cmv1-Mediated Resistance to CMV in Melon Can Be Overcome by Mixed Infections with Potyviruses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant, Insects and Virus Materials
2.2. Viral Inoculations
2.3. Sampling and RNA Extraction
2.4. Quantification of CMV Viral Load by RT-qPCR
2.5. Virus Detection by Reverse Transcriptase-PCR
3. Results
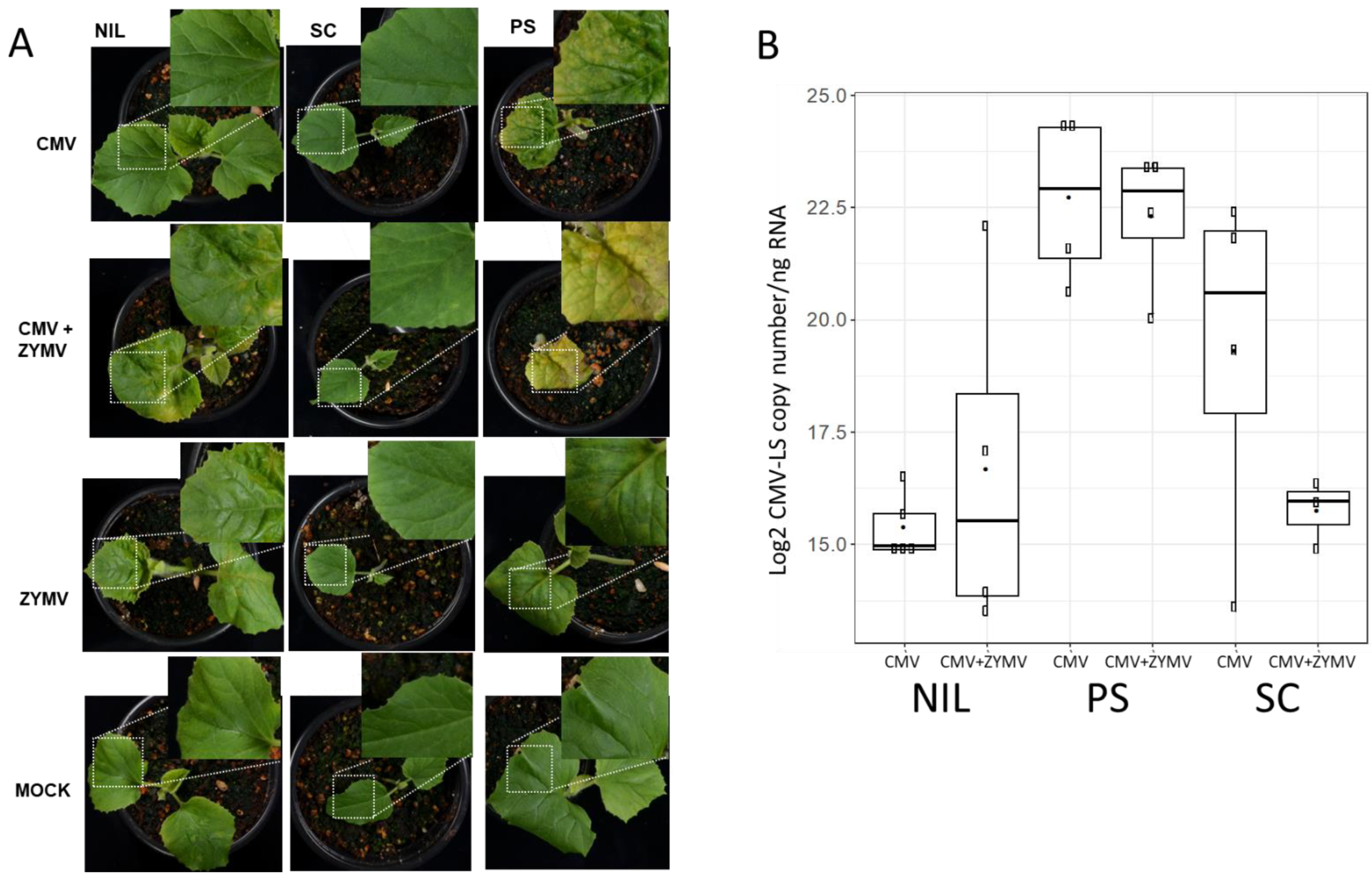
3.1. Mixed Infections Allow CMV-LS to Overcome cmv1-Controlled Resistance without Increasing Its Accumulation in the Inoculated Cotyledons
3.2. cmv1 Resistance Is Compromised in Mixed Infections Initiated by Aphid Inoculation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 2004, 19, 535–544. [Google Scholar] [CrossRef]
- Moreno, A.B.; López-Moya, J.J. When Viruses Play Team Sports: Mixed Infections in Plants. Phytopathology 2020, 110, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Alcaide, C.; Rabadán, M.P.; Moreno-Pérez, M.G.; Gómez, P. Implications of mixed viral infections on plant disease ecology and evolution. Adv. Virus Res. 2020, 106, 145–169. [Google Scholar]
- Edwardson, J.R.; Christie, R.G. Cucumoviruses. In CRC Handbook of Viruses Infecting Legumes; Edwardson, J.R., Christie, R.G., Eds.; CRC Press: Boca Raton, FL, USA, 1991; pp. 293–319. [Google Scholar]
- Palukaitis, P.; Roossinck, M.J.; Dietzgen, R.G.; Francki, R.I.B. Cucumber mosaic virus. Adv. Virus Res. 1992, 41, 281–348. [Google Scholar]
- Perry, K.L.; Zhang, L.; Shintaku, M.H.; Palukaitis, P. Mapping Determinants in Cucumber mosaic virus for Transmission by Aphis gossypii. Virology 1994, 205, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Karchi, Z.; Cohen, S.; Govers, A. Inheritance of resistance to Cucumber Mosaic virus in melons. Phytopathology 1975, 65, 479–481. [Google Scholar] [CrossRef]
- Essafi, A.; Diaz-Pendon, J.A.; Moriones, E.; Monforte, A.J.; Garcia-Mas, J.; Martin-Hernandez, A.M. Dissection of the oligogenic resistance to Cucumber mosaic virus in the melon accession PI 161375. Theor. Appl. Genet. 2009, 118, 275–284. [Google Scholar] [CrossRef]
- Guiu-Aragonés, C.; Monforte, A.J.; Saladié, M.; Corrêa, R.X.; Garcia-Mas, J.; Martín-Hernández, A.M. The complex resistance to Cucumber mosaic cucumovirus (CMV) in the melon accession PI 161375 is governed by one gene and at least two quantitative trait loci. Mol. Breed. 2014, 34, 351–362. [Google Scholar] [CrossRef]
- Guiu-Aragonés, C.; Sánchez-Pina, M.A.; Díaz-Pendón, J.; Peña, E.J.; Heinlein, M.; Martín-Hernández, A.M. cmv1 is a gate for Cucumber mosaic virus transport from bundle sheath cells to phloem in melon. Mol. Plant Pathol. 2016, 17, 973–984. [Google Scholar] [CrossRef]
- Guiu-Aragonés, C.; Díaz-Pendón, J.A.; Martín-Hernández, A.M. Four sequence positions of the movement protein of Cucumber mosaic virus determine the virulence against cmv1-mediated resistance in melon. Mol. Plant Pathol. 2015, 16, 675–684. [Google Scholar] [CrossRef]
- Pascual, L.; Yan, J.; Pujol, M.; Monforte, A.J.; Picó, B.; Martín-Hernández, A.M. CmVPS41 Is a General Gatekeeper for Resistance to Cucumber mosaic virus Phloem Entry in Melon. Front. Plant Sci. 2019, 10, 1219. [Google Scholar] [CrossRef]
- Giner, A.; Pascual, L.; Bourgeois, M.; Gyetvai, G.; Rios, P.; Picó, B.; Troadec, C.; Bendahmane, A.; Garcia-Mas, J.; Martín-Hernández, A.M. A mutation in the melon Vacuolar Protein Sorting 41 prevents systemic infection of Cucumber mosaic virus. Sci. Rep. 2017, 7, 10471. [Google Scholar] [CrossRef]
- Balderhaar, H.J.; Ungermann, C. CORVET and HOPS tethering complexes—Coordinators of endosome and lysosome fusion. J. Cell Sci. 2013, 126, 1307–1316. [Google Scholar] [CrossRef] [PubMed]
- Real, N.; Villar, I.; Serrano, I.; Guiu-Aragonés, C.; Martín-Hernández, A.M. Mutations in CmVPS41 controlling resistance to Cucumber mosaic virus display specific subcellular localization. Plant Physiol. 2023, 191, 1596–1611. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Liu, J.; Zhong, S.; Gu, H.; Qu, L.J. AtVPS41-mediated endocytic pathway is essential for pollen tube-stigma interaction in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 6307–6312. [Google Scholar] [CrossRef]
- Jiang, D.; He, Y.; Zhou, X.; Cao, Z.; Pang, L.; Zhong, S.; Jiang, L.; Li, R. Arabidopsis HOPS subunit VPS41 carries out plant-specific roles in vacuolar transport and vegetative growth. Plant Physiol. 2022, 189, 1416–1434. [Google Scholar] [CrossRef]
- Wang, Y.; Gaba, V.; Yang, J.; Palukaitis, P.; Gal-On, A. Characterization of Synergy Between Cucumber mosaic virus and Potyviruses in Cucurbit Hosts. Phytopathology 2002, 92, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, M.; Koizumi, E.; Noguchi, M.; Sueda, K.; Shimura, H.; Ishikawa, N.; Matsuura, H.; Ohshima, K.; Natsuaki, T.; Kuwata, S.; et al. Infection dynamics in viral spread and interference under the synergism between Cucumber mosaic virus and Turnip mosaic virus. Mol. Plant Microbe Interact. 2012, 25, 18–27. [Google Scholar] [CrossRef]
- Choi, S.K.; Yoon, J.Y.; Ryu, K.H.; Choi, J.K.; Palukaitis, P.; Park, W.M. Systemic movement of a movement-deficient strain of Cucumber mosaic virus in zucchini squash is facilitated by a cucurbit-infecting potyvirus. J. Gen. Virol. 2002, 83 Pt 12, 3173–3178. [Google Scholar] [CrossRef]
- Fukuzawa, N.; Itchoda, N.; Ishihara, T.; Goto, K.; Masuta, C.; Matsumura, T. HC-Pro, a potyvirus RNA silencing suppressor, cancels cycling of Cucumber mosaic virus in Nicotiana benthamiana plants. Virus Genes 2010, 40, 440–446. [Google Scholar] [CrossRef]
- Wang, Y.; Lee, K.; Gaba, V.; Wong, S.; Palukaitis, P.; Gal-On, A. Breakage of resistance to Cucumber mosaic virus by co-infection with Zucchini yellow mosaic virus: Enhancement of CMV accumulation independent of symptom expression. Arch. Virol. 2004, 149, 379–396. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, T.; Nobuhara, S.; Nishimura, M.; Ryang, B.S.; Naoe, M.; Matsumoto, T.; Kosaka, Y.; Ohki, S.T. The entry of cucumber mosaic virus into cucumber xylem is facilitated by co-infection with zucchini yellow mosaic virus. Arch. Virol. 2016, 161, 2683–2692. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Handa, K.; Palukaitis, P. Mapping local and systemic symptom determinants of Cucumber mosaic cucumovirus in tobacco. J. Gen. Virol. 1994, 75, 3185–3191. [Google Scholar] [CrossRef]
- Gal-On, A.; Meiri, E.; Huet, H.; Hua, W.J.; Raccah, B.; Gaba, V. Particle bombardment drastically increases the infectivity of cloned DNA of zucchini yellow mosaic potyvirus. J. Gen. Virol. 1995, 76 Pt 12, 3223–3227. [Google Scholar] [CrossRef] [PubMed]
- Desbiez, C.; Chandeysson, C.; Lecoq, H.; Moury, B. A simple, rapid and efficient way to obtain infectious clones of potyviruses. J. Virol. Methods 2012, 183, 94–97. [Google Scholar] [CrossRef]
- Domingo-Calap, M.L.; Moreno, A.B.; Díaz Pendón, J.A.; Moreno, A.; Fereres, A.; López-Moya, J.J. Assessing the Impact on Virus Transmission and Insect Vector Behavior of a Viral Mixed Infection in Melon. Phytopathology 2020, 110, 174–186. [Google Scholar] [CrossRef]
- Arazi, T.; Shiboleth, Y.M.; Gal-On, A. A nonviral peptide can replace the entire N terminus of zucchini yellow mosaic potyvirus coat protein and permits viral systemic infection. J. Virol. 2001, 75, 6329–6336. [Google Scholar] [CrossRef]
- Gal-On, A.; Antignus, Y.; Rosner, A.; Raccah, B. Infectious in vitro RNA transcripts derived from cloned cDNA of the cucurbit potyvirus, zucchini yellow mosaic virus. J. Gen. Virol. 1991, 72 Pt 11, 2639–2643. [Google Scholar] [CrossRef]
- Mascia, T.; Cillo, F.; Fanelli, V.; Finetti-Sialer, M.M.; De Stradis, A.; Palukaitis, P.; Gallitelli, D. Characterization of the interactions between Cucumber mosaic virus and Potato virus Y in mixed infections in tomato. Mol. Plant Microbe Interact. 2010, 23, 1514–1524. [Google Scholar] [CrossRef]
- Ding, X.; Shintaku, M.H.; Carter, S.A.; Nelson, R.S. Invasion of minor veins of tobacco leaves inoculated with Tobacco mosaic virus mutants defective in phloem-dependent movement. Proc. Natl. Acad. Sci. USA 1996, 93, 11155–11160. [Google Scholar] [CrossRef]
- Goodrick, B.; Kuhn, C.; Hussey, R. Restricted systemic movement of Cowpea chlorotic mottle virus in soybean with nonnecrotic resistance. Phytopathology 1991, 81, 1246–1431. [Google Scholar] [CrossRef]
- Wang, H.-L.; Wang, Y.; Giesman-Cookmeyer, D.; Lommel, S.A.; Lucas, W.J. Mutations in Viral Movement Protein Alter Systemic Infection and Identify an Intercellular Barrier to Entry into the Phloem Long-Distance Transport System. Virology 1998, 245, 75–89. [Google Scholar] [CrossRef]
- Wintermantel, W.; Banerjee, N.; Oliver, J.; Paolillo, D.J.; Zaitlin, M. Cucumber mosaic virus is restricted from entering minor veins in transgenic tobacco exhibiting replicase-mediated resistance. Virology 1997, 231, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Pirone, T.P. Quantity of Virus Required for Aphid Transmission of a Potyvirus. Phytopathology 1988, 78, 104. [Google Scholar] [CrossRef]
- Moury, B.; Fabre, F.; Senoussi, R. Estimation of the number of virus particles transmitted by an insect vector. Proc. Natl. Acad. Sci. USA 2007, 104, 17891–17896. [Google Scholar] [CrossRef]
- Michalakis, Y.; Blanc, S. The Curious Strategy of Multipartite Viruses. Annu. Rev. Virol. 2020, 7, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Leeks, A.; Young, P.G.; Turner, P.E.; Wild, G.; West, S.A. Cheating leads to the evolution of multipartite viruses. PLoS Biol. 2023, 21, e3002092. [Google Scholar] [CrossRef] [PubMed]
- Hipper, C.; Brault, V.; Ziegler-Graff, V.; Revers, F. Viral and cellular factors involved in phloem transport of plant viruses. Front. Plant Sci. 2013, 4, 154. [Google Scholar] [CrossRef] [PubMed]
- Agaoua, A.; Rittener, V.; Troadec, C.; Desbiez, C.; Bendahmane, A.; Moquet, F.; Dogimont, C. A single substitution in Vacuolar protein sorting 4 is responsible for resistance to Watermelon mosaic virus in melon. J. Exp. Bot. 2022, 73, 4008–4021. [Google Scholar] [CrossRef]
- Amano, M.; Mochizuki, A.; Kawagoe, Y.; Iwahori, K.; Niwa, K.; Svoboda, J.; Maeda, T.; Imura, Y. High-resolution mapping of zym, a recessive gene for Zucchini yellow mosaic virus resistance in cucumber. Theor. Appl. Genet. 2013, 126, 2983–2993. [Google Scholar] [CrossRef]
- Palukaitis, P.; Kaplan, I.B. Synergy of Virus Accumulation and Pathology in Transgenic Plants Expressing Viral Sequences; Springer: Berlin/Heidelberg, Germany, 1997; pp. 77–84. [Google Scholar]
- Poolpol, P.; Inouye, T. Enhancement of Cucumber mosaic virus Multiplication by Zucchini Yellow Mosaic Virus in Doubly Infected Cucumber Plants. Jpn. J. Phytopathol. 1986, 52, 22–30. [Google Scholar] [CrossRef]
- Zwart, M.P.; Elena, S.F. Modeling multipartite virus evolution: The genome formula facilitates rapid adaptation to heterogeneous environments(†). Virus Evol. 2020, 6, veaa022. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giordano, A.; Ferriol, I.; López-Moya, J.J.; Martín-Hernández, A.M. cmv1-Mediated Resistance to CMV in Melon Can Be Overcome by Mixed Infections with Potyviruses. Viruses 2023, 15, 1792. https://doi.org/10.3390/v15091792
Giordano A, Ferriol I, López-Moya JJ, Martín-Hernández AM. cmv1-Mediated Resistance to CMV in Melon Can Be Overcome by Mixed Infections with Potyviruses. Viruses. 2023; 15(9):1792. https://doi.org/10.3390/v15091792
Chicago/Turabian StyleGiordano, Andrea, Inmaculada Ferriol, Juan José López-Moya, and Ana Montserrat Martín-Hernández. 2023. "cmv1-Mediated Resistance to CMV in Melon Can Be Overcome by Mixed Infections with Potyviruses" Viruses 15, no. 9: 1792. https://doi.org/10.3390/v15091792





