, i.e., around 5 × 1010 EV/mL fetal plasma [60,61], we examined their potential proviral role on hCMV transmission towards the fetal brain, by performing similar experiments using human neural stem cells (NSCs) with different sources of sEVs. In this regard, NSCs, which are permissive for hCMV infection, constitute a particularly relevant model for studying the consequences of hCMV infection on fetal neural progenitors [7,27]. As observed with MRC5 cells, sEVs from hCMV-infected HIPECs promote a significant enhancement of hCMV infection of NCSs as compared to sEVs from NI HIPECs, with a significant mean increase of 42% (Figure 5a). To get closer to physiological conditions, an ex vivo model of first trimester placental histocultures, that we have previously developed [37,62], was next infected by hCMV and used for sEV isolation. Recent data obtained from this model showed a modification of sEV surface markers upon hCMV infection and suggested a proviral role for placental sEVs [25]. Again, sEVs produced by infected placental histocultures significantly potentiated hCMV infection of NSCs in comparison to sEVs secreted by NI histocultures, with a mean increase of 48% (Figure 5b).
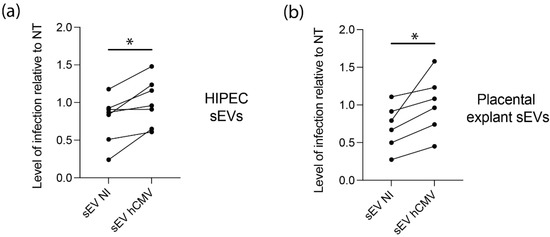
Figure 5.
Effect of sEVs from different sources on neural stem cells permissiveness for hCMV. NSCs were incubated with sEVs (200 sEVs per cell) prepared from HIPECs (a) or ex vivo first trimester placental histoculture (b), then infected by hCMV at a MOI of 3. Then, 24 h upon infection, expression of IE antigen was assessed by immunofluorescence. Each dot is an independent experiment and corresponds to the mean of the counting of 10 fields, with around 70 cells counted, i.e., around 700 cells per dot. n = 3 to 7 independent experiments. HIPECs or placental explants were either non-infected (sEV NI) or hCMV-infected (sEV hCMV). Quantification of the percentage of IE positive cells was carried out and normalized by the percentage of infection of cells infected by hCMV without any sEV (NT). Since sEVs used in functional assays were prepared each time in parallel between non-infected or hCMV-infected cytotrophoblasts and placental explants from a given batch, statistical analysis was done by pairing the results between sEV NI and sEV hCMV for each independent experiment. ns, non-significant; *, p < 0.05 by paired t-test.
4. Discussion
Although the role of placental sEVs during normal and pathological pregnancy is extensively studied [18,20,23,24], the impact and consequences of hCMV infection on placental sEVs are far to be deciphered, in particular at the beginning of pregnancy. We recently described that infection of first trimester ex vivo placental histocultures modified sEV surface markers, suggesting a potential proviral role of the sEVs [25]. However, the difficulty of purifying high sEV amounts from this model hampered the possibility to conduct a study combining exhaustive analysis of sEV composition and biological function. In this work, we used a combination of a cytotrophoblast cell line deriving from first trimester placenta [26] and ex vivo placental histocultures to isolate sEVs and examine the impact of hCMV infection on their composition and function.
Despite a large number of publications describing a role of EVs on pregnancy regulation, the nature of examined EVs is often not precisely stated, and confusion remains about the subtypes of EVs studied. Here, we focused our study on sEVs, considering their importance in communication between mother and fetus and the fact that they are often dysregulated during pregnancy pathologies [18,21,23]. Using the gold-standard method based on differential ultracentrifugation followed by density gradient ultracentrifugation, sEVs devoid of viral particles were systematically isolated in a rigorous manner [63]. Their characterization by a combination of complementary approaches revealed that they harbor exosome-like features in terms of size, structure, and presence of canonical EV markers [30], but their endosomal origin cannot formally be stated, since we did not examine the mechanisms underlying the biogenesis of sEVs in the present study. Instead, we combined proteomic and functional studies and demonstrated that sEVs from hCMV infected cells present an altered protein content and facilitate the infection of naive recipient fetal cells.
We found that hCMV infection of HIPECs increased the yield of sEV production. However, this increase was counterbalanced by a reduced cell growth upon infection. Hence, the global quantity of sEVs harvested from hCMV- or mock-infected HIPECs remained similar between both conditions. We thus choose to use similar amounts of sEVs from NI or infected HIPECs in functional experiments. However, it would be interesting to examine whether in pregnant women infected by hCMV, placental sEV amount would be modified in maternal and fetal blood compared to non-infected pregnant women.
Interestingly, most of the viral proteins identified in our proteomic study have not been described in the other proteomic study done of sEVs secreted from hCMV-infected MRC5 cells [51]. Conversely, some viral proteins identified by Turner et al., such as IR11, IRL12, and US14, were not found in our study. One plausible explanation is that, at relatively early time points within the replication cycle (48 h to 72 h upon infection in our present study), the content of sEVs secreted—highly enriched in viral envelop, capsid, tegument, and immediate early proteins—may prime the neighboring cells for viral dissemination and spread. In contrast, at latter time point (5 days post infection in Turner et al.’s study), the sEV protein cargo could more serve immuno-evasion functions, by expressing, for example, the viral Fc-gamma receptor homologue IR11/gp34 [51]. Hence, it seems that composition of sEV secreted by hCMV-infected cells evolves with time, depending on their state of infection and the step of hCMV replication cycle.
In addition to the viral proteins carried by HIPEC sEVs, sEV composition for proteins of cellular origin was also altered upon hCMV infection. By using IPA to analyze biological pathway, the term “autophagy” was the first over-represented in sEVs from infected HIPECs, likely reflecting the autophagy activation induced by hCMV in host cells at the very early times of infection [42,43,44]. Importantly, TRS1 and IRS1, described to antagonize autophagy at latter time points of the viral cycle [42,43], were also found in sEVs, which may consequently inhibit the induction of autophagy in recipient cells upon sEV uptake. Thy-1, which plays an important role for favoring hCMV entry into cells by macropinocytosis [40,41], was also highly over-represented in sEVs from infected cells and we can imagine that it may be deposited via sEVs on recipient cell membranes to promote hCMV entry. Hence, all the elements brought by the proteomic analysis of sEVs from infected HIPECs indicate that they were prone to potentially facilitate viral infection of recipient cells. Interestingly, a previous bead-based flow cytometry analysis carried out in our team showed a modification in placental sEV surface markers [25]. Although the modifications of the surface expression pattern obtained in that study were different from those in the present one (Supplementary Figure S3), certainly due to the nature of the markers expressed by placental tissues compared with the HIPEC, they already suggested the hypothesis of a proviral role of placental sEVs upon hCMV infection [25].
By examining the function of sEVs from NI or hCMV-infected HIPECs, we observed that sEVs hCMV enhanced further infection of MRC5 cells. Such proviral properties of sEVs produced by infected cells have already been described for other Herpesviridae [64,65] and very recently for hCMV [52]—although not in a placental context—and confirm a general role of sEVs in modulating viral transmission for many viral families [66,67]. Placental sEVs are found both in maternal and fetal sides at high quantities as soon as the early beginning of pregnancy [60,61], representing 45% of total fetal plasma EVs (around 5 × 1010 EV/mL fetal plasma). We thus hypothesized that they may facilitate hCMV dissemination not only in the placental tissues but also towards the fetus, notably in the fetal brain since at this stage of development the blood brain barrier is highly permeable, by enhancing hCMV infectivity in fetal neural cells. Indeed, we observed that sEVs prepared from HIPECs or first trimester placental explants enhanced hCMV infection of human NSCs.
Even if further studies are now needed to confirm the hypothesis of a role of placental sEVs in the transmission of hCMV to the fetal brain, in particular to examine whether placental sEVs may exhibit a facilitating action of viral infection in vivo, our study suggests that placental sEVs could be important players of hCMV dissemination towards the fetus during congenital infection.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v14092030/s1, Figure S1: Model system; Figure S2: Wide-field transmission electron microscopy; Figure S3: Multiplex bead-based flow cytometry comparison of sEV prepared from non-infected of hCMV-infected HIPECs; Figure S4: Wide field immuno-electron microscopy anti CD63; Figure S5: Complement to proteomic analysis; Figure S6: sEV internalization by MRC5 at 2 h post-incubation; Table S1: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD029146; Table S2: Bibliography references for establishment of Figure 2d.
Author Contributions
Conceptualization, C.E.M. and M.B. (Mathilde Bergamelli); methodology, C.E.M., M.B. (Mathilde Bergamelli), O.B.-S., N.M. and G.D.; validation, M.B. (Mathilde Bergamelli), H.M., J.-M.M., J.I., I.H., G.R., M.M., O.B.-S., G.D. and C.E.M.; formal analysis, M.B. (Mathilde Bergamelli), H.M., I.H., M.M., O.B.-S., Y.A., G.D. and C.E.M.; resources, T.F., A.B., J.I., G.R., M.B. (Mélinda Bénard), M.G., G.C., Y.T.L.G. and N.M.; data curation, M.B. (Mathilde Bergamelli), H.M., J.-M.M., I.H. and M.M.; writing—original draft preparation, C.E.M. and G.D. with the help of M.B. (Mathilde Bergamelli), Y.A. and O.B.-S.; writing—review and editing, C.E.M.; supervision, C.E.M.; project administration, C.E.M.; funding acquisition, C.E.M., G.R. and O.B.-S. All authors have read and agreed to the published version of the manuscript.
Funding
This project has received financial support from the French Biomedicine Agency, and institutional grants from Inserm, CNRS, and Toulouse 3 University. This project is part of the doctorate thesis of Mathilde Bergamelli, who was funded by the Ministry of Education and Research (MESR). The TEM experiments were performed on PICT-IBiSA, Institut Curie, Paris, member of the France-BioImaging national research infrastructure, and were supported by the French National Research Agency through the “Investments for the Future” program (France-BioImaging, ANR-11-INSB-04), supported by the CelTisPhyBio Labex (N° ANR-11-LB0038) part of the IDEX PSL (N°ANR-10-IDEX-0001-02 PSL). The proteomic part of this project was supported in part by the Région Occitanie, European funds (Fonds Européens de Développement Régional, FEDER), Toulouse Métropole, and by the French Ministry of Research with the Investissement d’Avenir Infrastructures Nationales en Biologie et Santé program (ProFI, Proteomics French Infrastructure project, ANR-10-INBS-08).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki. The use of NSCs from human embryonic stem cells was approved by the French authorities (Agence de la Biomédecine, authorization number SASB0920178S). The biological resource center Germethèque (BB-0033-00081; declaration: DC-2014-2202; authorization: AC-2015-2350) obtained the written consent from each patient (CPP.2.15.27) for the use of human samples and their associated data. For first trimester placenta explants, the steering committee of Germethèque gave its approval for the realization of this study on Feb 5 2019. The hosting request made to Germethèque bears the number 20190201 and its contract is referenced under the number 19 155C.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [35] partner repository with the dataset identifier PXD029146.
Acknowledgments
The authors warmly thank C. Sinzger for VHL/E strain of hCMV. The authors greatly thank C. Chouquet, B. Rauwel, M. Mouysset, L. Battut, C. Mengelle, S. Lhomme, F. Chauvrier, P. Verdy, and N. Kopf, as well as the whole ViNeDys team, for their technical assistance and their numerous advice and discussions which allowed the progress of this work. The authors also thank AL. Iscache, V. Duplan-Eche, and F. L’Faqihi-Olive, from the cytometry facility of Infinity, as well as S. Allart and D. Daviaud, from the imaging facility of Infinity, and finally, the GeT technical service of Infinity. The authors thank the medical and paramedical staff of the gynecology unit at Paule de Viguier Hospital, the patients who agreed to participate in the study, as well as L. Bujan and M. Aubry, from the Germethèque. The authors finally warmly thank E. Haanappel from the Institute of Pharmacology and Structural Biology, who granted the authors access to the Nanosight device, and Audrey Esclatine and Daniel Gonzalez-Dunia for critical reading of the manuscript and insightful comments.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Cannon, M.J.; Schmid, D.S.; Hyde, T.B. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev. Med. Virol. 2010, 20, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Leruez-Ville, M.; Foulon, I.; Pass, R.; Ville, Y. Cytomegalovirus infection during pregnancy: State of the science. Am. J. Obstet. Gynecol. 2020, 223, 330–349. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.; Maidji, E.; Fisher, S.J.; McDonagh, S.; Tabata, T. HCMV persistence in the population: Potential transplacental transmission. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Pereira, L.; Tabata, T.; Petitt, M.; Fang-Hoover, J. Congenital cytomegalovirus infection undermines early development and functions of the human placenta. Placenta 2017, 59, S8–S16. [Google Scholar] [CrossRef] [PubMed]
- Tabata, T.; Petitt, M.; Fang-Hoover, J.; Rivera, J.; Nozawa, N.; Shiboski, S.; Inoue, N.; Pereira, L. Cytomegalovirus Impairs Cytotrophoblast-Induced Lymphangiogenesis and Vascular Remodeling in an In Vivo Human Placentation Model. Am. J. Pathol. 2012, 181, 1540–1559. [Google Scholar] [CrossRef]
- Tabata, T.; Petitt, M.; Fang-Hoover, J.; Zydek, M.; Pereira, L. Persistent Cytomegalovirus Infection in Amniotic Membranes of the Human Placenta. Am. J. Pathol. 2016, 186, 2970–2986. [Google Scholar] [CrossRef]
- Rolland, M.; Li, X.; Sellier, Y.; Martin, H.; Perez-Berezo, T.; Rauwel, B.; Benchoua, A.; Bessières, B.; Aziza, J.; Cenac, N.; et al. PPARgamma Is Activated during Congenital Cytomegalovirus Infection and Inhibits Neuronogenesis from Human Neural Stem Cells. PLoS Pathog. 2016, 12, e1005547. [Google Scholar] [CrossRef]
- Rolland, M.; Martin, H.; Bergamelli, M.; Sellier, Y.; Bessières, B.; Aziza, J.; Benchoua, A.; Leruez-Ville, M.; Gonzalez-Dunia, D.; Chavanas, S. Human cytomegalovirus infection is associated with increased expression of the lissencephaly gene PAFAH1B1 encoding LIS1 in neural stem cells and congenitally infected brains. J. Pathol. 2021, 254, 92–102. [Google Scholar] [CrossRef]
- Sarker, S.; Scholz-Romero, K.; Perez, A.; Illanes, S.E.; Mitchell, M.D.; Rice, G.E.; Salomon, C. Placenta-derived exosomes continuously increase in maternal circulation over the first trimester of pregnancy. J. Transl. Med. 2014, 12, 204. [Google Scholar] [CrossRef]
- Luo, S.-S.; Ishibashi, O.; Ishikawa, G.; Ishikawa, T.; Katayama, A.; Mishima, T.; Takizawa, T.; Shigihara, T.; Goto, T.; Izumi, A.; et al. Human Villous Trophoblasts Express and Secrete Placenta-Specific MicroRNAs into Maternal Circulation via Exosomes. Biol. Reprod. 2009, 81, 717–729. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Ishida, Y.; Zhao, D.; Ohkuchi, A.; Kuwata, T.; Yoshitake, H.; Yuge, K.; Takizawa, T.; Matsubara, S.; Suzuki, M.; Saito, S.; et al. Maternal peripheral blood natural killer cells incorporate placenta-associated microRNAs during pregnancy. Int. J. Mol. Med. 2015, 35, 1511–1524. [Google Scholar] [CrossRef] [PubMed]
- Chaiwangyen, W.; Murrieta-Coxca, J.M.; Favaro, R.R.; Photini, S.M.; Gutiérrez-Samudio, R.N.; Schleussner, E.; Markert, U.R.; Morales-Prieto, D.M. MiR-519d-3p in Trophoblastic Cells: Effects, Targets and Transfer to Allogeneic Immune Cells via Extracellular Vesicles. Int. J. Mol. Sci. 2020, 21, 3458. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Pinilla-Macua, I.; Ouyang, Y.; Sadovsky, E.; Kajiwara, K.; Sorkin, A.; Sadovsky, Y. Internalization of trophoblastic small extracellular vesicles and detection of their miRNA cargo in P-bodies. J. Extracell. Vesicles 2020, 9, 1812261. [Google Scholar] [CrossRef]
- Salomon, C.; Yee, S.; Scholz-Romero, K.; Kobayashi, M.; Vaswani, K.; Kvaskoff, D.; Illanes, S.; Mitchell, M.; Rice, G.E. Extravillous trophoblast cells-derived exosomes promote vascular smooth muscle cell migration. Front. Pharmacol. 2014, 5, 175. [Google Scholar] [CrossRef]
- Sadovsky, Y.; Ouyang, Y.; Powell, J.S.; Li, H.; Mouillet, J.-F.; Morelli, A.E.; Sorkin, A.; Margolis, L. Placental small extracellular vesicles: Current questions and investigative opportunities. Placenta 2020, 102, 34–38. [Google Scholar] [CrossRef]
- Chiarello, D.I.; Salsoso, R.; Toledo, F.; Mate, A.; Vázquez, C.M.; Sobrevia, L. Foetoplacental communication via extracellular vesicles in normal pregnancy and preeclampsia. Mol. Asp. Med. 2017, 60, 69–80. [Google Scholar] [CrossRef]
- Mitchell, M.D.; Peiris, H.N.; Kobayashi, M.; Koh, Y.Q.; Duncombe, G.; Illanes, S.E.; Rice, G.E.; Salomon, C. Placental exosomes in normal and complicated pregnancy. Am. J. Obstet. Gynecol. 2015, 213, S173–S181. [Google Scholar] [CrossRef]
- Salomon, C.; Rice, G.E. Role of Exosomes in Placental Homeostasis and Pregnancy Disorders. Prog. Mol. Biol. Transl. Sci. 2017, 145, 163–179. [Google Scholar]
- Salomon, C.; Scholz-Romero, K.; Sarker, S.; Sweeney, E.; Kobayashi, M.; Correa, P.; Longo, S.; Duncombe, G.; Mitchell, M.D.; Rice, G.E.; et al. Gestational Diabetes Mellitus Is Associated with Changes in the Concentration and Bioactivity of Placenta-Derived Exosomes in Maternal Circulation across Gestation. Diabetes 2015, 65, 598–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, J.; Menon, R. Placental exosomes: A proxy to understand pregnancy complications. Am. J. Reprod. Immunol. 2017, 79, e12788. [Google Scholar] [CrossRef] [PubMed]
- Malnou, C.; Umlauf, D.; Mouysset, M.; Cavaillé, J. Imprinted MicroRNA Gene Clusters in the Evolution, Development, and Functions of Mammalian Placenta. Front. Genet. 2019, 9, 706. [Google Scholar] [CrossRef] [PubMed]
- Bergamelli, M.; Martin, H.; Bénard, M.; Ausseil, J.; Mansuy, J.-M.; Hurbain, I.; Mouysset, M.; Groussolles, M.; Cartron, G.; le Gac, Y.T.; et al. Human Cytomegalovirus Infection Changes the Pattern of Surface Markers of Small Extracellular Vesicles Isolated from First Trimester Placental Long-Term Histocultures. Front. Cell Dev. Biol. 2021, 9, 689122. [Google Scholar] [CrossRef]
- Pavan, L.; Tarrade, A.; Hermouet, A.; Delouis, C.; Titeux, M.; Vidaud, M.; Thérond, P.; Evain-Brion, D.; Fournier, T. Human invasive trophoblasts transformed with simian virus 40 provide a new tool to study the role of PPARgamma in cell invasion process. Carcinogenesis 2003, 24, 1325–1336. [Google Scholar] [CrossRef]
- Boissart, C.; Nissan, X.; Giraud-Triboult, K.; Peschanski, M.; Benchoua, A. miR-125 potentiates early neural specification of human embryonic stem cells. Development 2012, 139, 1247–1257. [Google Scholar] [CrossRef]
- Stegmann, C.; Rothemund, F.; Laib Sampaio, K.; Adler, B.; Sinzger, C. The N Terminus of Human Cytomegalovirus Glycoprotein O Is Important for Binding to the Cellular Receptor PDGFRalpha. J. Virol. 2019, 93, e00138-19. [Google Scholar] [CrossRef]
- Mengelle, C.; Sandres-Sauné, K.; Mansuy, J.; Haslé, C.; Boineau, J.; Izopet, J. Performance of a completely automated system for monitoring CMV DNA in plasma. J. Clin. Virol. 2016, 79, 25–31. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- EV-TRACK Consortium; Van Deun, J.; Mestdagh, P.; Agostinis, P.; Akay, Ö.; Anand, S.; Anckaert, J.; Martinez, Z.A.; Baetens, T.; Beghein, E.; et al. EV-TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods 2017, 14, 228–232. [Google Scholar] [CrossRef]
- Hurbain, I.; Romao, M.; Bergam, P.; Heiligenstein, X.; Raposo, G. Analyzing Lysosome-Related Organelles by Electron Microscopy. In Lysosomes; Humana Press: New York, NY, USA, 2017; Volume 1594, pp. 43–71. [Google Scholar] [CrossRef]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Bouyssié, D.; Hesse, A.-M.; Mouton-Barbosa, E.; Rompais, M.; Macron, C.; Carapito, C.; De Peredo, A.G.; Couté, Y.; Dupierris, V.; Burel, A.; et al. Proline: An efficient and user-friendly software suite for large-scale proteomics. Bioinformatics 2020, 36, 3148–3155. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Bonchev, D. A survey of current software for network analysis in molecular biology. Hum. Genom. 2010, 4, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Lopez, H.; Benard, M.; Saint-Aubert, E.; Baron, M.; Martin, H.; Al Saati, T.; Plantavid, M.; Duga-Neulat, I.; Berrebi, A.; Cristini, C.; et al. Novel model of placental tissue explants infected by cytomegalovirus reveals different permissiveness in early and term placentae and inhibition of indoleamine 2,3-dioxygenase activity. Placenta 2011, 32, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Leghmar, K.; Cenac, N.; Rolland, M.; Martin, H.; Rauwel, B.; Bertrand-Michel, J.; Le Faouder, P.; Bénard, M.; Casper, C.; Davrinche, C.; et al. Cytomegalovirus Infection Triggers the Secretion of the PPARgamma Agonists 15-Hydroxyeicosatetraenoic Acid (15-HETE) and 13-Hydroxyoctadecadienoic Acid (13-HODE) in Human Cytotrophoblasts and Placental Cultures. PLoS ONE 2015, 10, e0132627. [Google Scholar] [CrossRef]
- Rauwel, B.; Mariamé, B.; Martin, H.; Nielsen, R.; Allart, S.; Pipy, B.; Mandrup, S.; Devignes, M.D.; Evain-Brion, D.; Fournier, T.; et al. Activation of Peroxisome Proliferator-Activated Receptor Gamma by Human Cytomegalovirus for De Novo Replication Impairs Migration and Invasiveness of Cytotrophoblasts from Early Placentas. J. Virol. 2010, 84, 2946–2954. [Google Scholar] [CrossRef]
- Li, Q.; Fischer, E.; Cohen, J.I. Cell Surface THY-1 Contributes to Human Cytomegalovirus Entry via a Macropinocytosis-like Process. J. Virol. 2016, 90, 9766–9781. [Google Scholar] [CrossRef]
- Li, Q.; Wilkie, A.R.; Weller, M.; Liu, X.; Cohen, J.I. THY-1 Cell Surface Antigen (CD90) Has an Important Role in the Initial Stage of Human Cytomegalovirus Infection. PLoS Pathog. 2015, 11, e1004999. [Google Scholar] [CrossRef]
- Chaumorcel, M.; Lussignol, M.; Mouna, L.; Cavignac, Y.; Fahie, K.; Cotte-Laffitte, J.; Geballe, A.; Brune, W.; Beau, I.; Codogno, P.; et al. The Human Cytomegalovirus Protein TRS1 Inhibits Autophagy via Its Interaction with Beclin 1. J. Virol. 2012, 86, 2571–2584. [Google Scholar] [CrossRef] [Green Version]
- Mouna, L.; Hernandez, E.; Bonte, D.; Brost, R.; Amazit, L.; Delgui, L.R.; Brune, W.; Geballe, A.P.; Beau, I.; Esclatine, A. Analysis of the role of autophagy inhibition by two complementary human cytomegalovirus BECN1/Beclin 1-binding proteins. Autophagy 2016, 12, 327–342. [Google Scholar] [CrossRef] [PubMed]
- Taisne, C.; Lussignol, M.; Hernandez, E.; Moris, A.; Mouna, L.; Esclatine, A. Human cytomegalovirus hijacks the autophagic machinery and LC3 homologs in order to optimize cytoplasmic envelopment of mature infectious particles. Sci. Rep. 2019, 9, 4560. [Google Scholar] [CrossRef] [PubMed]
- Karniely, S.; Weekes, M.P.; Antrobus, R.; Rorbach, J.; van Haute, L.; Umrania, Y.; Smith, D.L.; Stanton, R.J.; Minczuk, M.; Lehner, P.J.; et al. Human Cytomegalovirus Infection Upregulates the Mitochondrial Transcription and Translation Machineries. mBio 2016, 7, e00029-16. [Google Scholar] [CrossRef] [PubMed]
- Federspiel, J.D.; Cook, K.C.; Kennedy, M.A.; Venkatesh, S.S.; Otter, C.J.; Hofstadter, W.A.; Beltran, P.M.J.; Cristea, I.M. Mitochondria and Peroxisome Remodeling across Cytomegalovirus Infection Time Viewed through the Lens of Inter-ViSTA. Cell Rep. 2020, 32, 107943. [Google Scholar] [CrossRef]
- Combs, J.A.; Norton, E.B.; Saifudeen, Z.R.; Bentrup, K.H.Z.; Katakam, P.V.; Morris, C.A.; Myers, L.; Kaur, A.; Sullivan, D.E.; Zwezdaryk, K.J. Human Cytomegalovirus Alters Host Cell Mitochondrial Function during Acute Infection. J. Virol. 2020, 94, e01183-19. [Google Scholar] [CrossRef]
- Monk, C.H.; Zwezdaryk, K.J. Host Mitochondrial Requirements of Cytomegalovirus Replication. Curr. Clin. Microbiol. Rep. 2020, 7, 115–123. [Google Scholar] [CrossRef]
- Seo, J.-Y.; Yaneva, R.; Hinson, E.R.; Cresswell, P. Human Cytomegalovirus Directly Induces the Antiviral Protein Viperin to Enhance Infectivity. Science 2011, 332, 1093–1097. [Google Scholar] [CrossRef]
- Zicari, S.; Arakelyan, A.; Palomino, R.A.Ñ.; Fitzgerald, W.; Vanpouille, C.; Lebedeva, A.; Schmitt, A.; Bomsel, M.; Britt, W.; Margolis, L. Human cytomegalovirus-infected cells release extracellular vesicles that carry viral surface proteins. Virology 2018, 524, 97–105. [Google Scholar] [CrossRef]
- Turner, D.L.; Korneev, D.V.; Purdy, J.G.; De Marco, A.; Mathias, R.A. The host exosome pathway underpins biogenesis of the human cytomegalovirus virion. eLife 2020, 9, e58288. [Google Scholar] [CrossRef]
- Streck, N.T.; Zhao, Y.; Sundstrom, J.M.; Buchkovich, N.J. Human Cytomegalovirus Utilizes Extracellular Vesicles to Enhance Virus Spread. J. Virol. 2020, 94, e00609-20. [Google Scholar] [CrossRef]
- Mocarski, E.S.; Kemble, G.W.; Lyle, J.M.; Greaves, R.F. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA 1996, 93, 11321–11326. [Google Scholar] [CrossRef] [PubMed]
- Nevels, M.; Paulus, C.; Shenk, T. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc. Natl. Acad. Sci. USA 2004, 101, 17234–17239. [Google Scholar] [CrossRef] [PubMed]
- Wiebusch, L.; Hagemeier, C. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G(1). J. Virol. 1999, 73, 9274–9283. [Google Scholar] [CrossRef]
- Cristea, I.M.; Moorman, N.J.; Terhune, S.S.; Cuevas, C.D.; O’Keefe, E.S.; Rout, M.P.; Chait, B.T.; Shenk, T. Human Cytomegalovirus pUL83 Stimulates Activity of the Viral Immediate-Early Promoter through Its Interaction with the Cellular IFI16 Protein. J. Virol. 2010, 84, 7803–7814. [Google Scholar] [CrossRef]
- Arcangeletti, M.-C.; Rodighiero, I.; Mirandola, P.; De Conto, F.; Covan, S.; Germini, D.; Razin, S.; Dettori, G.; Chezzi, C. Cell-cycle-dependent localization of human cytomegalovirus UL83 phosphoprotein in the nucleolus and modulation of viral gene expression in human embryo fibroblasts in vitro. J. Cell. Biochem. 2010, 112, 307–317. [Google Scholar] [CrossRef]
- Lukashchuk, V.; McFarlane, S.; Everett, R.D.; Preston, C.M. Human Cytomegalovirus Protein pp71 Displaces the Chromatin-Associated Factor ATRX from Nuclear Domain 10 at Early Stages of Infection. J. Virol. 2008, 82, 12543–12554. [Google Scholar] [CrossRef]
- Fu, Y.-Z.; Su, S.; Gao, Y.-Q.; Wang, P.-P.; Huang, Z.-F.; Hu, M.-M.; Luo, W.-W.; Li, S.; Luo, M.-H.; Wang, Y.-Y.; et al. Human Cytomegalovirus Tegument Protein UL82 Inhibits STING-Mediated Signaling to Evade Antiviral Immunity. Cell Host Microbe 2017, 21, 231–243. [Google Scholar] [CrossRef]
- Liu, H.; Kang, M.; Wang, J.; Blenkiron, C.; Lee, A.; Wise, M.; Chamley, L.; Chen, Q. Estimation of the burden of human placental micro- and nano-vesicles extruded into the maternal blood from 8 to 12 weeks of gestation. Placenta 2018, 72–73, 41–47. [Google Scholar] [CrossRef]
- Miranda, J.; Paules, C.; Nair, S.; Lai, A.; Palma, C.; Scholz-Romero, K.; Rice, G.E.; Gratacos, E.; Crispi, F.; Salomon, C. Placental exosomes profile in maternal and fetal circulation in intrauterine growth restriction—Liquid biopsies to monitoring fetal growth. Placenta 2018, 64, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Benard, M.; Straat, K.; Omarsdottir, S.; Leghmari, K.; Bertrand, J.; Davrinche, C.; Duga-Neulat, I.; Söderberg-Nauclér, C.; Rahbar, A.; Casper, C. Human cytomegalovirus infection induces leukotriene B4 and 5-lipoxygenase expression in human placentae and umbilical vein endothelial cells. Placenta 2014, 35, 345–350. [Google Scholar] [CrossRef]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3.22. [Google Scholar] [CrossRef] [PubMed]
- Bello-Morales, R.; López-Guerrero, J.A. Extracellular vesicles in herpes viral spread and immune evasion. Front. Microbiol. 2018, 9, 2572. [Google Scholar] [CrossRef] [PubMed]
- Cone, A.S.; York, S.B.; Meckes, D.G., Jr. Extracellular Vesicles in Epstein-Barr Virus Pathogenesis. Curr. Clin. Microbiol. Rep. 2019, 6, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Lenassi, M.; Cagney, G.; Liao, M.; Vaupotic, T.; Bartholomeeusen, K.; Cheng, Y.; Krogan, N.J.; Plemenitas, A.; Peterlin, B.M. HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic 2010, 11, 110–122. [Google Scholar] [CrossRef]
- Grünvogel, O.; Colasanti, O.; Lee, J.-Y.; Klöss, V.; Belouzard, S.; Reustle, A.; Esser-Nobis, K.; Hesebeck-Brinckmann, J.; Mutz, P.; Hoffmann, K.; et al. Secretion of Hepatitis C Virus Replication Intermediates Reduces Activation of Toll-like Receptor 3 in Hepatocytes. Gastroenterology 2018, 154, 2237–2251.e16. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).