In-Depth Analysis of the Re-Emergence of Respiratory Syncytial Virus at a Tertiary Care Hospital in Germany in the Summer of 2021 after the Alleviation of Non-Pharmaceutical Interventions Due to the SARS-CoV-2 Pandemic
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Clinical Data
2.2. Meteorological Data
2.3. Nucleic Acid (NA) Extraction and RSV Detection
2.4. RSV Genotyping and Phylogenetic Analysis
2.5. Statistical Analysis
3. Results
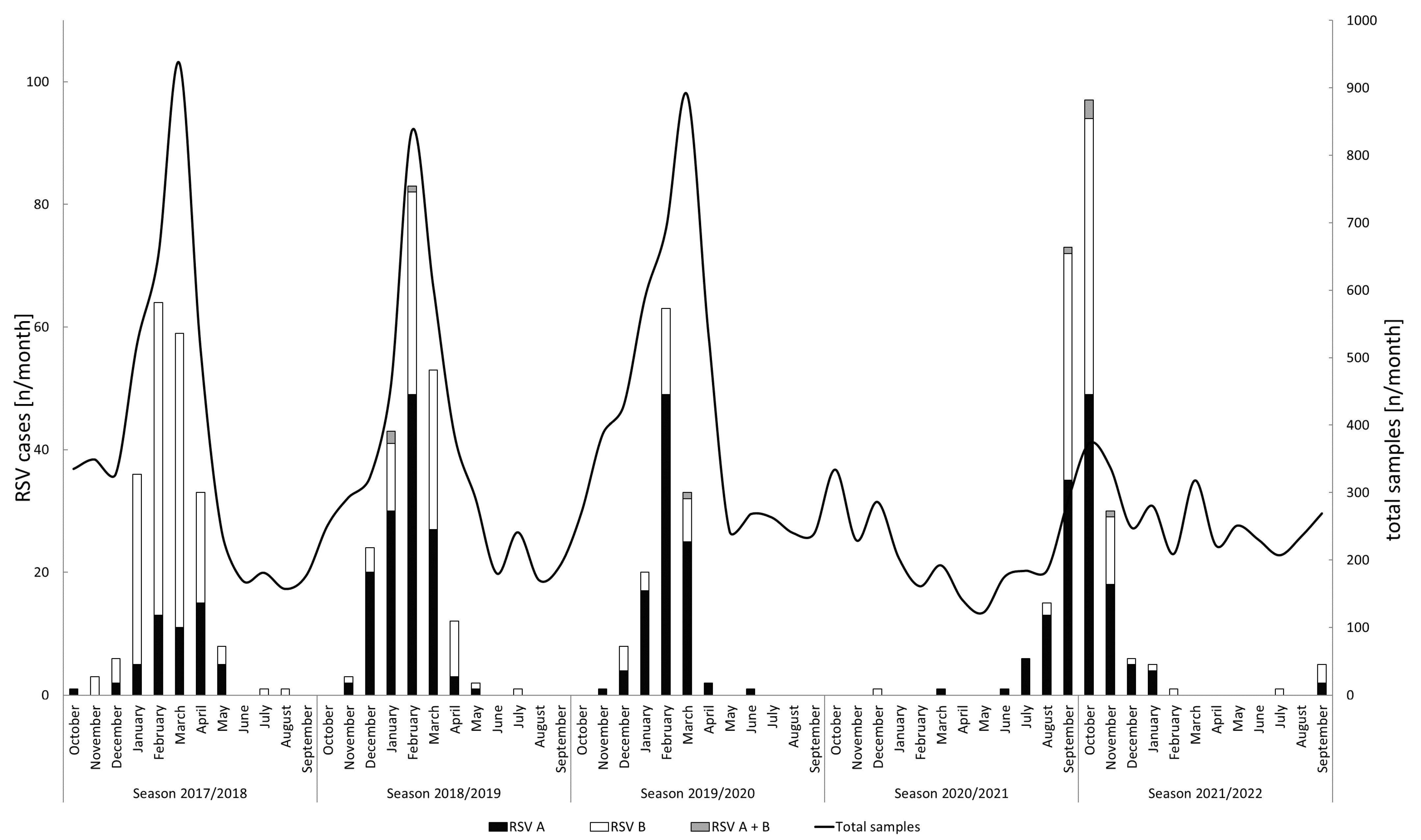
3.1. RSV Detection and Seasonality
3.2. Phylogenetic Analysis
3.3. Study Population and Clinical Features
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Borchers, A.T.; Chang, C.; Gershwin, M.E.; Gershwin, L.J. Respiratory syncytial virus—A comprehensive review. Clin. Rev. Allergy Immunol. 2013, 45, 331–379. [Google Scholar] [CrossRef]
- Krause, C.I. The ABCs of RSV. Nurse Pract. 2018, 43, 20–26. [Google Scholar] [CrossRef] [PubMed]
- El Kholy, A.A.; Mostafa, N.A.; El-Sherbini, S.A.; Ali, A.A.; Ismail, R.I.; Magdy, R.I.; Hamdy, M.S.; Soliman, M.S. Morbidity and outcome of severe respiratory syncytial virus infection. Pediatr. Int. 2013, 55, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Jartti, T.; Smits, H.H.; Bønnelykke, K.; Bircan, O.; Elenius, V.; Konradsen, J.R.; Maggina, P.; Makrinioti, H.; Stokholm, J.; Hedlin, G.; et al. Bronchiolitis needs a revisit: Distinguishing between virus entities and their treatments. Allergy 2019, 74, 40–52. [Google Scholar] [CrossRef] [Green Version]
- Hall, C.B.; Long, C.E.; Schnabel, K.C. Respiratory syncytial virus infections in previously healthy working adults. Clin. Infect. Dis. 2001, 33, 792–796. [Google Scholar] [CrossRef]
- Geevarghese, B.; Weinberg, A. Cell-mediated immune responses to respiratory syncytial virus infection: Magnitude, kinetics, and correlates with morbidity and age. Hum. Vaccin. Immunother. 2014, 10, 1047–1056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffiths, C.; Drews, S.J.; Marchant, D.J. Respiratory Syncytial Virus: Infection, Detection, and New Options for Prevention and Treatment. Clin. Microbiol. Rev. 2017, 30, 277–319. [Google Scholar] [CrossRef] [Green Version]
- Tabor, D.E.; Fernandes, F.; Langedijk, A.C.; Wilkins, D.; Lebbink, R.J.; Tovchigrechko, A.; Ruzin, A.; Kragten-Tabatabaie, L.; Jin, H.; Esser, M.T.; et al. Global Molecular Epidemiology of Respiratory Syncytial Virus from the 2017-2018 INFORM-RSV Study. J. Clin. Microbiol. 2020, 59, e01828-20. [Google Scholar] [CrossRef]
- Trovão, N.S.; Khuri-Bulos, N.; Tan, Y.; Puri, V.; Shilts, M.H.; Halpin, R.A.; Fedorova, N.B.; Nelson, M.I.; Halasa, N.; Das, S.R. Molecular characterization of respiratory syncytial viruses circulating in a paediatric cohort in Amman, Jordan. Microb. Genom. 2021, 7, 1–13. [Google Scholar] [CrossRef]
- Tripp, R.A.; Power, U.F.; Openshaw, P.J.M.; Kauvar, L.M. Respiratory Syncytial Virus: Targeting the G Protein Provides a New Approach for an Old Problem. J. Virol. 2018, 92, e01302-17. [Google Scholar] [CrossRef] [Green Version]
- Hammitt, L.L.; Dagan, R.; Yuan, Y.; Baca Cots, M.; Bosheva, M.; Madhi, S.A.; Muller, W.J.; Zar, H.J.; Brooks, D.; Grenham, A.; et al. Nirsevimab for Prevention of RSV in Healthy Late-Preterm and Term Infants. N. Engl. J. Med. 2022, 386, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Jiang, H.; Wan, Z.; Li, S.; Li, Y.; Wang, W.; Jin, X.; Li, Y.; Zhang, C. Evolutionary dynamics of group A and B respiratory syncytial virus in China, 2009–2018. Arch. Virol. 2021, 166, 2407–2418. [Google Scholar] [CrossRef]
- Yu, J.-M.; Fu, Y.-H.; Peng, X.-L.; Zheng, Y.-P.; He, J.-S. Genetic diversity and molecular evolution of human respiratory syncytial virus A and B. Sci. Rep. 2021, 11, 12941. [Google Scholar] [CrossRef] [PubMed]
- Goya, S.; Galiano, M.; Nauwelaers, I.; Trento, A.; Openshaw, P.J.; Mistchenko, A.S.; Zambon, M.; Viegas, M. Toward unified molecular surveillance of RSV: A proposal for genotype definition. Influenza Other Respir. Viruses 2020, 14, 274–285. [Google Scholar] [CrossRef]
- Muñoz-Escalante, J.C.; Comas-García, A.; Bernal-Silva, S.; Noyola, D.E. Respiratory syncytial virus B sequence analysis reveals a novel early genotype. Sci. Rep. 2021, 11, 3452. [Google Scholar] [CrossRef]
- Muñoz-Escalante, J.C.; Comas-García, A.; Bernal-Silva, S.; Robles-Espinoza, C.D.; Gómez-Leal, G.; Noyola, D.E. Respiratory syncytial virus A genotype classification based on systematic intergenotypic and intragenotypic sequence analysis. Sci. Rep. 2019, 9, 20097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altman, G.; Ahuja, J.; Monrad, J.T.; Dhaliwal, G.; Rogers-Smith, C.; Leech, G.; Snodin, B.; Sandbrink, J.B.; Finnveden, L.; Norman, A.J.; et al. A dataset of non-pharmaceutical interventions on SARS-CoV-2 in Europe. Sci. Data 2022, 9, 145. [Google Scholar] [CrossRef]
- Davison, A.J.; Dolan, A.; Akter, P.; Addison, C.; Dargan, D.J.; Alcendor, D.J.; McGeoch, D.J.; Hayward, G.S. The human cytomegalovirus genome revisited: Comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 2003, 84, 17–28. [Google Scholar] [CrossRef] [Green Version]
- Golke, P.; Hönemann, M.; Bergs, S.; Liebert, U.G. Human Rhinoviruses in Adult Patients in a Tertiary Care Hospital in Germany: Molecular Epidemiology and Clinical Significance. Viruses 2021, 13, 2027. [Google Scholar] [CrossRef]
- Laux, G.; Perricaudet, M.; Farrell, P.J. A spliced Epstein-Barr virus gene expressed in immortalized lymphocytes is created by circularization of the linear viral genome. EMBO J. 1988, 7, 769–774. [Google Scholar] [CrossRef]
- Gompels, U.A.; Nicholas, J.; Lawrence, G.; Jones, M.; Thomson, B.J.; Martin, M.E.; Efstathiou, S.; Craxton, M.; Macaulay, H.A. The DNA sequence of human herpesvirus-6: Structure, coding content, and genome evolution. Virology 1995, 209, 29–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deutscher Wetterdienst. Mean Monthly Temperature Station 2932. Available online: https://www.dwd.de/EN/climate_environment/cdc/clis/clis_node.html (accessed on 15 November 2022).
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iezadi, S.; Gholipour, K.; Azami-Aghdash, S.; Ghiasi, A.; Rezapour, A.; Pourasghari, H.; Pashazadeh, F. Effectiveness of non-pharmaceutical public health interventions against COVID-19: A systematic review and meta-analysis. PLoS ONE 2021, 16, e0260371. [Google Scholar] [CrossRef]
- Bundesministerium der Justiz. Gesetzestext IfSG [German]. Available online: http://www.gesetze-im-internet.de/ifsg/index.html (accessed on 21 January 2023).
- Bundesministerium für Gesundheit. COVID-19-SchG [German]. Available online: https://www.bundesgesundheitsministerium.de/service/gesetze-und-verordnungen/detail/gesetz-zur-staerkung-des-schutzes-der-bevoelkerung-und-insbesondere-vulnerabler-personengruppen-vor-covid-19.html (accessed on 21 January 2023).
- Robert Koch Institut. Zusammenstellung IfSG [German]. Available online: https://www.rki.de/DE/Content/Infekt/IfSG/Gesetze/gesetze_node.html (accessed on 21 January 2023).
- State Government of Saxony. Regulations Saxony [German]. Available online: https://www.coronavirus.sachsen.de/archiv-der-abgelaufenen-amtlichen-bekanntmachungen-7295.html (accessed on 21 January 2022).
- Jørgensen, F.; Bor, A.; Rasmussen, M.S.; Lindholt, M.F.; Petersen, M.B. Pandemic fatigue fueled political discontent during the COVID-19 pandemic. Proc. Natl. Acad. Sci. USA 2022, 119, e2201266119. [Google Scholar] [CrossRef]
- Messacar, K.; Baker, R.E.; Park, S.W.; Nguyen-Tran, H.; Cataldi, J.R.; Grenfell, B. Preparing for uncertainty: Endemic paediatric viral illnesses after COVID-19 pandemic disruption. Lancet 2022, 400, 1663–1665. [Google Scholar] [CrossRef] [PubMed]
- Broberg, E.K.; Waris, M.; Johansen, K.; Snacken, R.; Penttinen, P. Seasonality and geographical spread of respiratory syncytial virus epidemics in 15 European countries, 2010 to 2016. Euro Surveill. 2018, 23, 17–00284. [Google Scholar] [CrossRef] [Green Version]
- Teirlinck, A.C.; Broberg, E.K.; Stuwitz Berg, A.; Campbell, H.; Reeves, R.M.; Carnahan, A.; Lina, B.; Pakarna, G.; Bøås, H.; Nohynek, H.; et al. Recommendations for respiratory syncytial virus surveillance at the national level. Eur. Respir. J. 2021, 58, 2003766. [Google Scholar] [CrossRef]
- Vos, L.M.; Teirlinck, A.C.; Lozano, J.E.; Vega, T.; Donker, G.A.; Im Hoepelman, A.; Bont, L.J.; Oosterheert, J.J.; Meijer, A. Use of the moving epidemic method (MEM) to assess national surveillance data for respiratory syncytial virus (RSV) in the Netherlands, 2005 to 2017. Euro Surveill. 2019, 24, 1800469. [Google Scholar] [CrossRef] [Green Version]
- Cai, W.; Dürrwald, R.; Biere, B.; Schweiger, B.; Haas, W.; Wolff, T.; Buda, S.; Reiche, J. Determination of respiratory syncytial virus epidemic seasons by using 95% confidence interval of positivity rates, 2011-2021, Germany. Influenza Other Respir. Viruses 2022, 16, 854–857. [Google Scholar] [CrossRef]
- Nawrocki, J.; Olin, K.; Holdrege, M.C.; Hartsell, J.; Meyers, L.; Cox, C.; Powell, M.; Cook, C.V.; Jones, J.; Robbins, T.; et al. The Effects of Social Distancing Policies on Non-SARS-CoV-2 Respiratory Pathogens. Open Forum Infect. Dis. 2021, 8, ofab133. [Google Scholar] [CrossRef] [PubMed]
- Baertl, S.; Pietsch, C.; Maier, M.; Hönemann, M.; Bergs, S.; Liebert, U.G. Enteroviruses in Respiratory Samples from Paediatric Patients of a Tertiary Care Hospital in Germany. Viruses 2021, 13, 882. [Google Scholar] [CrossRef] [PubMed]
- Midgley, S.E.; Benschop, K.; Dyrdak, R.; Mirand, A.; Bailly, J.-L.; Bierbaum, S.; Buderus, S.; Böttcher, S.; Eis-Hübinger, A.-M.; Hönemann, M.; et al. Co-circulation of multiple enterovirus D68 subclades, including a novel B3 cluster, across Europe in a season of expected low prevalence, 2019/20. Euro Surveill. 2020, 25, 1900749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hönemann, M.; Martin, D.; Pietsch, C.; Maier, M.; Bergs, S.; Bieck, E.; Liebert, U.G. Influenza B virus infections in Western Saxony, Germany in three consecutive seasons between 2015 and 2018: Analysis of molecular and clinical features. Vaccine 2019, 37, 6550–6557. [Google Scholar] [CrossRef]
- Petrova, V.N.; Russell, C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018, 16, 47–60. [Google Scholar] [CrossRef]
- Martin, D.; Hönemann, M.; Liebert, U.G. Dynamics of nosocomial parainfluenza virus type 3 and influenza virus infections at a large German University Hospital between 2012 and 2019. Diagn. Microbiol. Infect. Dis. 2021, 99, 115244. [Google Scholar] [CrossRef]
- Eden, J.-S.; Sikazwe, C.; Xie, R.; Deng, Y.-M.; Sullivan, S.G.; Michie, A.; Levy, A.; Cutmore, E.; Blyth, C.C.; Britton, P.N.; et al. Off-season RSV epidemics in Australia after easing of COVID-19 restrictions. Nat. Commun. 2022, 13, 2884. [Google Scholar] [CrossRef]
- Foley, D.A.; Phuong, L.K.; Peplinski, J.; Lim, S.M.; Lee, W.H.; Farhat, A.; Minney-Smith, C.A.; Martin, A.C.; Mace, A.O.; Sikazwe, C.T.; et al. Examining the interseasonal resurgence of respiratory syncytial virus in Western Australia. Arch. Dis. Child. 2022, 107, e7. [Google Scholar] [CrossRef]
- Halabi, K.C.; Saiman, L.; Zachariah, P. The Epidemiology of Respiratory Syncytial Virus in New York City during the Coronavirus Disease-2019 Pandemic Compared with Previous Years. J. Pediatr. 2022, 242, 242–244.e1. [Google Scholar] [CrossRef]
- Kuitunen, I.; Artama, M.; Haapanen, M.; Renko, M. Respiratory virus circulation in children after relaxation of COVID-19 restrictions in fall 2021-A nationwide register study in Finland. J. Med. Virol. 2022, 94, 4528–4532. [Google Scholar] [CrossRef]
- Kuitunen, I.; Artama, M.; Haapanen, M.; Renko, M. Record high parainfluenza season in children after relaxation of COVID-19 restrictions in fall 2021-A nationwide register study in Finland. Influenza Other Respir. Viruses 2022, 16, 613–616. [Google Scholar] [CrossRef] [PubMed]
- Dhanasekaran, V.; Sullivan, S.; Edwards, K.M.; Xie, R.; Khvorov, A.; Valkenburg, S.A.; Cowling, B.J.; Barr, I.G. Human seasonal influenza under COVID-19 and the potential consequences of influenza lineage elimination. Nat. Commun. 2022, 13, 1721. [Google Scholar] [CrossRef] [PubMed]
- Peret, T.C.; Hall, C.B.; Hammond, G.W.; Piedra, P.A.; Storch, G.A.; Sullender, W.M.; Tsou, C.; Anderson, L.J. Circulation patterns of group A and B human respiratory syncytial virus genotypes in 5 communities in North America. J. Infect. Dis. 2000, 181, 1891–1896. [Google Scholar] [CrossRef] [Green Version]
- Houspie, L.; Lemey, P.; Keyaerts, E.; Reijmen, E.; Vergote, V.; Vankeerberghen, A.; Vaeyens, F.; de Beenhouwer, H.; van Ranst, M. Circulation of HRSV in Belgium: From multiple genotype circulation to prolonged circulation of predominant genotypes. PLoS ONE 2013, 8, e60416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, G.; Zhu, R.; Qian, Y.; Deng, J.; Zhao, L.; Sun, Y.; Wang, F. Genetic variation in attachment glycoprotein genes of human respiratory syncytial virus subgroups a and B in children in recent five consecutive years. PLoS ONE 2013, 8, e75020. [Google Scholar] [CrossRef]
- Tabatabai, J.; Thielen, A.; Lehners, N.; Daeumer, M.; Schnitzler, P. Respiratory syncytial virus A in haematological patients with prolonged shedding: Premature stop codons and deletion of the genotype ON1 72-nucleotide-duplication in the attachment G gene. J. Clin. Virol. 2018, 98, 10–17. [Google Scholar] [CrossRef]
- Botosso, V.F.; Zanotto, P.M.d.A.; Ueda, M.; Arruda, E.; Gilio, A.E.; Vieira, S.E.; Stewien, K.E.; Peret, T.C.T.; Jamal, L.F.; Pardini, M.I.d.M.C.; et al. Positive selection results in frequent reversible amino acid replacements in the G protein gene of human respiratory syncytial virus. PLoS Pathog. 2009, 5, e1000254. [Google Scholar] [CrossRef]
- Eshaghi, A.; Duvvuri, V.R.; Lai, R.; Nadarajah, J.T.; Li, A.; Patel, S.N.; Low, D.E.; Gubbay, J.B. Genetic variability of human respiratory syncytial virus A strains circulating in Ontario: A novel genotype with a 72 nucleotide G gene duplication. PLoS ONE 2012, 7, e32807. [Google Scholar] [CrossRef] [Green Version]
- Trento, A.; Galiano, M.; Videla, C.; Carballal, G.; García-Barreno, B.; Melero, J.A.; Palomo, C. Major changes in the G protein of human respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. J. Gen. Virol. 2003, 84, 3115–3120. [Google Scholar] [CrossRef]
- Reicherz, F.; Xu, R.Y.; Abu-Raya, B.; Majdoubi, A.; Michalski, C.; Golding, L.; Stojic, A.; Vineta, M.; Granoski, M.; Cieslak, Z.; et al. Waning Immunity Against Respiratory Syncytial Virus During the Coronavirus Disease 2019 Pandemic. J. Infect. Dis. 2022, 226, 2064–2068. [Google Scholar] [CrossRef]
- Pons-Salort, M.; Grassly, N.C. Serotype-specific immunity explains the incidence of diseases caused by human enteroviruses. Science 2018, 361, 800–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barclay, W.S.; al-Nakib, W.; Higgins, P.G.; Tyrrell, D.A. The time course of the humoral immune response to rhinovirus infection. Epidemiol. Infect. 1989, 103, 659–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stensballe, L.G.; Ravn, H.; Kristensen, K.; Meakins, T.; Aaby, P.; Simoes, E.A.F. Seasonal variation of maternally derived respiratory syncytial virus antibodies and association with infant hospitalizations for respiratory syncytial virus. J. Pediatr. 2009, 154, 296–298. [Google Scholar] [CrossRef] [PubMed]
- Falsey, A.R.; Cameron, A.; Branche, A.R.; Walsh, E.E. Perturbations in Respiratory Syncytial Virus Activity during the SARS-CoV-2 Pandemic. J. Infect. Dis. 2022, 227, 83–86. [Google Scholar] [CrossRef]
- Agha, R.; Avner, J.R. Delayed Seasonal RSV Surge Observed During the COVID-19 Pandemic. Pediatrics 2021, 148, 2021052089. [Google Scholar] [CrossRef]
- Foley, D.A.; Yeoh, D.K.; Minney-Smith, C.A.; Martin, A.C.; Mace, A.O.; Sikazwe, C.T.; Le, H.; Levy, A.; Moore, H.C.; Blyth, C.C. The Interseasonal Resurgence of Respiratory Syncytial Virus in Australian Children Following the Reduction of Coronavirus Disease 2019-Related Public Health Measures. Clin. Infect. Dis. 2021, 73, e2829–e2830. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Blau, D.M.; Caballero, M.T.; Feikin, D.R.; Gill, C.J.; Madhi, S.A.; Omer, S.B.; Simões, E.A.F.; Campbell, H.; et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in children younger than 5 years in 2019: A systematic analysis. Lancet 2022, 399, 2047–2064. [Google Scholar] [CrossRef]
- Ikäheimo, T.M. Cardiovascular diseases, cold exposure and exercise. Temperature 2018, 5, 123–146. [Google Scholar] [CrossRef]
- Lu, P.; Xia, G.; Zhao, Q.; Xu, R.; Li, S.; Guo, Y. Temporal trends of the association between ambient temperature and hospitalisations for cardiovascular diseases in Queensland, Australia from 1995 to 2016: A time-stratified case-crossover study. PLoS Med. 2020, 17, e1003176. [Google Scholar] [CrossRef]
- Hartmann, K.; Liese, J.G.; Kemmling, D.; Prifert, C.; Weißbrich, B.; Thilakarathne, P.; Diels, J.; Weber, K.; Streng, A. Clinical Burden of Respiratory Syncytial Virus in Hospitalized Children Aged ≤5 Years (INSPIRE Study). J. Infect. Dis. 2022, 226, 386–395. [Google Scholar] [CrossRef]
- Meyer, M.; Ruebsteck, E.; Eifinger, F.; Klein, F.; Oberthuer, A.; van Koningsbruggen-Rietschel, S.; Huenseler, C.; Weber, L.T. Morbidity of Respiratory Syncytial Virus (RSV) Infections: RSV Compared With Severe Acute Respiratory Syndrome Coronavirus 2 Infections in Children Aged 0-4 Years in Cologne, Germany. J. Infect. Dis. 2022, 226, 2050–2053. [Google Scholar] [CrossRef] [PubMed]
- Perniciaro, S.; van der Linden, M.; Weinberger, D.M. Reemergence of Invasive Pneumococcal Disease in Germany During the Spring and Summer of 2021. Clin. Infect. Dis. 2022, 75, 1149–1153. [Google Scholar] [CrossRef] [PubMed]
- Makrinioti, H.; Hasegawa, K.; Lakoumentas, J.; Xepapadaki, P.; Tsolia, M.; Castro-Rodriguez, J.A.; Feleszko, W.; Jartti, T.; Johnston, S.L.; Bush, A.; et al. The role of respiratory syncytial virus- and rhinovirus-induced bronchiolitis in recurrent wheeze and asthma-A systematic review and meta-analysis. Pediatr. Allergy Immunol. 2022, 33, e13741. [Google Scholar] [CrossRef] [PubMed]
- Bergroth, E.; Aakula, M.; Elenius, V.; Remes, S.; Piippo-Savolainen, E.; Korppi, M.; Piedra, P.A.; Bochkov, Y.A.; Gern, J.E.; Camargo, C.A.; et al. Rhinovirus Type in Severe Bronchiolitis and the Development of Asthma. J. Allergy Clin. Immunol. Pract. 2020, 8, 588–595.e4. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; McLellan, J.S.; Kallewaard, N.L.; Ulbrandt, N.D.; Palaszynski, S.; Zhang, J.; Moldt, B.; Khan, A.; Svabek, C.; McAuliffe, J.M.; et al. A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants. Sci. Transl. Med. 2017, 9, eaaj1928. [Google Scholar] [CrossRef] [PubMed]
- Gunatilaka, A.; Giles, M.L. Maternal RSV vaccine development. Where to from here? Hum. Vaccin. Immunother. 2021, 17, 4542–4548. [Google Scholar] [CrossRef]




| Season 2017/2018 | Season 2018/2019 | Season 2019/2020 | Season 2021/2022 | Total | p-Value | |||
|---|---|---|---|---|---|---|---|---|
| age group | ||||||||
| <18 | [% (n/total)] | 71.4 (152/213) | 68.3 (151/221) | 69.5 (89/128) | 88.9 (208/234) | 75.4 (600/796) |  | <0.001 |
| ≥18 | [% (n/total)] | 28.6 (61/213) | 31.7 (70/221) | 30.5 (39/128) | 11.1 (26/234) | 24.6 (196/796) | ||
| gender | ||||||||
| male | [% (n/total)] | 62.9 (134/213) | 50.7 (112/221) | 62.5 (80/128) | 54.7 (128/234) | 57.0 (454/796) |  | n.s. |
| female | [% (n/total)] | 37.1 (79/213) | 49.3 (109/221) | 37.5 (48/128) | 45.3 (106/234) | 43.0 (342/796) | ||
| RV species | ||||||||
| RSV-A | [% (n/total)] | 24.9 (53/213) | 59.7 (132/221) | 77.3 (99/128) | 56.0 (131/234) | 52.1 (415/796) |  | <0.001 |
| RSV-B | [% (n/total)] | 75.1 (160/213) | 38.9 (86/221) | 21.9 (28/128) | 41.9 (98/234) | 46.9 (373/796) | ||
| mixed | [% (n/total)] | 0 (0/213) | 1.4 (3/221) | 0.8 (1/128) | 2.1 (5/234) | 1.1 (9/796) |
| Pre-Pandemic | 2021/2022 | Total | p-Value | |||
|---|---|---|---|---|---|---|
| study population | ||||||
| female | [% (n/total)] | 41.6 (163/392) | 45.6 (95/208) | 43.0 (258/600) |  | n.s. |
| male | [% (n/total)] | 58.4 (229/392) | 54.3 (113/208) | 57.0 (342/600) | ||
| age [years] | [mean ± SD] | 1.2 ± 2.3 | 1.4 ± 1.98 | 1.3 ± 2.20 | n.s. | |
| gestational week at birth | [mean ± SD] | 38.1 ± 3.64 | 38 ± 3.68 | 38.1 ± 3.66 | n.s. | |
| preterm birth | [% (n/total)] | 19.2 (61/318) | 20.3 (36/177) | 19.6 (97/495) | n.s. | |
| siblings | [% (n/total)] | 62.1 (231/372) | 74.1 (143/193) | 66.2 (374/565) | 0.004 | |
| twins | [% (n/total)] | 4.6 (17/373) | 3.6 (7/193) | 4.2 (24/566) | n.s. | |
| caesarean section | [% (n/total)] | 16.0 (60/375) | 17.0 (32/188) | 16.3 (92/563) | n.s. | |
| RSV prophylaxis | [% (n/total)] | 3.1 (12/392) | 3.4 (7/208) | 3.2 (19/600) | n.s. | |
| inpatients | [% (n/total)] | 97.2 (381/392) | 93.3 (194/208) | 95.8 (575/600) |  | 0.022 |
| outpatients | [% (n/total)] | 2.8 (11/392) | 6.7 (14/208) | 4.2 (25/600) | ||
| length of hospital stay [days] | [mean ± SD] | 6.6 ± 7.99 | 6.1 ± 8.19 | 6.5 ± 8.06 | n.s. | |
| comorbidities and risk factors | ||||||
| asthma | [% (n/total)] | 1.0 (4/392) | 1.0 (2/207) | 1.5 (9/599) | n.s. | |
| COPD | [% (n/total)] | 0 (0/392) | 0 (0/207) | 0 (0/599) | n.s. | |
| lung transplant | [% (n/total)] | 0 (0/392) | 0 (0/207) | 0 (0/599) | n.s. | |
| chronic kidney failure | [% (n/total)] | 0.3 (1/392) | 0 (0/207) | 0.2 (1/599) | n.s. | |
| cardiac failure | [% (n/total)] | 1.8 (7/392) | 1.9 (4/207) | 1.8 (11/599) | n.s. | |
| arterial hypertension | [% (n/total)] | 0.5 (2/392) | 1.0 (2/207) | 0.7 (4/599) | n.s. | |
| cardiovascular diseases | [% (n/total)] | 0.5 (2/392) | 1.4 (3/207) | 0.8 (5/599) | n.s. | |
| diabetes | [% (n/total)] | 0.8 (3/392) | 0.5 (1/207) | 0.7 (4/599) | n.s. | |
| malignancy | [% (n/total)] | 2.0 (8/392) | 1.4 (3/207) | 1.8 (11/599) | n.s. | |
| immunosuppression | [% (n/total)] | 1.8 (7/392) | 2.4 (5/207) | 2.0 (12/599) | n.s. | |
| clinical presentation and features | ||||||
| nosocomial | [% (n/total)] | 0.2 (1/391) | 0.4 (1/207) | 0.3 (2/598) | n.s. | |
| symptomatic | [% (n/total)] | 97.4 (380/390) | 94.2 (195/207) | 96.3 (575/597) | 0.046 | |
| fever | [% (n/total)] | 40.3 (157/389) | 29.1 (60/206) | 36.4 (217/595) | 0.007 | |
| URTI | [% (n/total)] | 74.4 (268/360) | 76.0 (140/184) | 75.0 (408/544) | n.s. | |
| pharyngitis | [% (n/total)] | 68.2 (237/347) | 72.1 (127/176) | 69.5 (364/523) | n.s. | |
| LRTI | [% (n/total)] | 82.0 (321/391) | 83.0 (171/206) | 82.4 (492/597) | n.s. | |
| bronchiolitis/bronchitis | [% (n/total)] | 65.2 (255/391) | 70.8 (146/206) | 67.1 (401/597) | n.s. | |
| pneumonia | [% (n/total)] | 21.4 (84/391) | 14.4 (30/207) | 19.0 (114/598) | 0.038 | |
| respiratory failure | [% (n/total)] | 57.2 (224/391) | 61.1 (126/206) | 58.6 (350/597) | n.s. | |
| ICU stay | [% (n/total)] | 12.0 (47/391) | 7.6 (16/208) | 10.5 (63/599) | n.s. | |
| length of ICU stay [days] | [mean ± SD] | 9.3 ± 11.14 | 7.4 ± 6.39 | 8.8 ± 10.06 | n.s. | |
| assisted ventilation | [% (n/total)] | 2.0 (8/390) | 1.4 (3/208) | 1.8 (11/599) | n.s. | |
| oxygen supply | [% (n/total)] | 59.7 (233/390) | 57.9 (120/207) | 59.1 (353/597) | n.s. | |
| inhalation | [% (n/total)] | 84.1 (328/390) | 80.6 (167/207) | 82.9 (495/597) | n.s. | |
| adrenalin | [% (n/total)] | 7.2 (28/385) | 5.5 (11/200) | 6.6 (39/585) | n.s. | |
| salbutamol | [% (n/total)] | 70.5 (275/390) | 64.7 (134/207) | 69.9 (409/585) | n.s. | |
| ipratropium bromide bromide | [% (n/total)] | 17.6 (68/385) | 17.5 (35/200) | 17.6 (103/585) | n.s. | |
| budesonide | [% (n/total)] | 15.5 (60/385) | 8.5 (17/200) | 13.1 (77/585) | 0.016 | |
| methylxanthine administration | [% (n/total)] | 0.7 (3/390) | 0.4 (1/207) | 0.6 (4/597) | n.s. | |
| syst. prednisolone administration | [% (n/total)] | 34.8 (136/390) | 31.8 (66/207) | 33.8 (202/597) | n.s. | |
| co-infections | [% (n/total)] | 38.7 (152/392) | 25.9 (54/208) | 34.3 (206/600) | 0.001 | |
| bacterial | [% (n/total)] | 9.9 (39/392) | 4.8 (10/208) | 8.1 (49/600) |  | 0.006 |
| viral | [% (n/total)] | 25.0 (98/392) | 19.2 (40/208) | 23.0 (138/600) | ||
| fungal | [% (n/total)] | 0 (0/392) | 0.4 (1/208) | 0.1 (1/600) | ||
| combined | [% (n/total)] | 3.8 (15/392) | 1.4 (3/208) | 3.0 (18/600) | ||
| in-hospital mortality | [% (n/total)] | 0 (0/392) | 0 (0/208) | 0 (0/600) | n.s. |
| Pre-Pandemic | 2021/2022 | Total | p-Value | |||
|---|---|---|---|---|---|---|
| study population | ||||||
| female | [% (n/total)] | 42.9 (73/170) | 42.3 (11/26) | 42.8 (84/196) |  | n.s. |
| male | [% (n/total)] | 57.1 (97/170) | 57.6 (15/26) | 57.1 (112/196) | ||
| age [years] | [mean ± SD] | 64.7 ± 16.98 | 47.07 ± 17.82 | 62.4 ± 18.10 | <0.001 | |
| inpatients | [% (n/total)] | 79.4 (135/170) | 76.0 (19/25) | 78.9 (154/195) |  | n.s. |
| outpatients | [% (n/total)] | 20.6 (35/170) | 24.0 (6/25) | 21.0 (41/195) | ||
| length of hospital stay [days] | [mean ± SD] | 18.5 ± 15.62 | 17 ± 22.71 | 18.3 ± 16.54 | n.s. | |
| comorbidities and risk factors | ||||||
| asthma | [% (n/total)] | 5.3 (9/168) | 8.3 (2/24) | 5.7 (11/192) | n.s. | |
| COPD | [% (n/total)] | 17.9 (30/168) | 12.5 (3/24) | 17.1 (33/192) | n.s. | |
| lung transplant | [% (n/total)] | 4.7 (8/170) | 3.8 (1/26) | 4.5 (9/196) | n.s. | |
| chronic kidney failure | [% (n/total)] | 39.9 (67/168) | 20.8 (5/24) | 37.5 (72/192) | n.s. | |
| cardiac failure | [% (n/total)] | 19.0 (32/168) | 8.3 (2/24) | 17.7 (34/192) | n.s. | |
| arterial hypertension | [% (n/total)] | 58.9 (99/168) | 37.5 (9/24) | 56.2 (108/192) | 0.048 | |
| cardiovascular diseases | [% (n/total)] | 61.9 (104/168) | 37.5 (9/24) | 58.8 (113/192) | 0.023 | |
| diabetes | [% (n/total)] | 31.5 (53/168) | 29.1 (7/24) | 31.2 (60/192) | n.s. | |
| malignancy | [% (n/total)] | 35.7 (60/168) | 29.1 (7/24) | 34.8 (67/192) | n.s. | |
| immunosuppression | [% (n/total)] | 36.3 (61/168) | 50.0 (12/24) | 38.0 (73/192) | n.s. | |
| clinical presentation and features | ||||||
| nosocomial | [% (n/total)] | 4.7 (8/169) | 12.0 (3/25) | 5.6 (11/194) | n.s. | |
| symptomatic | [% (n/total)] | 78.7 (126/160) | 85.7 (18/21) | 79.5 (144/181) | n.s. | |
| fever | [% (n/total)] | 24.0 (38/158) | 21.0 (4/19) | 23.7 (42/177) | n.s. | |
| URTI | [% (n/total)] | 35.4 (28/79) | 33.3 (4/12) | 35.1 (32/91) | n.s. | |
| pharyngitis | [% (n/total)] | 11.2 (7/62) | 27.2 (3/11) | 13.6 (10/73) | n.s. | |
| LRTI | [% (n/total)] | 49.3 (78/158) | 63.1 (12/19) | 50.8 (90/177) | n.s. | |
| bronchitis | [% (n/total)] | 7.5 (12/158) | 15.7 (3/19) | 8.4 (15/177) | n.s. | |
| pneumonia | [% (n/total)] | 33.3 (53/159) | 42.1 (8/19) | 34.2 (61/178) | n.s. | |
| respiratory failure | [% (n/total)] | 28.9 (46/159) | 36.8 (7/19) | 29.7 (53/178) | n.s. | |
| ICU stay | [% (n/total)] | 25.8 (44/170) | 19.2 (5/26) | 25 (49/196) | n.s. | |
| length of ICU stay [days] | [mean ± SD] | 8.2 ± 11.91 | 3.5 ± 3.72 | 7.6 ± 11.3 | n.s. | |
| assisted ventilation | [% (n/total)] | 8.2 (14/170) | 7.6 (2/26) | 8.1 (16/196) | n.s. | |
| oxygen supply | [% (n/total)] | 23.8 (40/168) | 20.0 (5/25) | 23.3 (45/193) | n.s. | |
| inhalation | [% (n/total)] | 18.4 (31/168) | 16.6 (4/24) | 18.2 (35/192) | n.s. | |
| adrenalin | [% (n/total)] | 0 (0/166) | 0 (0/24) | 0 (0/190) | n.s. | |
| salbutamol | [% (n/total)] | 10.7 (18/168) | 4.1 (1/24) | 10.0 (19/190) | n.s. | |
| ipratropium bromide bromide | [% (n/total)] | 9.0 (15/166) | 4.1 (1/24) | 8.3 (16/190) | n.s. | |
| budesonide | [% (n/total)] | 0.6 (1/166) | 4.1 (1/24) | 1.0 (2/190) | n.s. | |
| syst. prednisolone administration | [% (n/total)] | 14.8 (25/168) | 12.5 (3/24) | 14.5 (28/192) | n.s. | |
| co-infection | [% (n/total)] | 21.1 (36/170) | 19.2 (5/26) | 20.9 (41/196) | n.s. | |
| bacterial | [% (n/total)] | 5.8 (10/170) | 7.6 (2/26) | 6.1 (12/196) |  | n.s. |
| viral | [% (n/total)] | 11.1 (19/170) | 11.5 (3/26) | 11.2 (22/196) | ||
| fungal | [% (n/total)] | 1.1 (2/170) | 0 (0/26) | 1.0 (2/196) | ||
| combined | [% (n/total)] | 2.9 (5/170) | 0 (0/26) | 2.5 (5/196) | ||
| in-hospital mortality | [% (n/total)] | 6.4 (11/170) | 3.8 (1/26) | 6.1 (12/196) | n.s. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hönemann, M.; Thiem, S.; Bergs, S.; Berthold, T.; Propach, C.; Siekmeyer, M.; Frille, A.; Wallborn, T.; Maier, M.; Pietsch, C. In-Depth Analysis of the Re-Emergence of Respiratory Syncytial Virus at a Tertiary Care Hospital in Germany in the Summer of 2021 after the Alleviation of Non-Pharmaceutical Interventions Due to the SARS-CoV-2 Pandemic. Viruses 2023, 15, 877. https://doi.org/10.3390/v15040877
Hönemann M, Thiem S, Bergs S, Berthold T, Propach C, Siekmeyer M, Frille A, Wallborn T, Maier M, Pietsch C. In-Depth Analysis of the Re-Emergence of Respiratory Syncytial Virus at a Tertiary Care Hospital in Germany in the Summer of 2021 after the Alleviation of Non-Pharmaceutical Interventions Due to the SARS-CoV-2 Pandemic. Viruses. 2023; 15(4):877. https://doi.org/10.3390/v15040877
Chicago/Turabian StyleHönemann, Mario, Stephanie Thiem, Sandra Bergs, Tom Berthold, Christian Propach, Manuela Siekmeyer, Armin Frille, Tillmann Wallborn, Melanie Maier, and Corinna Pietsch. 2023. "In-Depth Analysis of the Re-Emergence of Respiratory Syncytial Virus at a Tertiary Care Hospital in Germany in the Summer of 2021 after the Alleviation of Non-Pharmaceutical Interventions Due to the SARS-CoV-2 Pandemic" Viruses 15, no. 4: 877. https://doi.org/10.3390/v15040877
APA StyleHönemann, M., Thiem, S., Bergs, S., Berthold, T., Propach, C., Siekmeyer, M., Frille, A., Wallborn, T., Maier, M., & Pietsch, C. (2023). In-Depth Analysis of the Re-Emergence of Respiratory Syncytial Virus at a Tertiary Care Hospital in Germany in the Summer of 2021 after the Alleviation of Non-Pharmaceutical Interventions Due to the SARS-CoV-2 Pandemic. Viruses, 15(4), 877. https://doi.org/10.3390/v15040877





