Phenotypical Screening of an MMV Open Box Library and Identification of Compounds with Antiviral Activity against St. Louis Encephalitis Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Compounds
2.3. Viral Stock
2.4. CPE-Based Medium-Throughput Screening Assay
2.5. Confirmatory Assay
2.6. Viral Load Quantification
2.7. MTT Assay
2.8. Data Acquisition and Analysis
2.9. PDI Network Construction and BP and CC Pathway Enrichment Analysis
2.10. Statistical Analysis
3. Results
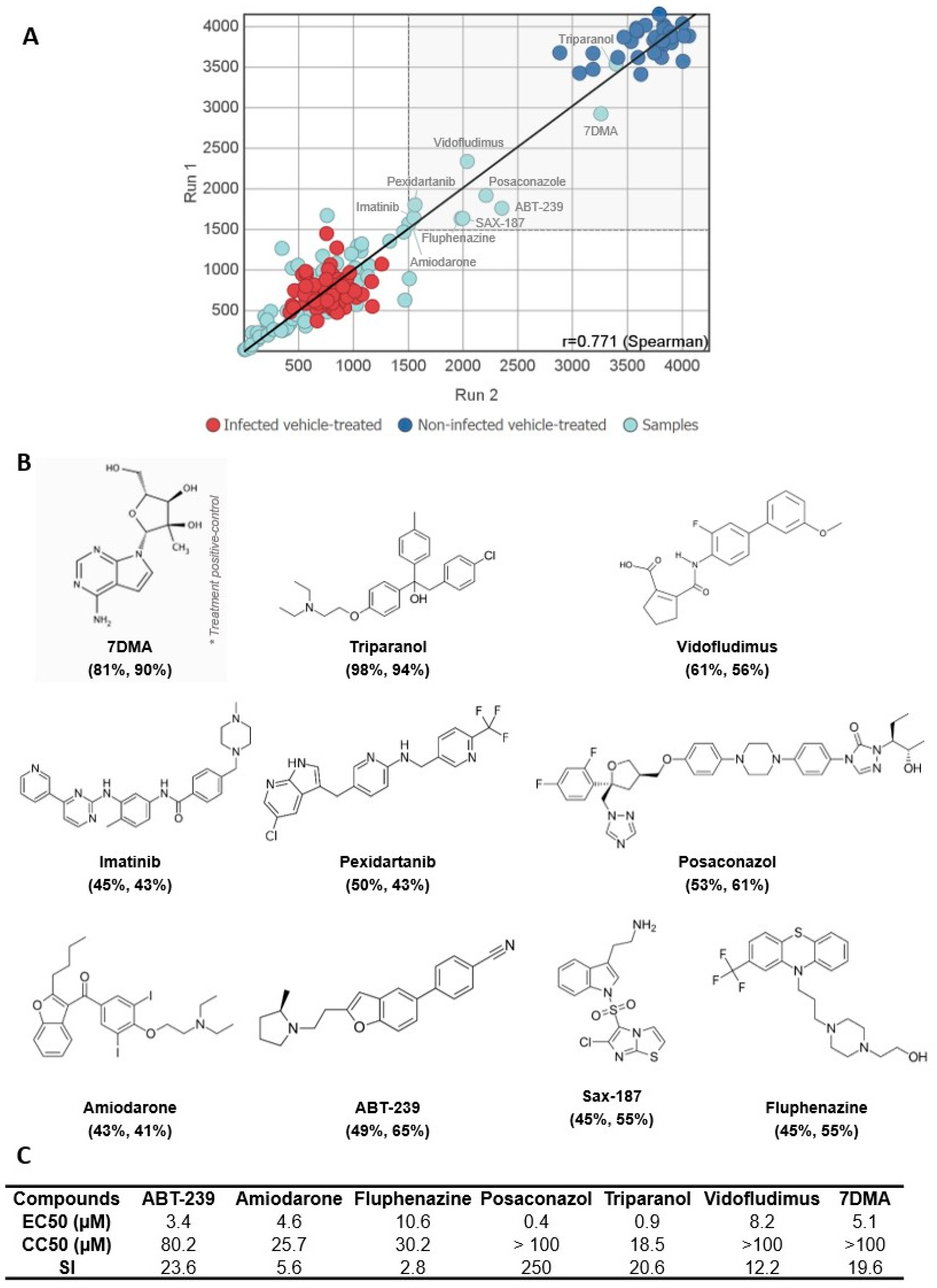
3.1. A Medium-Throughput Screening Pipeline for Identification of Bioactive Compounds against SLEV Infection
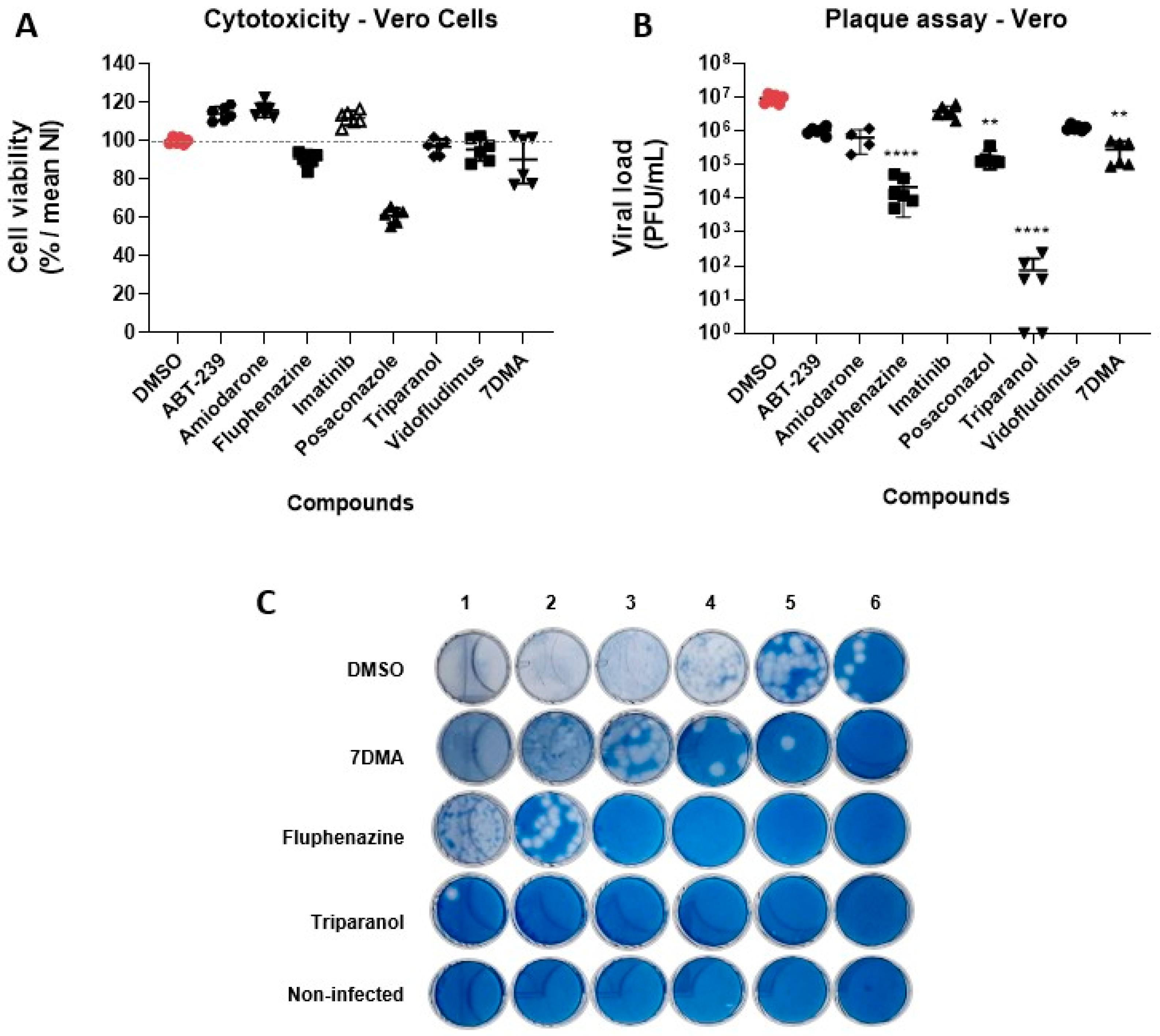
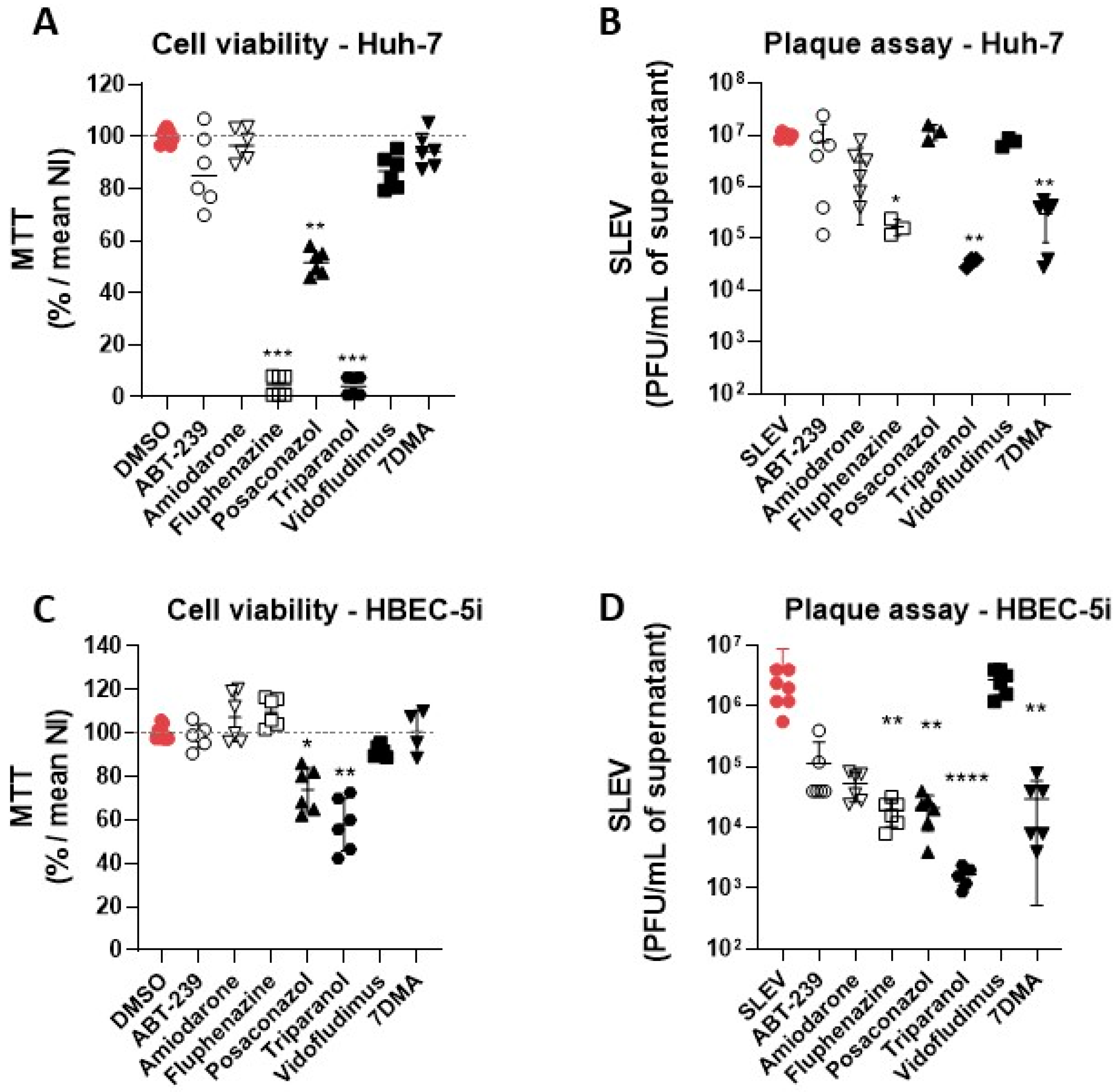
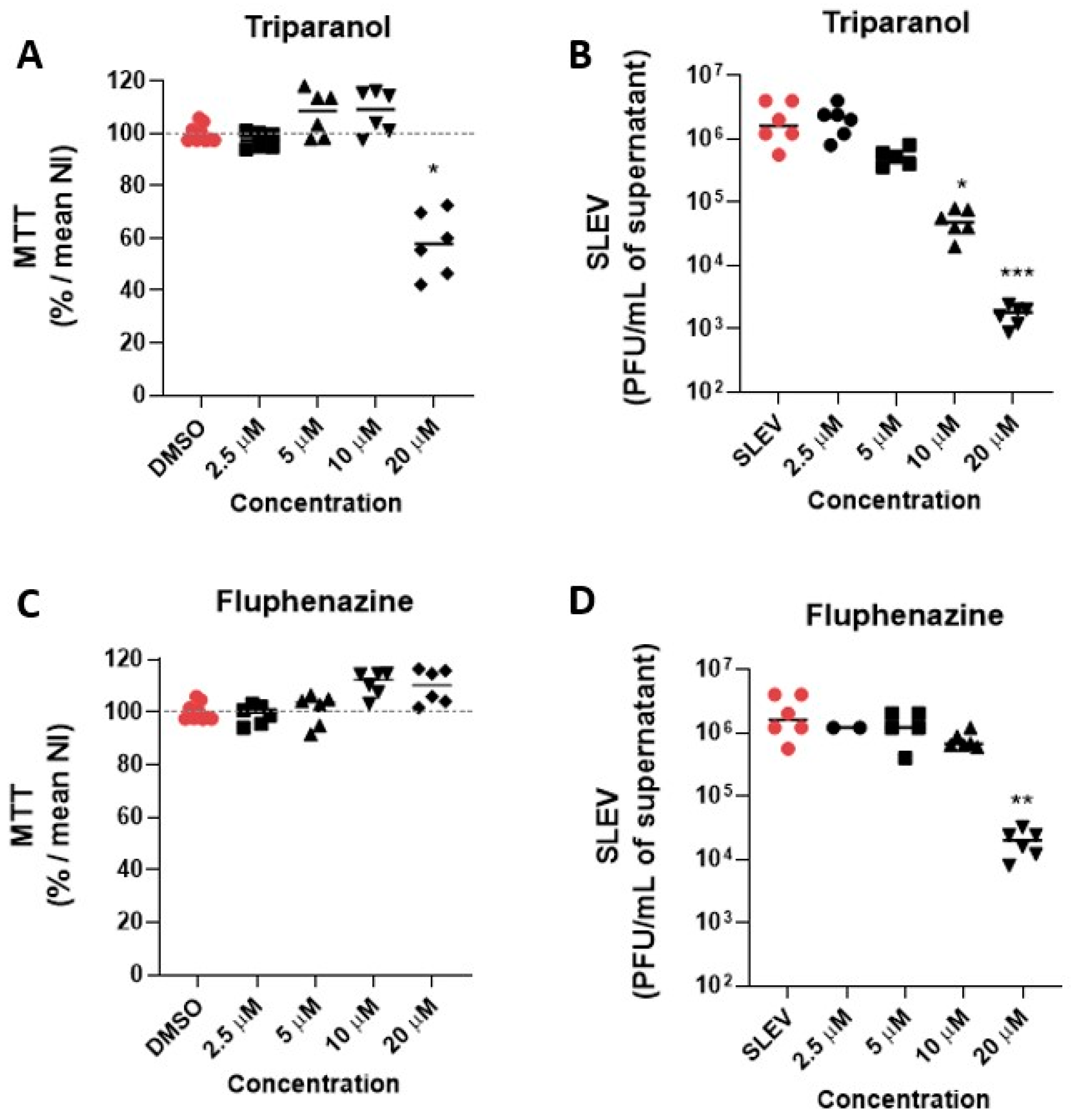
3.2. Hit Validation
3.3. Identification of Possible Host-Targeted Mechanisms of Action for Compounds with Antiviral Activity against SLEV
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chambers, T.J.; Diamond, M.S. Pathogenesis of flavivirus encephalitis. Adv. Virus Res. 2003, 60, 273–342. [Google Scholar] [CrossRef]
- Baillie, G.J.; Kolokotronis, S.O.; Waltari, E.; Maffei, J.G.; Kramer, L.D.; Perkins, S.L. Phylogenetic and evolutionary analyses of St. Louis encephalitis virus genomes. Mol. Phylogenet. Evol. 2008, 47, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Poidinger, M.; Hall, R.A.; Mackenzie, J.S. Molecular characterization of the Japanese encephalitis serocomplex of the flavivirus genus. Virology 1996, 218, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Barrows, N.J.; Campos, R.K.; Liao, K.C.; Prasanth, K.R.; Soto-Acosta, R.; Yeh, S.C.; Schott-Lerner, G.; Pompon, J.; Sessions, O.M.; Bradrick, S.S.; et al. Biochemistry and Molecular Biology of Flaviviruses. Chem. Rev. 2018, 118, 4448–4482. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.L.; Jones, C.T.; Rice, C.M. Architects of assembly: Roles of Flaviviridae non-structural proteins in virion morphogenesis. Nat. Rev. Microbiol. 2008, 6, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Diaz, A.; Coffey, L.L.; Burkett-Cadena, N.; Day, J.F. Reemergence of St. Louis Encephalitis Virus in the Americas. Emerg. Infect. Dis. 2018, 24, 2150–2157. [Google Scholar] [CrossRef]
- Kopp, A.; Gillespie, T.R.; Hobelsberger, D.; Estrada, A.; Harper, J.M.; Miller, R.A.; Eckerle, I.; Müller, M.A.; Podsiadlowski, L.; Leendertz, F.H.; et al. Provenance and geographic spread of St. Louis encephalitis virus. mBio 2013, 4, e00322-13. [Google Scholar] [CrossRef] [PubMed]
- Spinsanti, L.I.; Díaz, L.A.; Glatstein, N.; Arselán, S.; Morales, M.A.; Farías, A.A.; Fabbri, C.; Aguilar, J.J.; Ré, V.; Frías, M.; et al. Human outbreak of St. Louis encephalitis detected in Argentina, 2005. J. Clin. Virol. 2008, 42, 27–33. [Google Scholar] [CrossRef]
- McLean, R.G.; Bowen, G.S. Vertebrate hosts. In St. Louis Encephalitis; Monath, T.P., Ed.; American Public Health Association: Washington, DC, USA, 1980; pp. 381–450. [Google Scholar]
- Vasconcelos, P.F.; da Rosa, J.F.; da Rosa, A.P.; Dégallier, N.; Pinheiro, F.P.; Sá Filho, G.C. Epidemiology of encephalitis caused by arbovirus in the Brazilian Amazonia. Rev. Inst. Med. Trop Sao Paulo 1991, 33, 465–476. [Google Scholar] [CrossRef]
- Hervé, J.P.; Dégallier, N.; Travassos da Rosa, A.P.A.; Pinheiro, F.P.; Sá Filho, G.C. Aspectos ecológicos das arboviroses. In Instituto Evandro Chagas: 50 Anos de Contribuição às Ciências Biológicas e à Medicina Tropical; Fundação Serviços de Saúde Pública: Belém, Brazil, 1986; pp. 409–437. [Google Scholar]
- Rodrigues, S.G.; Nunes, M.R.; Casseb, S.M.; Prazeres, A.S.; Rodrigues, D.S.; Silva, M.O.; Cruz, A.C.; Tavares-Neto, J.C.; Vasconcelos, P.F. Molecular epidemiology of Saint Louis encephalitis virus in the Brazilian Amazon: Genetic divergence and dispersal. J. Gen. Virol. 2010, 91, 2420–2427. [Google Scholar] [CrossRef]
- Rosa, R.; Costa, E.A.; Marques, R.E.; Oliveira, T.S.; Furtini, R.; Bomfim, M.R.; Teixeira, M.M.; Paixão, T.A.; Santos, R.L. Isolation of saint louis encephalitis virus from a horse with neurological disease in Brazil. PLoS Negl. Trop Dis. 2013, 7, e2537. [Google Scholar] [CrossRef]
- Mondini, A.; Bronzoni, R.V.; Cardeal, I.L.; dos Santos, T.M.; Lázaro, E.; Nunes, S.H.; Silva, G.C.; Madrid, M.C.; Rahal, P.; Figueiredo, L.T.; et al. Simultaneous infection by DENV-3 and SLEV in Brazil. J. Clin. Virol. 2007, 40, 84–86. [Google Scholar] [CrossRef]
- Tsai, T.F.; Mitchell, C.J. St. Louis encephalitis. In The Arboviruses: Epidemiology and Ecology; Monath, T.P., Ed.; CRC Press: Boca Raton, FL, USA, 1988; pp. 113–143. [Google Scholar]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Zhang, J.H.; Chung, T.D.; Oldenburg, K.R. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen 1999, 4, 67–73. [Google Scholar] [CrossRef]
- Coimbra, L.D.; Borin, A.; Fontoura, M.; Gravina, H.D.; Nagai, A.; Shimizu, J.F.; Bispo-dos-Santos, K.; Granja, F.; Oliveira, P.S.L.; Franchini, K.G.; et al. Identification of Compounds with Antiviral Activity against SARS-CoV-2 in the MMV Pathogen Box Using a Phenotypic High-Throughput Screening Assay. Front. Virol. 2022, 2, 854363. [Google Scholar] [CrossRef]
- Zmurko, J.; Marques, R.E.; Schols, D.; Verbeken, E.; Kaptein, S.J.; Neyts, J. The Viral Polymerase Inhibitor 7-Deaza-2’-C-Methyladenosine Is a Potent Inhibitor of In Vitro Zika Virus Replication and Delays Disease Progression in a Robust Mouse Infection Model. PLoS Negl. Trop Dis. 2016, 10, e0004695. [Google Scholar] [CrossRef] [PubMed]
- Naciuk, F.F.; Nascimento, A.F.Z.; Rocha, R.P.F.; Rustiguel, J.K.; Coimbra, L.D.; Marques, R.E.; Bruder, M. Competing interests during the key N-glycosylation of 6-chloro-7-deaza-7-iodopurine for the synthesis of 7-deaza-2’-methyladenosine using Vorbrüggen conditions. Front. Chem. 2023, 11, 1163486. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, L.S.; Marques, R.E.; Jesus, A.M.; Almeida, R.P.; Teixeira, M.M. Zika crisis in Brazil: Challenges in research and development. Curr. Opin. Virol. 2016, 18, 76–81. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Figueiredo, P.; Stoffella-Dutra, A.G.; Barbosa Costa, G.; Silva de Oliveira, J.; Dourado Amaral, C.; Duarte Santos, J.; Soares Rocha, K.L.; Araújo Júnior, J.P.; Lacerda Nogueira, M.; Zazá Borges, M.A.; et al. Re-Emergence of Yellow Fever in Brazil during 2016–2019: Challenges, Lessons Learned, and Perspectives. Viruses 2020, 12, 1233. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Yip, C.C.; Tsang, J.O.; Tee, K.M.; Cai, J.P.; Chik, K.K.; Zhu, Z.; Chan, C.C.; Choi, G.K.; Sridhar, S.; et al. Differential cell line susceptibility to the emerging Zika virus: Implications for disease pathogenesis, non-vector-borne human transmission, and animal reservoirs. Emerg. Microbes Infect. 2016, 5, e93. [Google Scholar] [CrossRef]
- Leier, H.C.; Messer, W.B.; Tafesse, F.G. Lipids and pathogenic flaviviruses: An intimate union. PLoS Pathog. 2018, 14, e1006952. [Google Scholar] [CrossRef] [PubMed]
- Zerenturk, E.J.; Sharpe, L.J.; Ikonen, E.; Brown, A.J. Desmosterol and DHCR24: Unexpected new directions for a terminal step in cholesterol synthesis. Prog. Lipid Res. 2013, 52, 666–680. [Google Scholar] [CrossRef] [PubMed]
- Aizaki, H.; Morikawa, K.; Fukasawa, M.; Hara, H.; Inoue, Y.; Tani, H.; Saito, K.; Nishijima, M.; Hanada, K.; Matsuura, Y.; et al. Critical role of virion-associated cholesterol and sphingolipid in hepatitis C virus infection. J. Virol. 2008, 82, 5715–5724. [Google Scholar] [CrossRef] [PubMed]
- Rothwell, C.; Lebreton, A.; Young Ng, C.; Lim, J.Y.; Liu, W.; Vasudevan, S.; Labow, M.; Gu, F.; Gaither, L.A. Cholesterol biosynthesis modulation regulates dengue viral replication. Virology 2009, 389, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, J.M.; Khromykh, A.A.; Parton, R.G. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe 2007, 2, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Osuna-Ramos, J.F.; Reyes-Ruiz, J.M.; Del Ángel, R.M. The Role of Host Cholesterol during Flavivirus Infection. Front. Cell Infect. Microbiol. 2018, 8, 388. [Google Scholar] [CrossRef]
- Takano, T.; Tsukiyama-Kohara, K.; Hayashi, M.; Hirata, Y.; Satoh, M.; Tokunaga, Y.; Tateno, C.; Hayashi, Y.; Hishima, T.; Funata, N.; et al. Augmentation of DHCR24 expression by hepatitis C virus infection facilitates viral replication in hepatocytes. J. Hepatol. 2011, 55, 512–521. [Google Scholar] [CrossRef]
- Morgan, D.G.; Finch, C.E. [3H]Fluphenazine binding to brain membranes: Simultaneous measurement of D-1 and D-2 receptor sites. J. Neurochem. 1986, 46, 1623–1631. [Google Scholar] [CrossRef]
- Klutzny, S.; Lesche, R.; Keck, M.; Kaulfuss, S.; Schlicker, A.; Christian, S.; Sperl, C.; Neuhaus, R.; Mowat, J.; Steckel, M.; et al. Functional inhibition of acid sphingomyelinase by Fluphenazine triggers hypoxia-specific tumor cell death. Cell Death Dis. 2017, 8, e2709. [Google Scholar] [CrossRef]
- Kornhuber, J.; Tripal, P.; Reichel, M.; Mühle, C.; Rhein, C.; Muehlbacher, M.; Groemer, T.W.; Gulbins, E. Functional Inhibitors of Acid Sphingomyelinase (FIASMAs): A novel pharmacological group of drugs with broad clinical applications. Cell Physiol Biochem. 2010, 26, 9–20. [Google Scholar] [CrossRef]
- Grassmé, H.; Jendrossek, V.; Riehle, A.; von Kürthy, G.; Berger, J.; Schwarz, H.; Weller, M.; Kolesnick, R.; Gulbins, E. Host defense against Pseudomonas aeruginosa requires ceramide-rich membrane rafts. Nat. Med. 2003, 9, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Tani, H.; Shiokawa, M.; Kaname, Y.; Kambara, H.; Mori, Y.; Abe, T.; Moriishi, K.; Matsuura, Y. Involvement of ceramide in the propagation of Japanese encephalitis virus. J. Virol. 2010, 84, 2798–2807. [Google Scholar] [CrossRef]
- Chen, C.S.; Rosenwald, A.G.; Pagano, R.E. Ceramide as a modulator of endocytosis. J. Biol. Chem. 1995, 270, 13291–13297. [Google Scholar] [CrossRef] [PubMed]
- Smit, J.M.; Moesker, B.; Rodenhuis-Zybert, I.; Wilschut, J. Flavivirus cell entry and membrane fusion. Viruses 2011, 3, 160–171. [Google Scholar] [CrossRef]
- Chu, J.J.; Ng, M.L. Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J. Virol. 2004, 78, 10543–10555. [Google Scholar] [CrossRef]
- Kornhuber, J.; Muehlbacher, M.; Trapp, S.; Pechmann, S.; Friedl, A.; Reichel, M.; Mühle, C.; Terfloth, L.; Groemer, T.W.; Spitzer, G.M.; et al. Identification of novel functional inhibitors of acid sphingomyelinase. PLoS ONE 2011, 6, e23852. [Google Scholar] [CrossRef]
- Tummino, T.A.; Rezelj, V.V.; Fischer, B.; Fischer, A.; O’Meara, M.J.; Monel, B.; Vallet, T.; White, K.M.; Zhang, Z.; Alon, A.; et al. Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2. Science 2021, 373, 541–547. [Google Scholar] [CrossRef]
- Breiden, B.; Sandhoff, K. Emerging mechanisms of drug-induced phospholipidosis. Biol. Chem. 2021, 401, 31–46. [Google Scholar] [CrossRef]
- Sloskey, G.E. Amiodarone: A unique antiarrhythmic agent. Clin. Pharm. 1983, 2, 330–340. [Google Scholar] [PubMed]
- Lubic, S.P.; Nguyen, K.P.; Dave, B.; Giacomini, J.C. Antiarrhythmic agent amiodarone possesses calcium channel blocker properties. J. Cardiovasc. Pharmacol. 1994, 24, 707–714. [Google Scholar] [CrossRef]
- Schloer, S.; Brunotte, L.; Goretzko, J.; Mecate-Zambrano, A.; Korthals, N.; Gerke, V.; Ludwig, S.; Rescher, U. Targeting the endolysosomal host-SARS-CoV-2 interface by clinically licensed functional inhibitors of acid sphingomyelinase (FIASMA) including the antidepressant fluoxetine. Emerg. Microbes Infect. 2020, 9, 2245–2255. [Google Scholar] [CrossRef]
- Muehler, A.; Peelen, E.; Kohlhof, H.; Gröppel, M.; Vitt, D. Vidofludimus calcium, a next generation DHODH inhibitor for the Treatment of relapsing-remitting multiple sclerosis. Mult. Scler. Relat. Disord. 2020, 43, 102129. [Google Scholar] [CrossRef]
- Xiong, R.; Zhang, L.; Li, S.; Sun, Y.; Ding, M.; Wang, Y.; Zhao, Y.; Wu, Y.; Shang, W.; Jiang, X.; et al. Novel and potent inhibitors targeting DHODH are broad-spectrum antivirals against RNA viruses including newly-emerged coronavirus SARS-CoV-2. Protein Cell 2020, 11, 723–739. [Google Scholar] [CrossRef] [PubMed]
- Munier-Lehmann, H.; Vidalain, P.O.; Tangy, F.; Janin, Y.L. On dihydroorotate dehydrogenases and their inhibitors and uses. J. Med. Chem. 2013, 56, 3148–3167. [Google Scholar] [CrossRef] [PubMed]
- Luthra, P.; Naidoo, J.; Pietzsch, C.A.; De, S.; Khadka, S.; Anantpadma, M.; Williams, C.G.; Edwards, M.R.; Davey, R.A.; Bukreyev, A.; et al. Inhibiting pyrimidine biosynthesis impairs Ebola virus replication through depletion of nucleoside pools and activation of innate immune responses. Antivir. Res. 2018, 158, 288–302. [Google Scholar] [CrossRef]
- Wang, Q.Y.; Bushell, S.; Qing, M.; Xu, H.Y.; Bonavia, A.; Nunes, S.; Zhou, J.; Poh, M.K.; Florez de Sessions, P.; Niyomrattanakit, P.; et al. Inhibition of dengue virus through suppression of host pyrimidine biosynthesis. J. Virol. 2011, 85, 6548–6556. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.J.; Sharma, S.D.; Feng, J.Y.; Ray, A.S.; Smidansky, E.D.; Kireeva, M.L.; Cho, A.; Perry, J.; Vela, J.E.; Park, Y.; et al. Sensitivity of mitochondrial transcription and resistance of RNA polymerase II dependent nuclear transcription to antiviral ribonucleosides. PLoS Pathog. 2012, 8, e1003030. [Google Scholar] [CrossRef]
- Fox, G.B.; Esbenshade, T.A.; Pan, J.B.; Radek, R.J.; Krueger, K.M.; Yao, B.B.; Browman, K.E.; Buckley, M.J.; Ballard, M.E.; Komater, V.A.; et al. Pharmacological properties of ABT-239 [4-(2-{2-[(2R)-2-Methylpyrrolidinyl]ethyl}-benzofuran-5-yl)benzonitrile]: II. Neurophysiological characterization and broad preclinical efficacy in cognition and schizophrenia of a potent and selective histamine H3 receptor antagonist. J. Pharmacol. Exp. Ther. 2005, 313, 176–190. [Google Scholar] [CrossRef]
- Cowart, M.; Faghih, R.; Curtis, M.P.; Gfesser, G.A.; Bennani, Y.L.; Black, L.A.; Pan, L.; Marsh, K.C.; Sullivan, J.P.; Esbenshade, T.A.; et al. 4-(2-[2-(2(R)-methylpyrrolidin-1-yl)ethyl]benzofuran-5-yl)benzonitrile and related 2-aminoethylbenzofuran H3 receptor antagonists potently enhance cognition and attention. J. Med. Chem. 2005, 48, 38–55. [Google Scholar] [CrossRef]
- Zhao, J.F.; Ching, L.C.; Kou, Y.R.; Lin, S.J.; Wei, J.; Shyue, S.K.; Lee, T.S. Activation of TRPV1 prevents OxLDL-induced lipid accumulation and TNF-α-induced inflammation in macrophages: Role of liver X receptor α. Mediat. Inflamm. 2013, 2013, 925171. [Google Scholar] [CrossRef]
- Sanjai Kumar, P.; Nayak, T.K.; Mahish, C.; Sahoo, S.S.; Radhakrishnan, A.; De, S.; Datey, A.; Sahu, R.P.; Goswami, C.; Chattopadhyay, S.; et al. Inhibition of transient receptor potential vanilloid 1 (TRPV1) channel regulates chikungunya virus infection in macrophages. Arch. Virol. 2021, 166, 139–155. [Google Scholar] [CrossRef] [PubMed]
- Lepesheva, G.I.; Hargrove, T.Y.; Anderson, S.; Kleshchenko, Y.; Furtak, V.; Wawrzak, Z.; Villalta, F.; Waterman, M.R. Structural insights into inhibition of sterol 14alpha-demethylase in the human pathogen Trypanosoma cruzi. J. Biol. Chem. 2010, 285, 25582–25590. [Google Scholar] [CrossRef] [PubMed]
- Meutiawati, F.; Bezemer, B.; Strating, J.R.P.M.; Overheul, G.J.; Žusinaite, E.; van Kuppeveld, F.J.M.; van Cleef, K.W.R.; van Rij, R.P. Posaconazole inhibits dengue virus replication by targeting oxysterol-binding protein. Antivir. Res. 2018, 157, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Roberts, B.L.; Severance, Z.C.; Bensen, R.C.; Le-McClain, A.T.; Malinky, C.A.; Mettenbrink, E.M.; Nuñez, J.I.; Reddig, W.J.; Blewett, E.L.; Burgett, A.W.G. Differing activities of oxysterol-binding protein (OSBP) targeting anti-viral compounds. Antivir. Res. 2019, 170, 104548. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; He, S.; Martínez-Romero, C.; Kouznetsova, J.; Tawa, G.; Xu, M.; Shinn, P.; Fisher, E.; Long, Y.; Motabar, O.; et al. Synergistic drug combination effectively blocks Ebola virus infection. Antivir. Res. 2017, 137, 165–172. [Google Scholar] [CrossRef]
- Rhoden, E.; Ng, T.F.F.; Campagnoli, R.; Nix, W.A.; Konopka-Anstadt, J.; Selvarangan, R.; Briesach, L.; Oberste, M.S.; Weldon, W.C. Antifungal Triazole Posaconazole Targets an Early Stage of the Parechovirus A3 Life Cycle. Antimicrob. Agents Chemother. 2020, 64, 02372-19. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sotorilli, G.E.; Gravina, H.D.; de Carvalho, A.C.; Shimizu, J.F.; Fontoura, M.A.; Melo-Hanchuk, T.D.; Cordeiro, A.T.; Marques, R.E. Phenotypical Screening of an MMV Open Box Library and Identification of Compounds with Antiviral Activity against St. Louis Encephalitis Virus. Viruses 2023, 15, 2416. https://doi.org/10.3390/v15122416
Sotorilli GE, Gravina HD, de Carvalho AC, Shimizu JF, Fontoura MA, Melo-Hanchuk TD, Cordeiro AT, Marques RE. Phenotypical Screening of an MMV Open Box Library and Identification of Compounds with Antiviral Activity against St. Louis Encephalitis Virus. Viruses. 2023; 15(12):2416. https://doi.org/10.3390/v15122416
Chicago/Turabian StyleSotorilli, Giuliana Eboli, Humberto Doriguetto Gravina, Ana Carolina de Carvalho, Jacqueline Farinha Shimizu, Marina Alves Fontoura, Talita Diniz Melo-Hanchuk, Artur Torres Cordeiro, and Rafael Elias Marques. 2023. "Phenotypical Screening of an MMV Open Box Library and Identification of Compounds with Antiviral Activity against St. Louis Encephalitis Virus" Viruses 15, no. 12: 2416. https://doi.org/10.3390/v15122416
APA StyleSotorilli, G. E., Gravina, H. D., de Carvalho, A. C., Shimizu, J. F., Fontoura, M. A., Melo-Hanchuk, T. D., Cordeiro, A. T., & Marques, R. E. (2023). Phenotypical Screening of an MMV Open Box Library and Identification of Compounds with Antiviral Activity against St. Louis Encephalitis Virus. Viruses, 15(12), 2416. https://doi.org/10.3390/v15122416





