Novel P335-like Phage Resistance Arises from Deletion within Putative Autolysin yccB in Lactococcus lactis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Phages
| Biological Material | Relevant Characteristics | Reference |
|---|---|---|
| Bacteria | ||
| Lactococcus lactis | ||
| DGCC12699 | Phage-sensitive parental starter strain (DGCC12699 yccB accession number: OR544030) | This study |
| BIM2 | DGCC12699 yccB mutant | This study |
| BIM8 | DGCC12699 yccB mutant | This study |
| BIM19 | DGCC12699 yccB mutant | This study |
| BIM21 | DGCC12699 CWPS mutant | This study |
| BIM2 + pG9yccB | BIM2 transformant harboring pG9yccB | This study |
| BIM2 + pG9 | BIM2 transformant harboring empty pG9 vector | This study |
| BIM2ΔpG9yccB | BIM2 + pG9yccB after pG9yccB curing | This study |
| DGCC12699 + pG9RBP7893 | DGCC12699 transformant harboring pG9RBP7893 | This study |
| Escherichia coli | ||
| TG1 RepA+ | Plasmid propagation host; supE hsd∆5 thi ∆(kac-proAB) F’ (traD36 proAB+ lacIq lacZ∆M15) with chromosomal copy of pWV01 repA | [39] |
| Phages | ||
| D6867 | Propagation host DGCC12699; P335-like (accession number OR480986) | This study |
| D7138 | Propagation host DGCC12699; P335-like (accession number OR480987) | This study |
| D7893 | Propagation host BIM2; P335-like (accession number OR480988) | This study |
| D5694 | Propagation host DGCC12699; Teubervirus | This study |
| D6867-RBP7893 | Propagation host DGCC12699; D6867 Tal-RBP recombinant | This study |
| Plasmids | ||
| pGhost9 | Erythromycin resistance, temperature sensitive vector; aka pG9 | [40] |
| pG9yccB | pG9 + wild-type yccB from DGCC12699 | This study |
| pG9RBP7893 | pG9 + 3′ end of D7893 Tal-RBP | This study |
2.2. BIM Isolation
2.3. PCR, DNA Isolation, Sequencing, and Bioinformatic Analyses
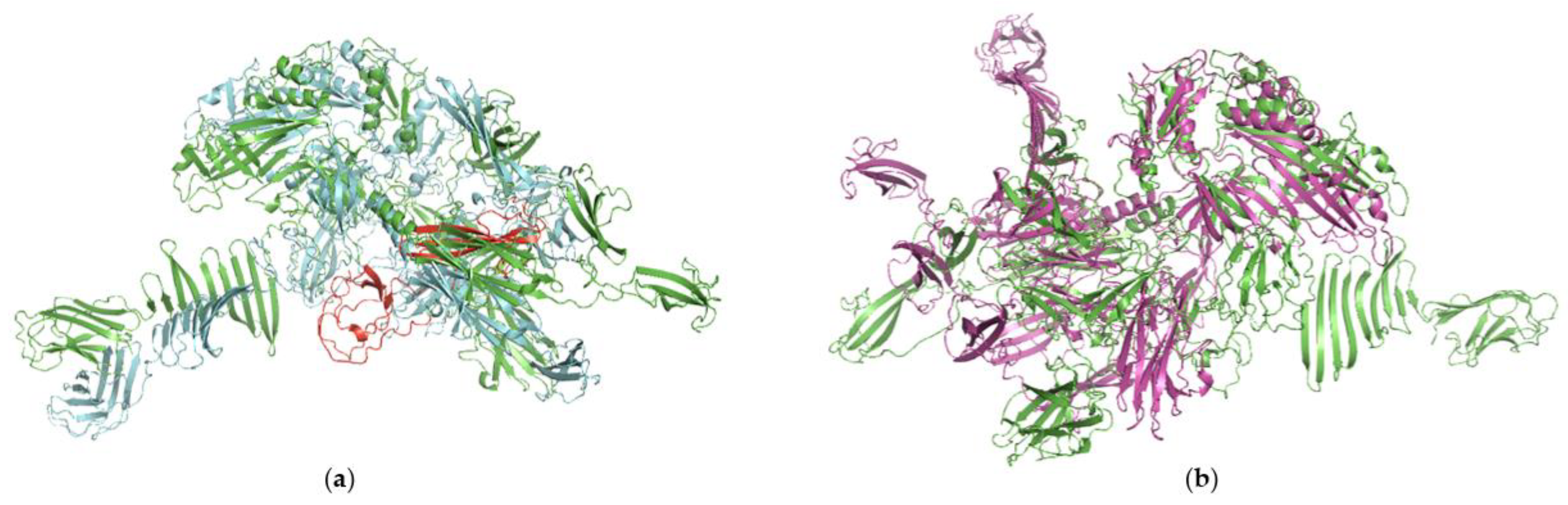
2.4. Protein Modeling
2.5. yccB Complementation and Phage Recombination
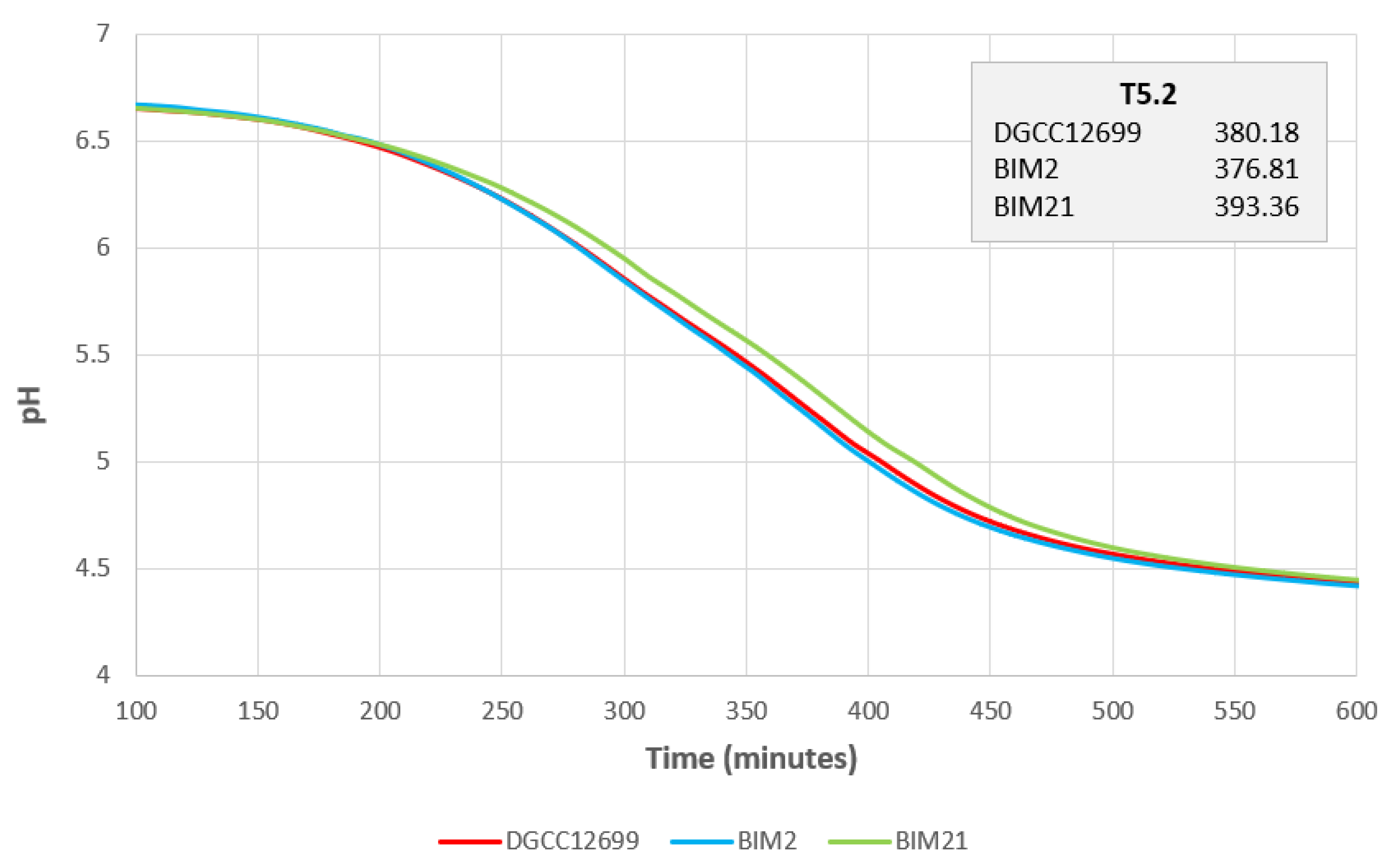
2.6. Acidification Profile
3. Results
3.1. BIM Isolation and Phage Resistance Characterization
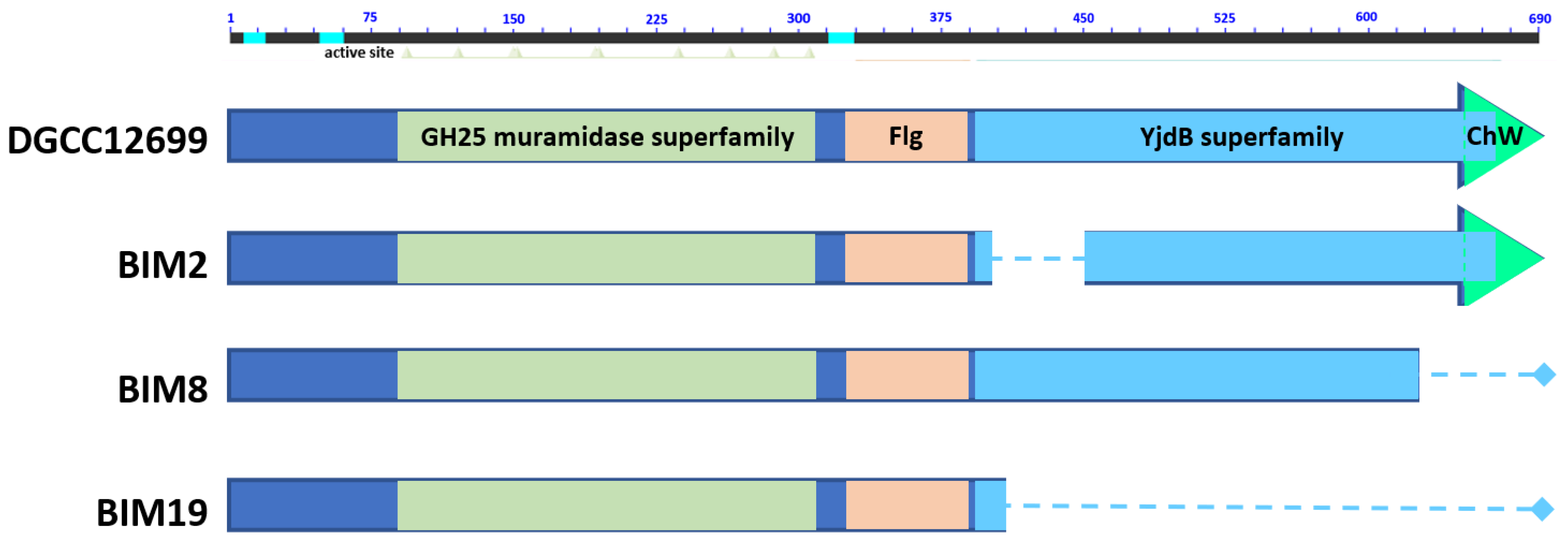
3.2. Identification of Lactococcal Mutations Conferring Phage Resistance
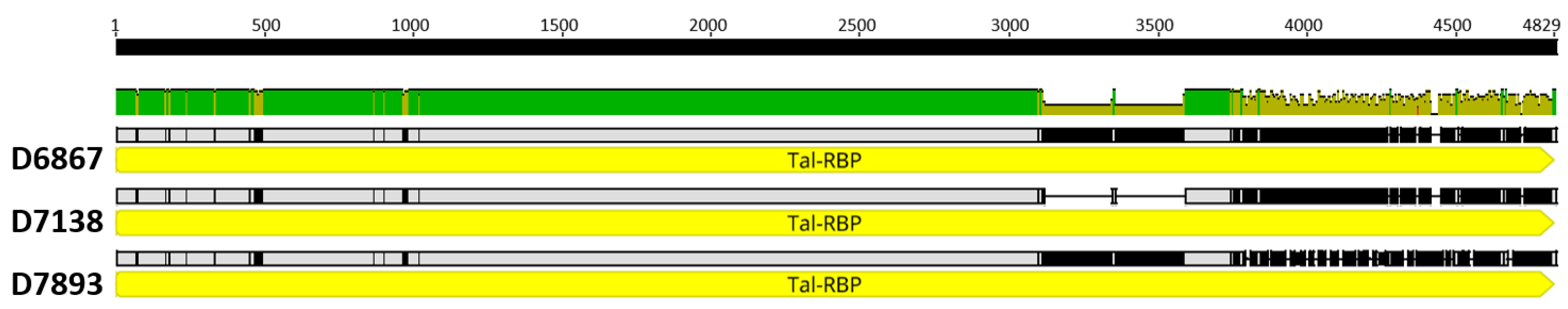
3.3. Phage Genome Analyses and D7893 Infectivity
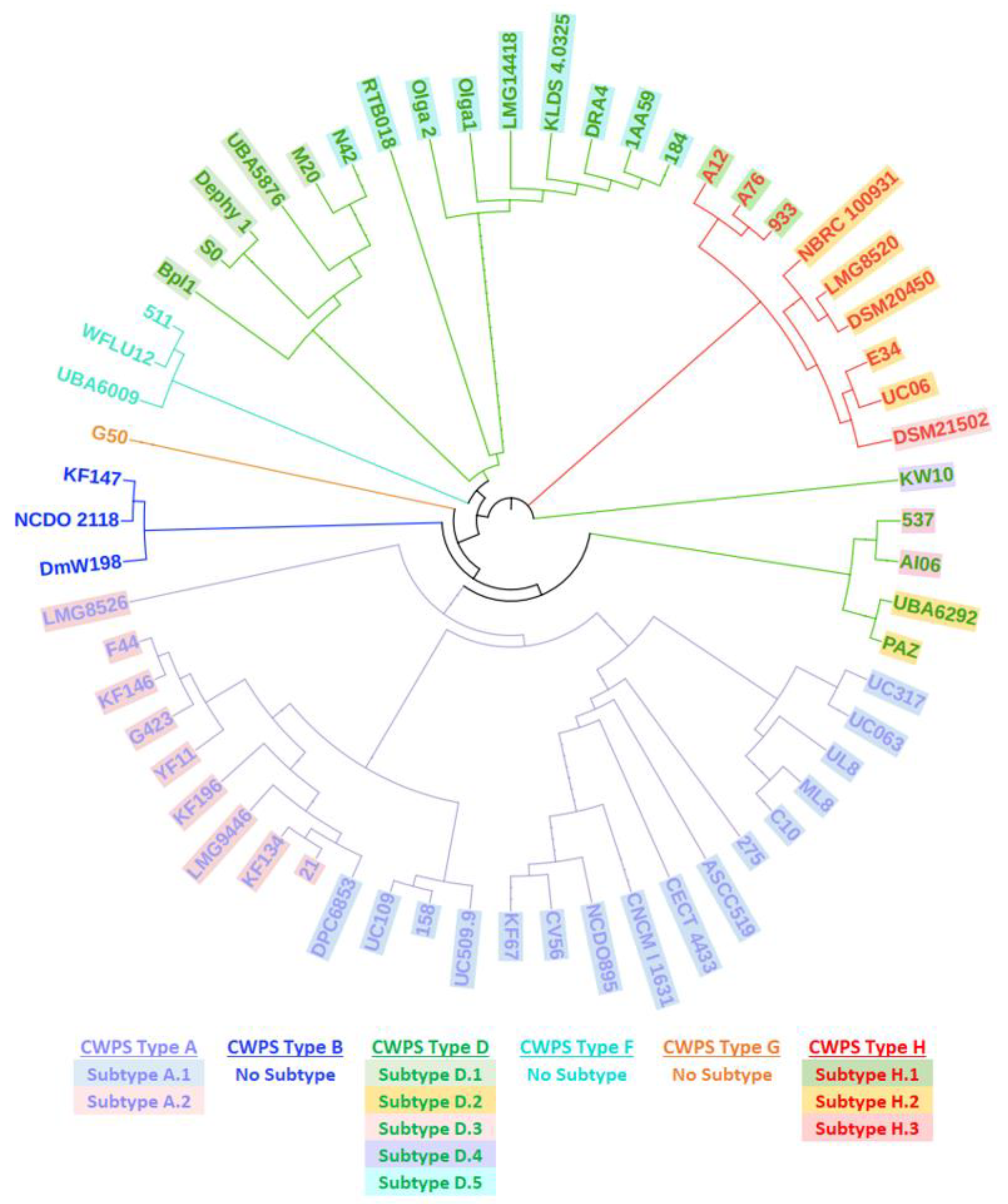
3.4. yccB–cwps Relationship
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| gBlock Name | Sequence (5′–3′) |
|---|---|
| D7893_RBP-Rec | TTCTGGAATGGGATTCAGGAACAACGGATAATCCACCTAAGGTTGGAGATACCATTATTGAT TTGGTAGGTAAGATTTATCAGATTACCAATGTAGTAGTTGATAGTACAATAGGTGGCGGCGG TCTATTCGACTATGGCCCAATGCTTACAAGTATCAAAGGCGCAGATGGTTCGCCGGGTAAG GACGGTATCGCTGGAAAAGATGGTGTTGGATTAAAAAGCACTATTATTACTTATGGGATTTC AGCATCAGATAGTACCATGCCGGGGACGTGGTCTCAAAATACACCAGCATTAGTTAAGGGA CAATGGTTCTGGACTAAAACACAGTGGACCTATACAGATAACAGCACTGAAACTGGTTATC AAAAAACATATATCTCTAAAGACGGAAATAACGGTCATGACGGAATTGCTGGTAAAGATG GTACTGGAATCAAAACTACGACCATTACATACGCAGGTTCAACAAGCGGAACGACAGCAC CAAATACTGGTTGGACTACCACAGTTCCGACAGTTGCAGCAGGTAATTACCTGTGGACTAA GACTGTTTGGGATTATACGGATAAGACCAGCGAGACAGGTTATTCAGTAGCTAAAATGGGA AATAATGGAACACCTGGTCCGCAAGGTCCTCCTGGAAGTAATGGTGATCCTGGTAAAACTG TTTCCAATACTGAGCCGACCACTCGATTTAAAGGCTTGACTTGGAAATACTCAGGTACGACT GACCTTACAGCGAGTGATAGAACAGTGATTAAGCCAAATACAGAGTATTACTATAATGGCA CTCATTGGGTGATTAACTATTTTAGCGTCAATAACTTTGCGGCTGAATCGATAACATCAGAT AAAATTGATGGTAAAAATTTAACAATTACTGATGGTGAGTTCATAAGCAAAACATCTAATG GCCCAGTTACAACCTCTACAGAAATCAAAGATAATCATATTGCAATTTCAAAGAAAGATGG AACTGTTAATACTAGAAATGATATAGCGCTTGATTCTGAACAAGGACTAGCTCAGAAATTTA CGAACATTAATACAGGATTCTACAGAACAGCTGGGATTAATTATCAAGGTCCATTCACAAG TGACTCAGATGGAAACTATGCTCAACTTACACCTCAAGGCACGAAGTTATCTACCGATGTTC CTTGGACCAAGCTTAGTTTGATGAATAATTTTAATGGAAATATTGAGTATGCAATTATCAAT GGGACTGTCTATATATCAGCGTCAGGAGTTGGTGTACCAGCAATGACTGCTGGTCAATGGA AGCAAGCGGCTCAATTGCCAACAGGAAGTTCAGCAATTCCAATTAGAGCAAATCGAATTGC AGCAGGAGATAGTGGAGATGGTCTAAGTTGGGCATTACTTTCTAATCAGGCTGGAGGAATA TTCATTCGATGCAGTGCTAATAAAGCACCAACACCTAACTTATTCAATGCCACATTACCATA TCCAATAGGATAAAAGGAGGAAAAATGGAAAAAGTCAATACAACGAATACAACAACTGAT ATCTTTGTCGGTGATAAGAATGTGGGTAATTTCACCCTTACAACGTTCAATAACGGAACAAT GAATGCAAATTTCATGATTAATGACCCTACAGCATTTCATGGCACGCCAGAAGCAGCTCAA GACCTAGCTAATTTGGTTAGCTCGGCAGTTAATCAGTCTAAAGCTTTGTTGGCTGATTTTGAA GCTAGTAAAGAATAGAAAGCAGGGGTTATGGAATTAGAACAACTTGTGGAACAGCATGAG GACAAACTCAAGCGGCACGATAAAGAATTATCTCGACTTAATGATATGTCAGTCGAAATAC AAAAGCAAATGAATGACGGATTGACTCGTGTGGATGAATCCAATCGCTTTTTAAGAGAACA GAATACTCGACAATCTGAACAGAATGCTCAAATACTACAAGCAGTTATCAAAGGTAATGAA AGCTCAGACGAAGTGGACAGAACGACACGGAT |
References
- Deveau, H.; Labrie, S.J.; Chopin, M.-C.; Moineau, S. Biodiversity and Classification of Lactococcal Phages. Appl. Environ. Microbiol. 2006, 72, 4338–4346. [Google Scholar] [CrossRef]
- Romero, D.A.; Magill, D.; Millen, A.; Horvath, P.; Fremaux, C. Dairy lactococcal and streptococcal phage–host interactions: An industrial perspective in an evolving phage landscape. FEMS Microbiol. Rev. 2020, 44, 909–932. [Google Scholar] [CrossRef] [PubMed]
- Coffey; Coakley; McGarry; Fitzgerald; Ross. Increasing phage resistance of cheese starters: A case study using Lactococcus lactis DPC4268. Lett. Appl. Microbiol. 2002, 26, 51–55. [Google Scholar] [CrossRef]
- King, W.R.; Collins, E.B.; Barrett, E.L. Frequencies of Bacteriophage-Resistant and Slow Acid-Producing Variants of Streptococcus cremoris. Appl. Environ. Microbiol. 1983, 45, 1481–1485. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.; Murphy, J.; Sinderen, D. Lactococcal 936-type phages and dairy fermentation problems: From detection to evolution and prevention. Front. Microbiol. 2012, 3, 335. [Google Scholar] [CrossRef] [PubMed]
- Labrie, S.; Moineau, S. Complete Genomic Sequence of Bacteriophage ul36: Demonstration of Phage Heterogeneity within the P335 Quasi-Species of Lactococcal Phages. Virology 2002, 296, 308–320. [Google Scholar] [CrossRef]
- Labrie, S.J.; Josephsen, J.; Neve, H.; Vogensen, F.K.; Moineau, S. Morphology, Genome Sequence, and Structural Proteome of Type Phage P335 from Lactococcus lactis. Appl. Environ. Microbiol. 2008, 74, 4636–4644. [Google Scholar] [CrossRef]
- Oliveira, J.; Mahony, J.; Hanemaaijer, L.; Kouwen, T.R.H.M.; van Sinderen, D. Biodiversity of bacteriophages infecting Lactococcus lactis starter cultures. J. Dairy. Sci. 2018, 101, 96–105. [Google Scholar] [CrossRef]
- Hill, C.; Miller, L.A.; Klaenhammer, T.R. In vivo genetic exchange of a functional domain from a type II A methylase between lactococcal plasmid pTR2030 and a virulent bacteriophage. J. Bacteriol. 1991, 173, 4363–4370. [Google Scholar] [CrossRef]
- Moineau, S.; Pandian, S.; Klaenhammer, T.R. Restriction/Modification Systems and Restriction Endonucleases Are More Effective on Lactococcal Bacteriophages That Have Emerged Recently in the Dairy Industry. Appl. Environ. Microbiol. 1993, 59, 197–202. [Google Scholar] [CrossRef]
- Labrie, S.J.; Moineau, S. Abortive Infection Mechanisms and Prophage Sequences Significantly Influence the Genetic Makeup of Emerging Lytic Lactococcal Phages. J. Bacteriol. 2007, 189, 5787. [Google Scholar] [CrossRef]
- Aucouturier, A.; Chain, F.; Langella, P.; Bidnenko, E. Characterization of a Prophage-Free Derivative Strain of Lactococcus lactis ssp. lactis IL1403 Reveals the Importance of Prophages for Phenotypic Plasticity of the Host. Front. Microbiol. 2018, 9, 2032. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, P.; Mahony, J.; Schweinlin, K.; Neve, H.; Franz, C.M.; van Sinderen, D. Assessing the functionality and genetic diversity of lactococcal prophages. Int. J. Food. Microbiol. 2018, 272, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Alexeeva, S.; Guerra Martínez, J.A.; Spus, M.; Smid, E.J. Spontaneously induced prophages are abundant in a naturally evolved bacterial starter culture and deliver competitive advantage to the host. BMC Microbiol. 2018, 18, 120. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Cruz, S.; Parlindungan, E.; Erazo Garzon, A.; Alqarni, M.; Lugli, G.A.; Ventura, M.; van Sinderen, D.; Mahony, J. Lysogenization of a Lactococcal Host with Three Distinct Temperate Phages Provides Homologous and Heterologous Phage Resistance. Microorganisms 2020, 8, 1685. [Google Scholar] [CrossRef] [PubMed]
- Dupont, K.; Janzen, T.; Vogensen, F.K.; Josephsen, J.; Stuer-Lauridsen, B. Identification of Lactococcus lactis Genes Required for Bacteriophage Adsorption. Appl. Environ. Microbiol. 2004, 70, 5825–5832. [Google Scholar] [CrossRef]
- Millen, A.M.; Romero, D.A.; Horvath, P.; Magill, D.; Simdon, L. Host-encoded, cell surface-associated exopolysaccharide required for adsorption and infection by lactococcal P335 phage subtypes. Front. Microbiol. 2022, 13, 971166. [Google Scholar] [CrossRef]
- Ainsworth, S.; Sadovskaya, I.; Vinogradov, E.; Courtin, P.; Guerardel, Y.; Mahony, J.; Grard, T.; Cambillau, C.; Chapot-Chartier, M.-P.; van Sinderen, D. Differences in Lactococcal Cell Wall Polysaccharide Structure Are Major Determining Factors in Bacteriophage Sensitivity. mBio 2014, 5, 10-1128. [Google Scholar] [CrossRef]
- Guérin, H.; Quénée, P.; Palussière, S.; Courtin, P.; André, G.; Péchoux, C.; Costache, V.; Mahony, J.; van Sinderen, D.; Kulakauskas, S.; et al. PBP2b Mutations Improve the Growth of Phage-Resistant Lactococcus cremoris Lacking Polysaccharide Pellicle. Appl. Environ. Microbiol. 2023, 89, e02103-22. [Google Scholar] [CrossRef]
- Mahony, J.; Oliveira, J.; Collins, B.; Hanemaaijer, L.; Lugli, G.A.; Neve, H.; Ventura, M.; Kouwen, T.R.; Cambillau, C.; van Sinderen, D. Genetic and functional characterisation of the lactococcal P335 phage-host interactions. BMC Genomics. 2017, 18, 146. [Google Scholar] [CrossRef]
- Theodorou, I.; Courtin, P.; Palussière, S.; Kulakauskas, S.; Bidnenko, E.; Péchoux, C.; Fenaille, F.; Penno, C.; Mahony, J.; van Sinderen, D.; et al. A dual-chain assembly pathway generates the high structural diversity of cell-wall polysaccharides in Lactococcus lactis. J. Biol. Chem. 2019, 294, 17612–17625. [Google Scholar] [CrossRef] [PubMed]
- Giesbers, C.A.P.; Fagan, J.; Parlindungan, E.; Palussière, S.; Courtin, P.; Lugli, G.A.; Ventura, M.; Kulakauskas, S.; Chapot-Chartier, M.P.; Mahony, J.; et al. Reduced synthesis of phospho-polysaccharide in Lactococcus as a strategy to evade phage infection. Int. J. Food Microbiol. 2023, 407, 110415. [Google Scholar] [CrossRef] [PubMed]
- Huard, C.; Miranda, G.; Wessner, F.; Bolotin, A.; Hansen, J.; Foster, S.J.; Chapot-Chartier, M.-P. Characterization of AcmB, an N-acetylglucosaminidase autolysin from Lactococcus lactis. Microbiology 2003, 149, 695–705. [Google Scholar] [CrossRef]
- Ouzari, H.; Cherif, A.; Mora, D. Autolytic phenotype of Lactococcus lactis strains isolated from traditional Tunisian dairy products. J. Appl. Microbiol. 2002, 92, 812–820. [Google Scholar] [CrossRef] [PubMed]
- Nakimbugwe, D.; Masschalck, B.; Deckers, D.; Callewaert, L.; Aertsen, A.; Michiels, C.W. Cell wall substrate specificity of six different lysozymes and lysozyme inhibitory activity of bacterial extracts. FEMS Microbiol. Lett. 2006, 259, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.J.; Verma, D.; Griswold, K.E.; Bailey-Kellogg, C. Building blocks and blueprints for bacterial autolysins. PLoS Comput. Biol. 2021, 17, e1008889. [Google Scholar] [CrossRef]
- Buist, G.; Karsens, H.; Nauta, A.; van Sinderen, D.; Venema, G.; Kok, J. Autolysis of Lactococcus lactis caused by induced overproduction of its major autolysin, AcmA. Appl. Environ. Microbiol. 1997, 63, 2722–2728. [Google Scholar] [CrossRef]
- Vollmer, W.; Joris, B.; Charlier, P.; Foster, S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 2008, 32, 259–286. [Google Scholar] [CrossRef]
- Huard, C.; Miranda, G.; Redko, Y.; Wessner, F.; Foster, S.J.; Chapot-Chartier, M.-P. Analysis of the Peptidoglycan Hydrolase Complement of Lactococcus lactis: Identification of a Third N-Acetylglucosaminidase, AcmC. Appl. Environ. Microbiol. 2004, 70, 3493–3499. [Google Scholar] [CrossRef]
- Bolotin, A.; Wincker, P.; Mauger, S.; Jaillon, O.; Malarme, K.; Weissenbach, J.; Ehrlich, S.D.; Sorokin, A. The Complete Genome Sequence of the Lactic Acid Bacterium Lactococcus lactis ssp. lactis IL1403. Genome. Res. 2001, 11, 731–753. [Google Scholar] [CrossRef]
- Oliveira, H.; Melo, L.D.R.; Santos, S.B.; Nóbrega, F.L.; Ferreira, E.C.; Cerca, N.; Azeredo, J.; Kluskens, L.D. Molecular Aspects and Comparative Genomics of Bacteriophage Endolysins. J. Virol. 2013, 87, 4558–4570. [Google Scholar] [CrossRef] [PubMed]
- Kenny, J.G.; McGrath, S.; Fitzgerald, G.F.; van Sinderen, D. Bacteriophage Tuc2009 Encodes a Tail-Associated Cell Wall-Degrading Activity. J. Bacteriol. 2004, 186, 3480–3491. [Google Scholar] [CrossRef] [PubMed]
- Frias, M.J.; Melo-Cristino, J.; Ramirez, M. The Autolysin LytA Contributes to Efficient Bacteriophage Progeny Release in Streptococcus pneumoniae. J. Bacteriol. 2009, 191, 5428–5440. [Google Scholar] [CrossRef] [PubMed]
- Dowah, A.S.A.; Clokie, M.R.J. Review of the nature, diversity and structure of bacteriophage receptor binding proteins that target Gram-positive bacteria. Biophys. Rev. 2018, 10, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Stockdale, S.R.; Mahony, J.; Courtin, P.; Chapot-Chartier, M.P.; van Pijkeren, J.P.; Britton, R.A.; Neve, H.; Heller, K.J.; Aideh, B.; Vogensen, F.K.; et al. The lactococcal phages Tuc2009 and TP901-1 incorporate two alternate forms of their tail fiber into their virions for infection specialization. J. Biol. Chem. 2013, 288, 5581–5590. [Google Scholar] [CrossRef]
- Terzaghi, B.E.; Sandine, W.E. Improved Medium for Lactic Streptococci and Their Bacteriophages. Appl. Microbiol. 1975, 29, 807–813. [Google Scholar] [CrossRef]
- Sandine, W.E. The Streptococci: Milk Products. In Bacterial Starter Cultures for Food; CRC Press: Boca Raton, FL, USA, 2018; pp. 5–20. [Google Scholar]
- Sanders, M.E.; Klaenhammer, T.R. Restriction and Modification in Group N Streptococci: Effect of Heat on Development of Modified Lytic Bacteriophage. Appl. Environ. Microbiol. 1980, 40, 500–506. [Google Scholar] [CrossRef]
- Duwat, P.; Cochu, A.; Ehrlich, S.D.; Gruss, A. Characterization of Lactococcus lactis UV-sensitive mutants obtained by ISS1 transposition. J. Bacteriol. 1997, 179, 4473–4479. [Google Scholar] [CrossRef]
- Maguin, E.; Prévost, H.; Ehrlich, S.D.; Gruss, A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 1996, 178, 931–935. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. Suppl. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Zhang, Y. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]
- Delano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crytallography 2002, 40, 82–92. [Google Scholar]
- Dower, W.J.; Miller, J.F.; Ragsdale, C.W. High efficiency transformation of E.coli by high voltage electroporation. Nucleic Acids Res. Suppl. 1988, 16, 6127–6145. [Google Scholar] [CrossRef]
- Holo, H.; Nes, I.F. High-Frequency Transformation, by Electroporation, of Lactococcus lactis subsp. cremoris Grown with Glycine in Osmotically Stabilized Media. Appl. Environ. Microbiol. 1989, 55, 3119–3123. [Google Scholar] [CrossRef]
- Tremblay, D.M.; Tegoni, M.; Spinelli, S.; Campanacci, V.; Blangy, S.; Huyghe, C.; Desmyter, A.; Labrie, S.; Moineau, S.; Cambillau, C. Receptor-Binding Protein of Lactococcus lactis Phages: Identification and Characterization of the Saccharide Receptor-Binding Site. J. Bacteriol. 2006, 188, 2400–2410. [Google Scholar] [CrossRef] [PubMed]
- Klotz, C.; Goh, Y.J.; O’Flaherty, S.; Johnson, B.; Barrangou, R. Deletion of S-Layer Associated Ig-Like Domain Protein Disrupts the Lactobacillus acidophilus Cell Surface. Front. Microbiol. 2020, 11, 345. [Google Scholar] [CrossRef] [PubMed]
- Bodelón, G.; Carmen Palomino, C.; Ángel Fernández, L. Immunoglobulin domains in Escherichia coli and other enterobacteria: From pathogenesis to applications in antibody technologies. FEMS Microbiol. Rev. 2013, 37, 204–250. [Google Scholar] [CrossRef]
- Hayes, S.; Mahony, J.; Vincentelli, R.; Ramond, L.; Nauta, A.; van Sinderen, D.; Cambillau, C. Ubiquitous Carbohydrate Binding Modules Decorate 936 Lactococcal Siphophage Virions. Viruses 2019, 11, 631. [Google Scholar] [CrossRef] [PubMed]
- Millen, A.M.; Magill, D.; Romero, D.; Simdon, L. Evolved distal tail protein of skunaviruses facilitates adsorption to exopolysaccharide-encoding lactococci. Microbiome. Res. Rep. 2023, 2, 26. [Google Scholar] [CrossRef]
- Mahony, J.; Frantzen, C.; Vinogradov, E.; Sadovskaya, I.; Theodorou, I.; Kelleher, P.; Chapot-Chartier, M.-P.; Cambillau, C.; Holo, H.; van Sinderen, D. The CWPS Rubik’s cube: Linking diversity of cell wall polysaccharide structures with the encoded biosynthetic machinery of selected Lactococcus lactis strains. Mol. Microbiol. 2020, 114, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Morita, T.; Nishino, R.; Aiba, H. Role of the terminator hairpin in the biogenesis of functional Hfq-binding sRNAs. RNA 2017, 239, 1419–1431. [Google Scholar] [CrossRef]

| Primer Name | Sequence (5′–3′) |
|---|---|
| VF-pg9 | GTGGACAGAACGACACGGAT |
| VR-pg9 | TCCTGAATCCCATTCCAGAA |
| CloneYccB_F | TTCTGGAATGGGATTCAGGACTACCACTATTCCCAAAGCATTTC |
| CloneYccB_R | ATCCGTGTCGTTCTGTCCACGCAATGGGAGATGATTGGAGAGG |
| RSF | TTCTGGAATGGGATTCAGGA |
| GC pG9vF | ATCCGTGTCGTTCTGTCCAC |
| D7893_TalCheck_F | CAATGCCACATTACCATATCC |
| pg9 + cm_R2 | AACTAGTGGATCCCCCGGGCTG |
| internalErmR-F | GCTGAATCGAGACTTGAGTG |
| internalErmR-R | GTCATCTATTCAACTTATCG |
| yccBNested_F | GCCAACTTTGTCTTCATACTCACC |
| yccBNested_R | GTATGTTTTGGAAGCCCTCAGG |
| yccB_F1 | CTCACATTATTTCACTAATACTCTTTTCTGG |
| yccB _F2 | CATGGATTGGAACAGATACAGCAACTTG |
| yccB _F3 | CAATACGATCTATACTACCGAGTTCAAGC |
| D6867tal_StartF | GGAAGCTGGTGGTCAGATTTG |
| D6867tal_F1 | CCAATACTGAGCCGACCACTC |
| D6867tal_EndR | GACCCATTCGTGAAGTCATTG |
| pG9yccB | pG9RBP7893 | ||
|---|---|---|---|
| Vector fragment | Forward primer | VF-pg9 | VF-pg9 |
| Reverse primer | VR-pg9 | VR-pg9 | |
| Template | pG9 | pG9 | |
| Insert fragment | Forward primer | CloneYccB_F | D7893_RBP-Rec (gBlock) |
| Reverse primer | CloneYccB_R | ||
| Template | DGCC12699 | ||
| Detection of TG1 RepA+ transformants | Forward primer | CloneYccB_F | D7893_TalCheck_F |
| Reverse primer | CloneYccB_R | pg9 + cmR_2 | |
| Detection of L. lactis transformants | Forward primer | RSF | D7893_TalCheck_F |
| Reverse primer | GC pG9vF | pg9 + cm_R2 |
| EOPs | D6867 Adsorption (%) | |||||
|---|---|---|---|---|---|---|
| D6867 | D7138 | D7893 | D5694 | D6867-RBP7893 | ||
| DGCC12699 | 1 | 1 | 1 | 1 | 1.03 ± 0.08 | 97.4 ± 1.3 |
| BIM2 | <9.09 × 10−8 | <2.00 × 10−7 | 0.75 ± 0.15 | 1.31 ± 0.20 | 1 | 82.3 ± 2.9 |
| BIM8 | <9.09 × 10−8 | <2.00 × 10−7 | 0.82 ± 0.19 | 1.16 ± 0.41 | -- | 87.8 ± 3.2 |
| BIM19 | <9.09 × 10−8 | <2.00 × 10−7 | 0.78 ± 0.18 | 0.91 ± 0.22 | -- | 87.5 ± 3.4 |
| BIM21 | <9.09 × 10−8 | <2.00 × 10−7 | <3.33 × 10−6 | 1.02 × 10−3 ± 7.76 × 10−4 | -- | 41.1 ± 4.4 |
| BIM2 + pG9yccB | 1.20 ± 0.20 | 0.98 ± 0.08 | -- | -- | -- | -- |
| BIM2 + pG9 | <9.09 × 10−8 | <2.00 × 10−7 | -- | -- | -- | -- |
| BIM2ΔpG9yccB | <9.09 × 10−8 | <2.00 × 10−7 | -- | -- | -- | -- |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seiler, J.; Millen, A.; Romero, D.A.; Magill, D.; Simdon, L. Novel P335-like Phage Resistance Arises from Deletion within Putative Autolysin yccB in Lactococcus lactis. Viruses 2023, 15, 2193. https://doi.org/10.3390/v15112193
Seiler J, Millen A, Romero DA, Magill D, Simdon L. Novel P335-like Phage Resistance Arises from Deletion within Putative Autolysin yccB in Lactococcus lactis. Viruses. 2023; 15(11):2193. https://doi.org/10.3390/v15112193
Chicago/Turabian StyleSeiler, Jenny, Anne Millen, Dennis A. Romero, Damian Magill, and Laura Simdon. 2023. "Novel P335-like Phage Resistance Arises from Deletion within Putative Autolysin yccB in Lactococcus lactis" Viruses 15, no. 11: 2193. https://doi.org/10.3390/v15112193
APA StyleSeiler, J., Millen, A., Romero, D. A., Magill, D., & Simdon, L. (2023). Novel P335-like Phage Resistance Arises from Deletion within Putative Autolysin yccB in Lactococcus lactis. Viruses, 15(11), 2193. https://doi.org/10.3390/v15112193







