Tailoring the Host Range of Ackermannviridae Bacteriophages through Chimeric Tailspike Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. DNA, Amino Acid, and Structural In Silico Comparisons
2.3. Phage Engineering
2.4. Phage Stock Preparation
2.5. Phage Spot Assay
2.6. Luciferase Reporter Phage Assays
3. Results
3.1. The Genome of SPTD1 Encodes a TSP Gene Cluster
3.2. Sequence Comparison of SPTD1’s Tailspike Proteins to Known Ackermanniridae Phages
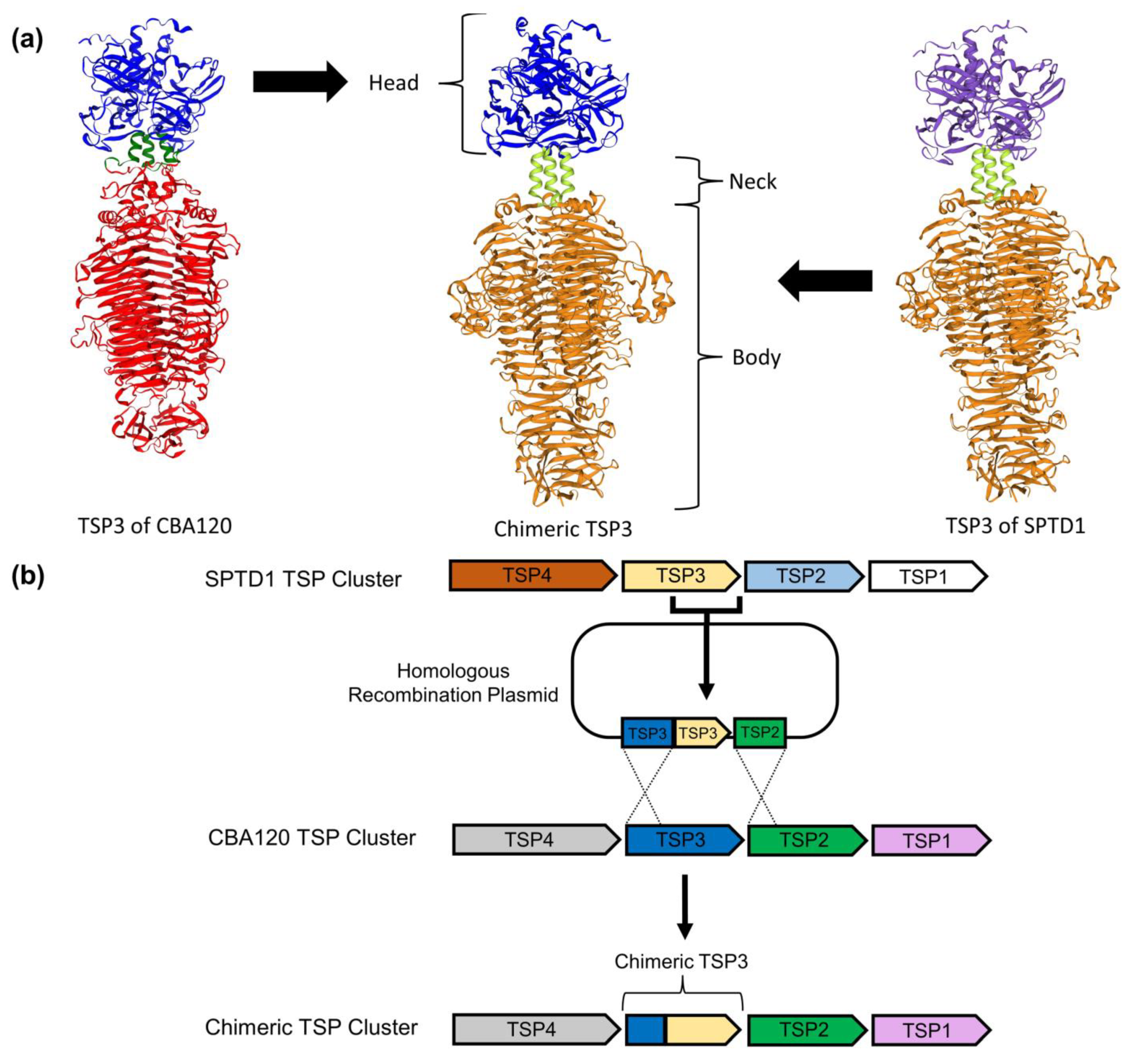
3.3. Generation of Chimeric Ackermannviridae Tailspike Proteins
3.4. Functional Characterization of TSP Chimeras
3.5. TSP3 Is Sufficient to Recapitulate SPTD1’s Known Salmonella Host Range
3.6. Expansion and Contraction of Host Range through Chimeric Tailspike Proteins
3.7. RBP-SPTD1-3’s Activity against Other O:3-Containing Salmonella Serovars
3.8. RBP-SPTD1-3 Is a Superior Luciferase Reporter for Salmonella Detection
3.9. RBP-SPTD1-3 Does Not Exhibit Cross-Reactivity with Other Gram-Negative Bacteria
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goodridge, L.D.; Bisha, B. Phage-based biocontrol strategies to reduce foodborne pathogens in foods. Bacteriophage 2011, 1, 130–137. [Google Scholar] [CrossRef] [Green Version]
- Cristobal-Cueto, P.; Garcia-Quintanilla, A.; Esteban, J.; Garcia-Quintanilla, M. Phages in Food Industry Biocontrol and Bioremediation. Antibiotics 2021, 10, 786. [Google Scholar] [CrossRef]
- Endersen, L.; O’Mahony, J.; Hill, C.; Ross, R.P.; McAuliffe, O.; Coffey, A. Phage therapy in the food industry. Annu. Rev. Food Sci. Technol. 2014, 5, 327–349. [Google Scholar] [CrossRef]
- Meile, S.; Kilcher, S.; Loessner, M.J.; Dunne, M. Reporter Phage-Based Detection of Bacterial Pathogens: Design Guidelines and Recent Developments. Viruses 2020, 12, 944. [Google Scholar] [CrossRef]
- Hall, M.P.; Unch, J.; Binkowski, B.F.; Valley, M.P.; Butler, B.L.; Wood, M.G.; Otto, P.; Zimmerman, K.; Vidugiris, G.; Machleidt, T.; et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 2012, 7, 1848–1857. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Coronel-Aguilera, C.P.; Romero, P.L.; Perry, L.; Minocha, U.; Rosenfield, C.; Gehring, A.G.; Paoli, G.C.; Bhunia, A.K.; Applegate, B. The Use of a Novel NanoLuc -Based Reporter Phage for the Detection of Escherichia coli O157:H7. Sci. Rep. 2016, 6, 33235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinkley, T.C.; Garing, S.; Singh, S.; Le Ny, A.M.; Nichols, K.P.; Peters, J.E.; Talbert, J.N.; Nugen, S.R. Reporter bacteriophage T7NLC utilizes a novel NanoLuc::CBM fusion for the ultrasensitive detection of Escherichia coli in water. Analyst 2018, 143, 4074–4082. [Google Scholar] [CrossRef] [Green Version]
- Alonzo, L.F.; Jain, P.; Hinkley, T.; Clute-Reinig, N.; Garing, S.; Spencer, E.; Dinh, V.T.T.; Bell, D.; Nugen, S.; Nichols, K.P.; et al. Rapid, sensitive, and low-cost detection of Escherichia coli bacteria in contaminated water samples using a phage-based assay. Sci. Rep. 2022, 12, 7741. [Google Scholar] [CrossRef] [PubMed]
- Erickson, S.; Gil, J.; Stach, J.; Hahn, W.; Nguyen, M.M. Validation of PhageDx Cronobacter Assay for the Identification of Cronobacter spp. in Powdered Infant Formula: AOAC Performance Tested MethodSM 051803. J. AOAC Int. 2021, 104, 1580–1592. [Google Scholar] [CrossRef]
- Nguyen, M.M.; Gil, J.; Brown, M.; Cesar Tondo, E.; Soraya Martins de Aquino, N.; Eisenberg, M.; Erickson, S. Accurate and sensitive detection of Salmonella in foods by engineered bacteriophages. Sci. Rep. 2020, 10, 17463. [Google Scholar] [CrossRef] [PubMed]
- Meile, S.; Sarbach, A.; Du, J.; Schuppler, M.; Saez, C.; Loessner, M.J.; Kilcher, S. Engineered Reporter Phages for Rapid Bioluminescence-Based Detection and Differentiation of Viable Listeria Cells. Appl. Environ. Microbiol. 2020, 86, e00442-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, M.M.; Gil, J.; Hahn, W.; Dong, T.; Paulson, J.; Stevens, D.; Zahn, H.; Zimmer, B.; Erickson, S. Validation of the PhageDx Listeria Assay for Detection of Listeria spp. on Stainless Steel and Ceramic Environmental Surfaces AOAC Performance Tested MethodSM 102005. J. AOAC Int. 2021, 104, 1609–1619. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Doval, C.; van Raaij, M.J. Structure of the receptor-binding carboxy-terminal domain of bacteriophage T7 tail fibers. Proc. Natl. Acad. Sci. USA 2012, 109, 9390–9395. [Google Scholar] [CrossRef] [Green Version]
- Xia, G.; Corrigan, R.M.; Winstel, V.; Goerke, C.; Grundling, A.; Peschel, A. Wall teichoic Acid-dependent adsorption of staphylococcal siphovirus and myovirus. J. Bacteriol. 2011, 193, 4006–4009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunne, M.; Denyes, J.M.; Arndt, H.; Loessner, M.J.; Leiman, P.G.; Klumpp, J. Salmonella Phage S16 Tail Fiber Adhesin Features a Rare Polyglycine Rich Domain for Host Recognition. Structure 2018, 26, 1573–1582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latka, A.; Leiman, P.G.; Drulis-Kawa, Z.; Briers, Y. Modeling the Architecture of Depolymerase-Containing Receptor Binding Proteins in Klebsiella Phages. Front. Microbiol. 2019, 10, 2649. [Google Scholar] [CrossRef]
- Nobrega, F.L.; Vlot, M.; de Jonge, P.A.; Dreesens, L.L.; Beaumont, H.J.E.; Lavigne, R.; Dutilh, B.E.; Brouns, S.J.J. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 2018, 16, 760–773. [Google Scholar] [CrossRef]
- Steinbacher, S.; Miller, S.; Baxa, U.; Budisa, N.; Weintraub, A.; Seckler, R.; Huber, R. Phage P22 tailspike protein: Crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage. J. Mol. Biol. 1997, 267, 865–880. [Google Scholar] [CrossRef]
- Seul, A.; Muller, J.J.; Andres, D.; Stettner, E.; Heinemann, U.; Seckler, R. Bacteriophage P22 tailspike: Structure of the complete protein and function of the interdomain linker. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 1336–1345. [Google Scholar] [CrossRef] [Green Version]
- Dunne, M.; Prokhorov, N.S.; Loessner, M.J.; Leiman, P.G. Reprogramming bacteriophage host range: Design principles and strategies for engineering receptor binding proteins. Curr. Opin. Biotechnol. 2021, 68, 272–281. [Google Scholar] [CrossRef]
- Dams, D.; Brondsted, L.; Drulis-Kawa, Z.; Briers, Y. Engineering of receptor-binding proteins in bacteriophages and phage tail-like bacteriocins. Biochem. Soc. Trans. 2019, 47, 449–460. [Google Scholar] [CrossRef]
- Dunne, M.; Rupf, B.; Tala, M.; Qabrati, X.; Ernst, P.; Shen, Y.; Sumrall, E.; Heeb, L.; Pluckthun, A.; Loessner, M.J.; et al. Reprogramming Bacteriophage Host Range through Structure-Guided Design of Chimeric Receptor Binding Proteins. Cell Rep. 2019, 29, 1336–1350. [Google Scholar] [CrossRef] [Green Version]
- Latka, A.; Lemire, S.; Grimon, D.; Dams, D.; Maciejewska, B.; Lu, T.; Drulis-Kawa, Z.; Briers, Y. Engineering the Modular Receptor-Binding Proteins of Klebsiella Phages Switches Their Capsule Serotype Specificity. mBio 2021, 12, e00455-21. [Google Scholar] [CrossRef]
- Gebhart, D.; Williams, S.R.; Scholl, D. Bacteriophage SP6 encodes a second tailspike protein that recognizes Salmonella enterica serogroups C2 and C3. Virology 2017, 507, 263–266. [Google Scholar] [CrossRef]
- Adriaenssens, E.M.; Ackermann, H.W.; Anany, H.; Blasdel, B.; Connerton, I.F.; Goulding, D.; Griffiths, M.W.; Hooton, S.P.; Kutter, E.M.; Kropinski, A.M.; et al. A suggested new bacteriophage genus: “Viunalikevirus”. Arch. Virol. 2012, 157, 2035–2046. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, I.; Osada, K.; Azam, A.H.; Asakawa, H.; Miyanaga, K.; Tanji, Y. The Presence of Two Receptor-Binding Proteins Contributes to the Wide Host Range of Staphylococcal Twort-Like Phages. Appl. Environ. Microbiol. 2016, 82, 5763–5774. [Google Scholar] [CrossRef] [Green Version]
- Plattner, M.; Shneider, M.M.; Arbatsky, N.P.; Shashkov, A.S.; Chizhov, A.O.; Nazarov, S.; Prokhorov, N.S.; Taylor, N.M.I.; Buth, S.A.; Gambino, M.; et al. Structure and Function of the Branched Receptor-Binding Complex of Bacteriophage CBA120. J. Mol. Biol. 2019, 431, 3718–3739. [Google Scholar] [CrossRef]
- Kutter, E.M.; Skutt-Kakaria, K.; Blasdel, B.; El-Shibiny, A.; Castano, A.; Bryan, D.; Kropinski, A.M.; Villegas, A.; Ackermann, H.W.; Toribio, A.L.; et al. Characterization of a ViI-like phage specific to Escherichia coli O157:H7. Virol. J. 2011, 8, 430. [Google Scholar] [CrossRef] [Green Version]
- Phothaworn, P.; Supokaivanich, R.; Lim, J.; Klumpp, J.; Imam, M.; Kutter, E.; Galyov, E.E.; Dunne, M.; Korbsrisate, S. Development of a broad-spectrum Salmonella phage cocktail containing Viunalike and Jerseylike viruses isolated from Thailand. Food Microbiol. 2020, 92, 103586. [Google Scholar] [CrossRef]
- Zhu, W.; Ding, Y.; Huang, C.; Wang, J.; Wang, J.; Wang, X. Genomic characterization of a novel bacteriophage STP55 revealed its prominent capacity in disrupting the dual-species biofilm formed by Salmonella Typhimurium and Escherichia coli O157: H7 strains. Arch. Microbiol. 2022, 204, 597. [Google Scholar] [CrossRef]
- Witte, S.; Huijboom, L.; Klamert, S.; van de Straat, L.; Hagens, S.; Fieseler, L.; de Vegt, B.T.; van Mierlo, J.T. Application of bacteriophages EP75 and EP335 efficiently reduces viable cell counts of Escherichia coli O157 on beef and vegetables. Food Microbiol. 2022, 104, 103978. [Google Scholar] [CrossRef]
- Islam, M.S.; Zhou, Y.; Liang, L.; Nime, I.; Yan, T.; Willias, S.P.; Mia, M.Z.; Bei, W.; Connerton, I.F.; Fischetti, V.A.; et al. Application of a Broad Range Lytic Phage LPST94 for Biological Control of Salmonella in Foods. Microorganisms 2020, 8, 247. [Google Scholar] [CrossRef] [Green Version]
- Thanh, N.C.; Nagayoshi, Y.; Fujino, Y.; Iiyama, K.; Furuya, N.; Hiromasa, Y.; Iwamoto, T.; Doi, K. Characterization and Genome Structure of Virulent Phage EspM4VN to Control Enterobacter sp. M4 Isolated from Plant Soft Rot. Front. Microbiol. 2020, 11, 885. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States--major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Grimont, P.A.D.; Weill, F.-X. Antigenic Formulae of the Salmonella Serovars, 9th ed.; WHO Collaborating Centre for Reference and Research on Salmonella; Pasteur Institute: Paris, France, 2007. [Google Scholar]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Needleman, S.B.; Wunsch, C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- Chen, C.; Bales, P.; Greenfield, J.; Heselpoth, R.D.; Nelson, D.C.; Herzberg, O. Crystal structure of ORF210 from E. coli O157:H1 phage CBA120 (TSP1), a putative tailspike protein. PLoS ONE 2014, 9, e93156. [Google Scholar] [CrossRef] [Green Version]
- Greenfield, J.; Shang, X.; Luo, H.; Zhou, Y.; Heselpoth, R.D.; Nelson, D.C.; Herzberg, O. Structure and tailspike glycosidase machinery of ORF212 from E. coli O157:H7 phage CBA120 (TSP3). Sci. Rep. 2019, 9, 7349. [Google Scholar] [CrossRef] [Green Version]
- Chao, K.L.; Shang, X.; Greenfield, J.; Linden, S.B.; Alreja, A.B.; Nelson, D.C.; Herzberg, O. Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N). Sci. Rep. 2022, 12, 2061. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Rose, A.S.; Hildebrand, P.W. NGL Viewer: A web application for molecular visualization. Nucleic Acids Res. 2015, 43, W576–W579. [Google Scholar] [CrossRef]
- Greenfield, J.; Shang, X.; Luo, H.; Zhou, Y.; Linden, S.B.; Heselpoth, R.D.; Leiman, P.G.; Nelson, D.C.; Herzberg, O. Structure and function of bacteriophage CBA120 ORF211 (TSP2), the determinant of phage specificity towards E. coli O157:H7. Sci. Rep. 2020, 10, 15402. [Google Scholar] [CrossRef]
- Sorensen, A.N.; Woudstra, C.; Sorensen, M.C.H.; Brondsted, L. Subtypes of tail spike proteins predicts the host range of Ackermannviridae phages. Comput. Struct. Biotechnol. J. 2021, 19, 4854–4867. [Google Scholar] [CrossRef]
- Witte, S.; Zinsli, L.V.; Gonzalez-Serrano, R.; Matter, C.I.; Loessner, M.J.; van Mierlo, J.T.; Dunne, M. Structural and functional characterization of the receptor binding proteins of Escherichia coli O157 phages EP75 and EP335. Comput. Struct. Biotechnol. J. 2021, 19, 3416–3426. [Google Scholar] [CrossRef]
- Walter, M.; Fiedler, C.; Grassl, R.; Biebl, M.; Rachel, R.; Hermo-Parrado, X.L.; Llamas-Saiz, A.L.; Seckler, R.; Miller, S.; van Raaij, M.J. Structure of the receptor-binding protein of bacteriophage det7: A podoviral tail spike in a myovirus. J. Virol. 2008, 82, 2265–2273. [Google Scholar] [CrossRef] [Green Version]
- Steinbacher, S.; Baxa, U.; Miller, S.; Weintraub, A.; Seckler, R.; Huber, R. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proc. Natl. Acad. Sci. USA 1996, 93, 10584–10588. [Google Scholar] [CrossRef]
- Kutter, E. Phage host range and efficiency of plating. In Methods in Molecular Biology; Bacteriophages; Humana Press: Totowa, NJ, USA, 2009; Volume 501, pp. 141–149. [Google Scholar] [CrossRef]
- Kipper, D.; Carroll, L.M.; Mascitti, A.K.; Streck, A.F.; Fonseca, A.S.K.; Ikuta, N.; Lunge, V.R. Genomic Characterization of Salmonella Minnesota Clonal Lineages Associated with Poultry Production in Brazil. Animals 2020, 10, 2043. [Google Scholar] [CrossRef]
- Broeker, N.K.; Roske, Y.; Valleriani, A.; Stephan, M.S.; Andres, D.; Koetz, J.; Heinemann, U.; Barbirz, S. Time-resolved DNA release from an O-antigen-specific Salmonella bacteriophage with a contractile tail. J. Biol. Chem. 2019, 294, 11751–11761. [Google Scholar] [CrossRef]
- Threlfall, E.J.; Ward, L.R.; Hampton, M.D.; Ridley, A.M.; Rowe, B.; Roberts, D.; Gilbert, R.J.; Van Someren, P.; Wall, P.G.; Grimont, P. Molecular fingerprinting defines a strain of Salmonella enterica serotype Anatum responsible for an international outbreak associated with formula-dried milk. Epidemiol. Infect. 1998, 121, 289–293. [Google Scholar] [CrossRef]
- Reeves, P.R.; Cunneen, M.M.; Liu, B.; Wang, L. Genetics and evolution of the Salmonella galactose-initiated set of o antigens. PLoS ONE 2013, 8, e69306. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Knirel, Y.A.; Feng, L.; Perepelov, A.V.; Senchenkova, S.N.; Reeves, P.R.; Wang, L. Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol. Rev. 2014, 38, 56–89. [Google Scholar] [CrossRef] [Green Version]
- Plawinska-Czarnak, J.; Wodz, K.; Kizerwetter-Swida, M.; Nowak, T.; Bogdan, J.; Kwiecinski, P.; Kwiecinski, A.; Anusz, K. Citrobacter braakii Yield False-Positive Identification as Salmonella, a Note of Caution. Foods 2021, 10, 2177. [Google Scholar] [CrossRef]
- Pilar, A.V.C.; Petronella, N.; Dussault, F.M.; Verster, A.J.; Bekal, S.; Levesque, R.C.; Goodridge, L.; Tamber, S. Similar yet different: Phylogenomic analysis to delineate Salmonella and Citrobacter species boundaries. BMC Genomics 2020, 21, 377. [Google Scholar] [CrossRef]
- Broeker, N.K.; Barbirz, S. Not a barrier but a key: How bacteriophages exploit host’s O-antigen as an essential receptor to initiate infection. Mol. Microbiol. 2017, 105, 353–357. [Google Scholar] [CrossRef] [Green Version]
- Hooton, S.P.; Timms, A.R.; Rowsell, J.; Wilson, R.; Connerton, I.F. Salmonella Typhimurium-specific bacteriophage PhiSH19 and the origins of species specificity in the Vi01-like phage family. Virol. J. 2011, 8, 498. [Google Scholar] [CrossRef]
- Chaudhary, N.; Singh, D.; Maurya, R.K.; Mohan, B.; Taneja, N. Complete Genome Sequence of Salmonella Phage vB_SenA_SM5, Active against Multidrug-Resistant Salmonella enterica Serovar Typhi Isolates. Microbiol. Resour. Announc. 2022, 11, e0030922. [Google Scholar] [CrossRef]
- Wong, C.W.Y.; Wang, S. Efficacy of Repeated Applications of Bacteriophages on Salmonella enterica-Infected Alfalfa Sprouts during Germination. Pathogens 2022, 11, 1156. [Google Scholar] [CrossRef] [PubMed]
- Newase, S.; Kapadnis, B.P.; Shashidhar, R. Isolation and Genome Sequence Characterization of Bacteriophage vB_SalM_PM10, a Cba120virus, Concurrently Infecting Salmonella enterica Serovars Typhimurium, Typhi, and Enteritidis. Curr. Microbiol. 2019, 76, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Kabanova, A.P.; Shneider, M.M.; Korzhenkov, A.A.; Bugaeva, E.N.; Miroshnikov, K.K.; Zdorovenko, E.L.; Kulikov, E.E.; Toschakov, S.V.; Ignatov, A.N.; Knirel, Y.A.; et al. Host Specificity of the Dickeya Bacteriophage PP35 Is Directed by a Tail Spike Interaction with Bacterial O-Antigen, Enabling the Infection of Alternative Non-pathogenic Bacterial Host. Front. Microbiol. 2018, 9, 3288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanegasaki, S.; Wright, A. Studies on the mechanism of phage adsorption: Interaction between phage epsilon15 and its cellular receptor. Virology 1973, 52, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Robbins, P.W.; Uchida, T. Studies on the chemical basis of the phage conversion of O-antigens in the E-group Salmonellae. Biochemistry 1962, 1, 323–335. [Google Scholar] [CrossRef]
- Kropinski, A.M.; Kovalyova, I.V.; Billington, S.J.; Patrick, A.N.; Butts, B.D.; Guichard, J.A.; Pitcher, T.J.; Guthrie, C.C.; Sydlaske, A.D.; Barnhill, L.M.; et al. The genome of epsilon15, a serotype-converting, Group E1 Salmonella enterica-specific bacteriophage. Virology 2007, 369, 234–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef]
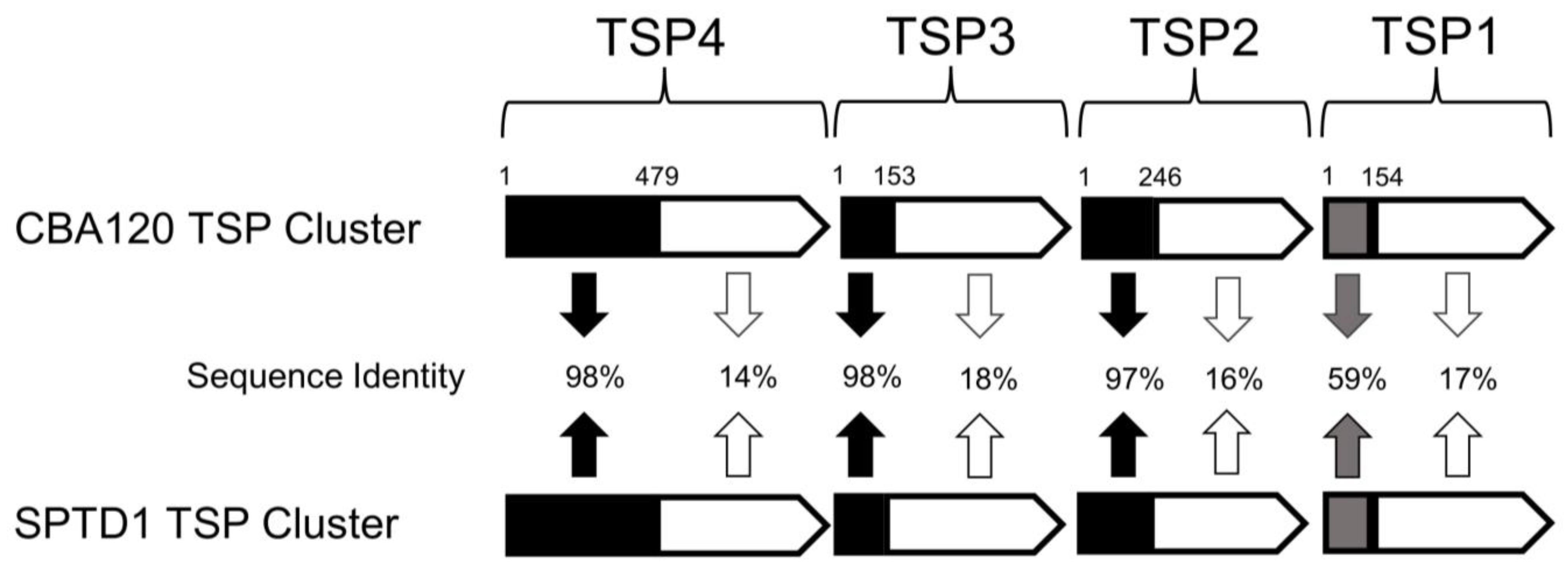
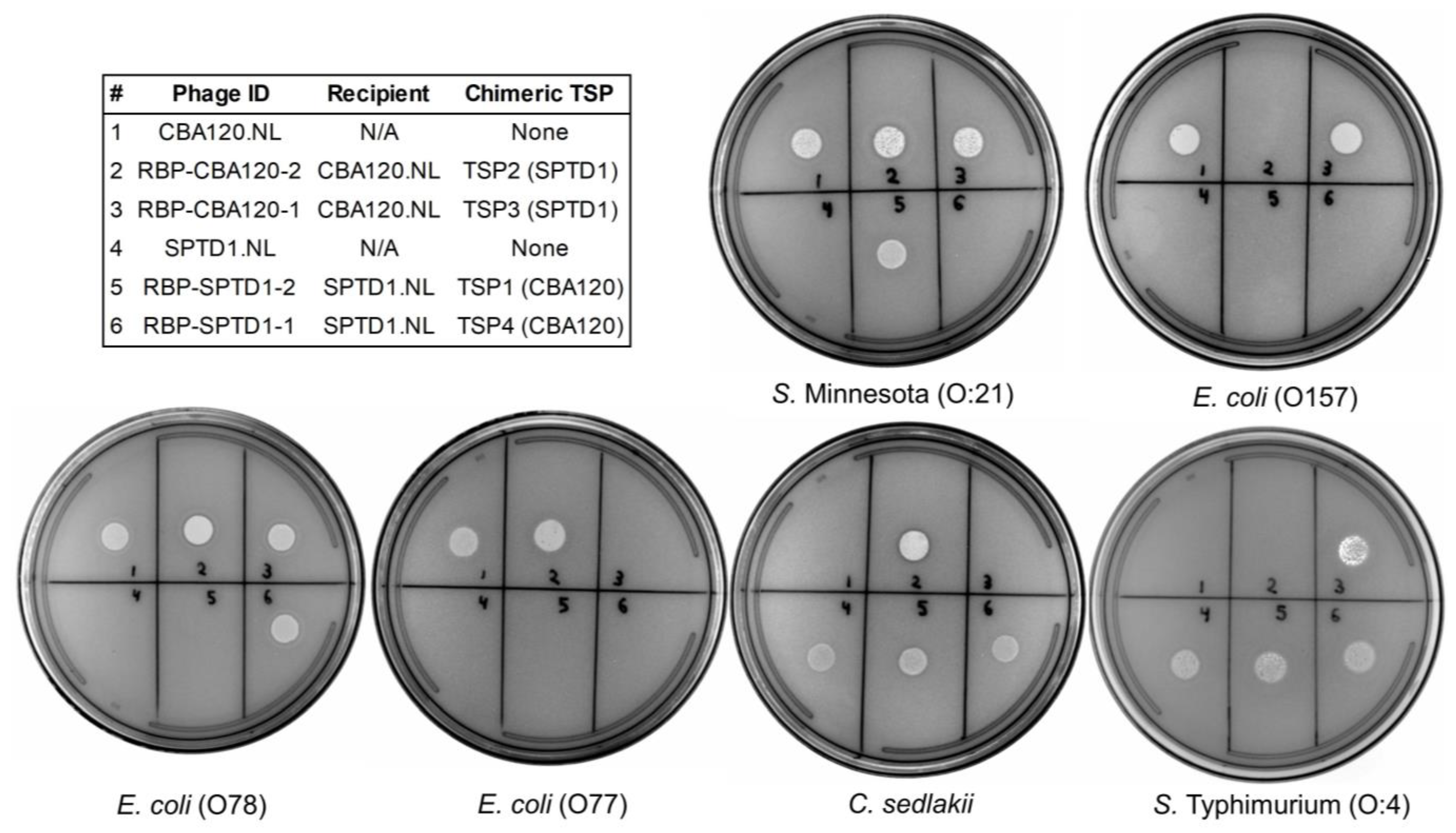
| Origin of C-Terminal Region of Each TSP 1 | Bacterial Species and Serogroups Targeted by Each TSP 2 | |||||||
|---|---|---|---|---|---|---|---|---|
| Phage ID | TSP1 | TSP2 | TSP3 | TSP4 | TSP1 | TSP2 3 | TSP3 4 | TSP4 |
| CBA120.NL | CBA120Native | CBA120Native | CBA120Native | CBA120Native | S. enterica O:21 | E. coli O157 | E. coli O77 | E. coli O78 |
| SPTD1.NL | SPTD1Native | SPTD1Native | SPTD1Native | SPTD1Native | - | C. sedlakii | S. enterica O:4, O:9 | - |
| RBP-CBA120-1 | CBA120Native | CBA120Native | SPTD1Chimera | CBA120Native | S. enterica O:21 | E. coli O157 | S. enterica O:4, O:9 | E. coli O78 |
| RBP-CBA120-2 | CBA120Native | SPTD1Chimera | CBA120Native | CBA120Native | S. enterica O:21 | C. sedlakii | E. coli O77 | E. coli O78 |
| RBP-SPTD1-1 | SPTD1Native | SPTD1Native | SPTD1Native | CBA120Chimera | - | C. sedlakii | S. enterica O:4, O:9 | E. coli O78 |
| RBP-SPTD1-2 | CBA120Chimera | SPTD1Native | SPTD1Native | SPTD1Native | S. enterica O:21 | C. sedlakii | S. enterica O:4, O:9 | - |
| RBP-SPTD1-3 | CBA120Chimera | Det7Chimera | SPTD1Native | SPTD1Native | S. enterica O:21 | S. enterica O:3, 10 | S. enterica O:4, O:9 | - |
| Spot Assay Result 2 | |||||
|---|---|---|---|---|---|
| Serogroup | S. Serovar 1 | CBA120.NL | RBP-CBA120-2 | RBP-CBA120-1 | SPTD1.NL |
| O:2 | Paratyphi A | Negative | Negative | Positive | Positive |
| O:4 | Abony | Negative | Negative | Positive | Positive |
| Agona | Negative | Negative | Positive | Positive | |
| Bispebjerg | Negative | Negative | Positive | Positive | |
| Brandenburg | Negative | Negative | Positive | Positive | |
| Chester | Negative | Negative | Positive | Positive | |
| Derby | Negative | Negative | Positive | Positive | |
| Heidelberg | Negative | Negative | Positive | Positive | |
| Kiambu | Negative | Negative | Positive | Positive | |
| Paratyphi B | Negative | Negative | Positive | Positive | |
| Saintpaul | Negative | Negative | Positive | Positive | |
| Sandiego | Negative | Negative | Positive | Positive | |
| Schwarzengrund | Negative | Negative | Positive | Positive | |
| Typhimurium 3 | Negative | Negative | Positive | Positive | |
| O:7 | Paratyphi C | Negative | Negative | Positive | Positive |
| O:8 | Newport | Negative | Negative | Positive | Positive |
| O:9 | Dublin | Negative | Negative | Positive | Positive |
| Enteritidis | Negative | Negative | Positive | Positive | |
| Gallinarum | Negative | Negative | Positive | Positive | |
| Javina | Negative | Negative | Positive | Positive | |
| Panama | Negative | Negative | Positive | Positive | |
| Typhi | Negative | Negative | Positive | Positive | |
| O:35 | Alachua | Negative | Negative | Positive | Positive |
| Spot Assay Result 2 | ||||
|---|---|---|---|---|
| Serogroup | S. Serovar 1 | Strain ID | SPTD1.NL | RBP-SPTD1-3 |
| O:1, 3, 19 | Liverpool | AUG365 | Negative | Negative |
| Senftenberg | 12004 | Negative | Negative | |
| 31072.1 | Negative | Negative | ||
| 43845 | Negative | Negative | ||
| 15106q | Negative | Negative | ||
| SARB59 | Negative | Negative | ||
| SEP160 | Negative | Positive | ||
| SL1315 | Negative | Negative | ||
| Taksony | 32133 | Negative | Negative | |
| O:3, 10 | Amsterdam | 41084 | Negative | Positive |
| Anatum 3 | 31064.1 | Negative | Positive | |
| DMSO13 | Negative | Negative | ||
| NOV091 | Negative | Negative | ||
| SARB2 | Negative | Positive | ||
| SLR 377 | Negative | Positive | ||
| Benfica | AUG071 | Negative | Positive | |
| Give | 9268 | Negative | Positive | |
| 63213 | Negative | Positive | ||
| Lexington | 11646 | Negative | Negative | |
| 9492-M | Negative | Negative | ||
| London | 43290 | Negative | Negative | |
| JUL218 | Negative | Positive | ||
| Meleagridis | 92 | Negative | Negative | |
| 11008.1 | Negative | Positive | ||
| FEB095 | Negative | Positive | ||
| Muenster | 31053 | Negative | Positive | |
| OCT084 | Negative | Positive | ||
| Uganda | 51278.2 | Negative | Positive | |
| Wagadugu | 53298 | Negative | Negative | |
| Weltevreden | BAA-2568 | Negative | Positive | |
| Relative Light Units 3 | |||
|---|---|---|---|
| Sample 1 | Serogroup 2 | SPTD1.NL | RBP-SPTD1-3 |
| Citrobacter braakii | - | 145 | 206 |
| Escherichia coli | O6 | 94 | 152 |
| O79 | 150 | 178 | |
| O111 | 242 | 255 | |
| O121 | 141 | 190 | |
| O145 | 165 | 209 | |
| Serratia marcescens | - | 105 | 213 |
| Shigella flexneri | - | 102 | 204 |
| Salmonella Typhimurium | O:4 | 801,150,016 | 1,007,500,000 |
| Media Control | - | 96 | 156 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gil, J.; Paulson, J.; Brown, M.; Zahn, H.; Nguyen, M.M.; Eisenberg, M.; Erickson, S. Tailoring the Host Range of Ackermannviridae Bacteriophages through Chimeric Tailspike Proteins. Viruses 2023, 15, 286. https://doi.org/10.3390/v15020286
Gil J, Paulson J, Brown M, Zahn H, Nguyen MM, Eisenberg M, Erickson S. Tailoring the Host Range of Ackermannviridae Bacteriophages through Chimeric Tailspike Proteins. Viruses. 2023; 15(2):286. https://doi.org/10.3390/v15020286
Chicago/Turabian StyleGil, Jose, John Paulson, Matthew Brown, Henriett Zahn, Minh M. Nguyen, Marcia Eisenberg, and Stephen Erickson. 2023. "Tailoring the Host Range of Ackermannviridae Bacteriophages through Chimeric Tailspike Proteins" Viruses 15, no. 2: 286. https://doi.org/10.3390/v15020286
APA StyleGil, J., Paulson, J., Brown, M., Zahn, H., Nguyen, M. M., Eisenberg, M., & Erickson, S. (2023). Tailoring the Host Range of Ackermannviridae Bacteriophages through Chimeric Tailspike Proteins. Viruses, 15(2), 286. https://doi.org/10.3390/v15020286






