Non-Polio Enterovirus C Replicate in Both Airway and Intestine Organotypic Cultures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Viruses
2.2. Next-Generation Sequencing
2.3. Primary Fetal Airway Epithelial Cell Isolation
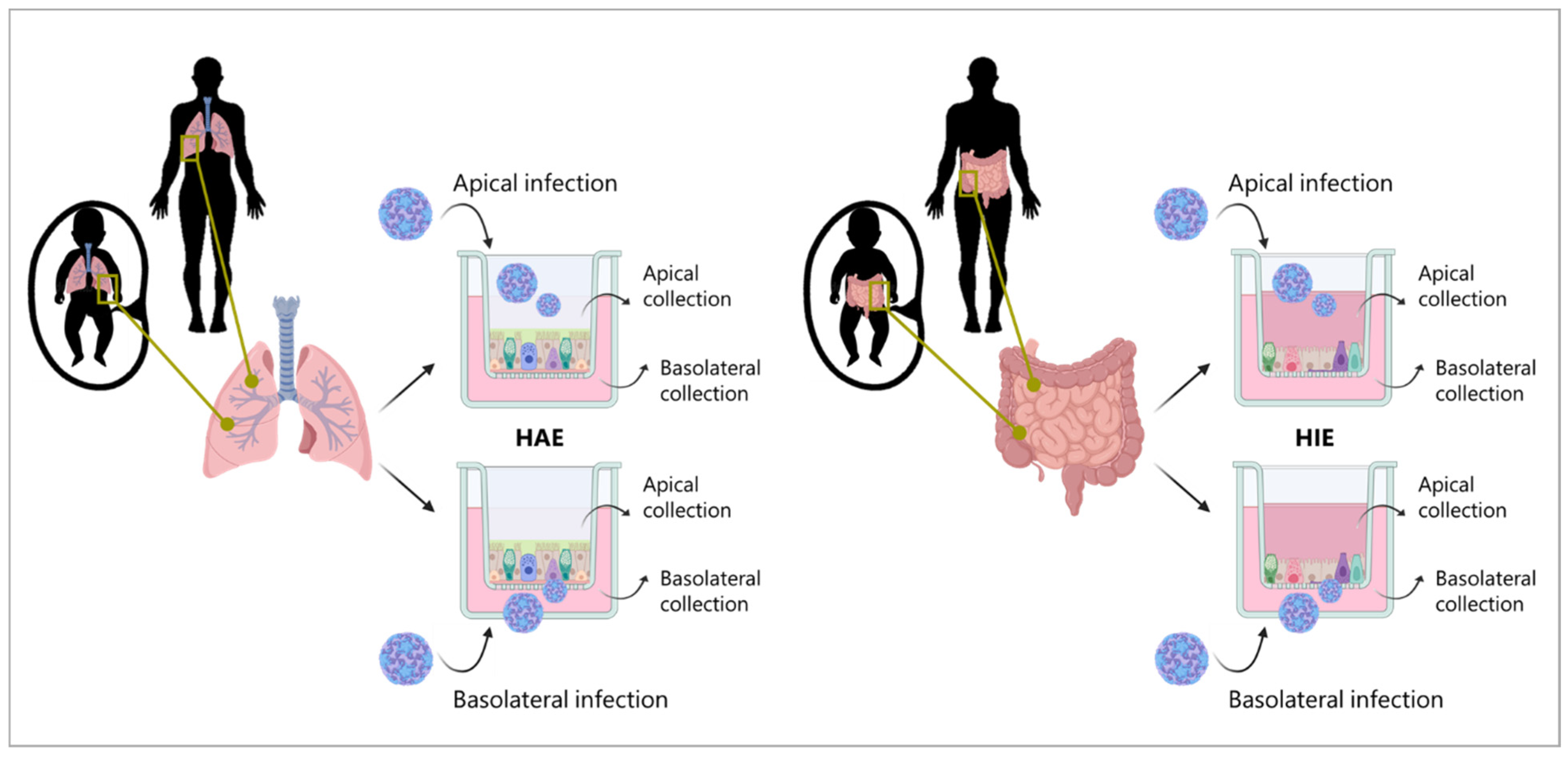
2.4. Fetal and Adult Human Airway Epithelial (HAE) Cultures
2.5. Fetal Small Intestinal Cultures
2.6. Fetal and Adult Human Intestinal Epithelial (HIE) Cultures
2.7. HAE and HIE Infection
2.8. Quantitative RT-qPCR to Determine Viral Copy Numbers
2.9. Infectious Viral Particle Estimation via TCID50
2.10. Immunofluorescence Staining
2.11. Statistical Analysis
3. Results
3.1. Generating Stocks of Clinical Isolates of CVA-13, CVA-20, and EV-C99
3.2. CVA-13, CVA-20, and EV-C99 Replicate in the Human Airway Epithelial (HAE) Model
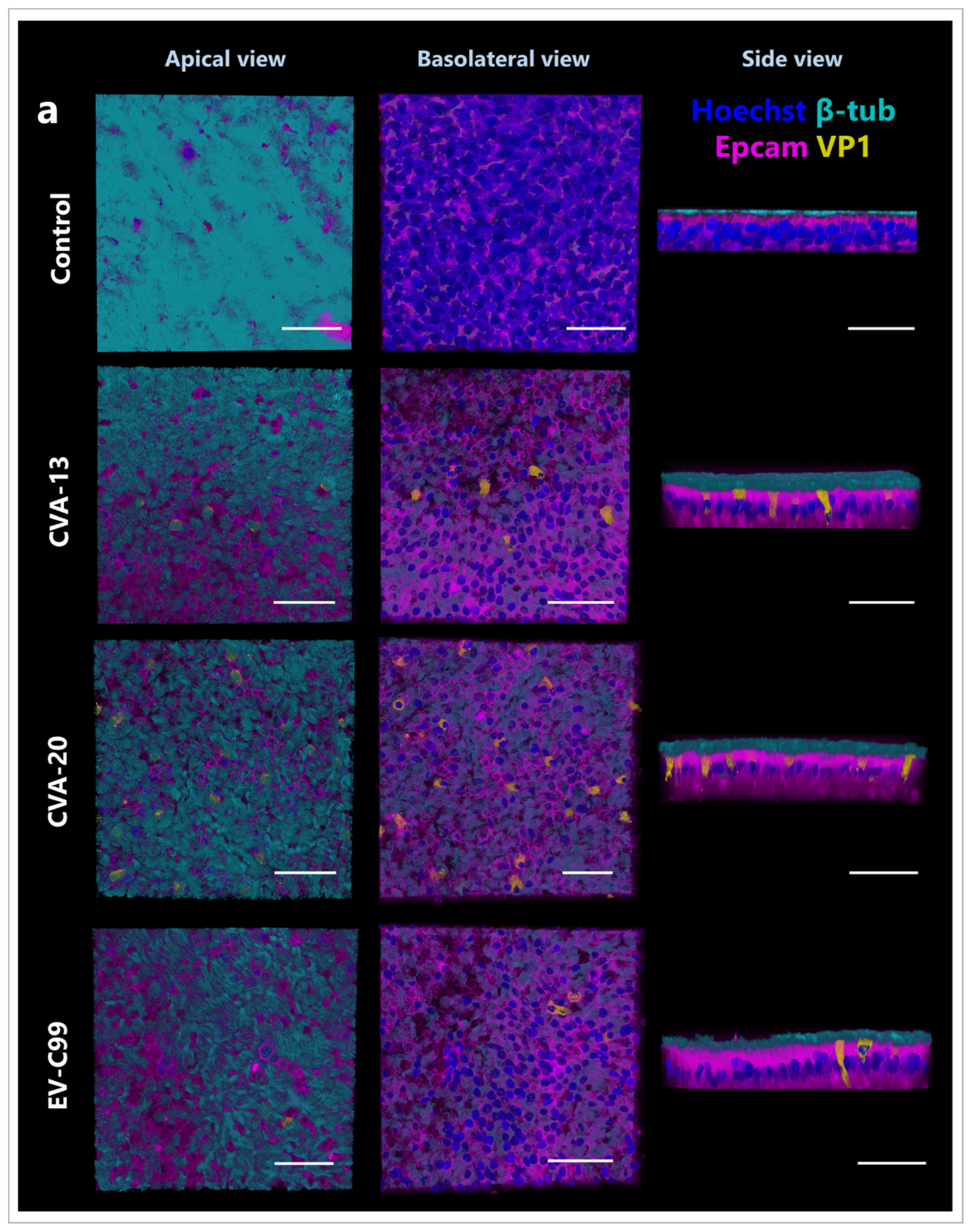
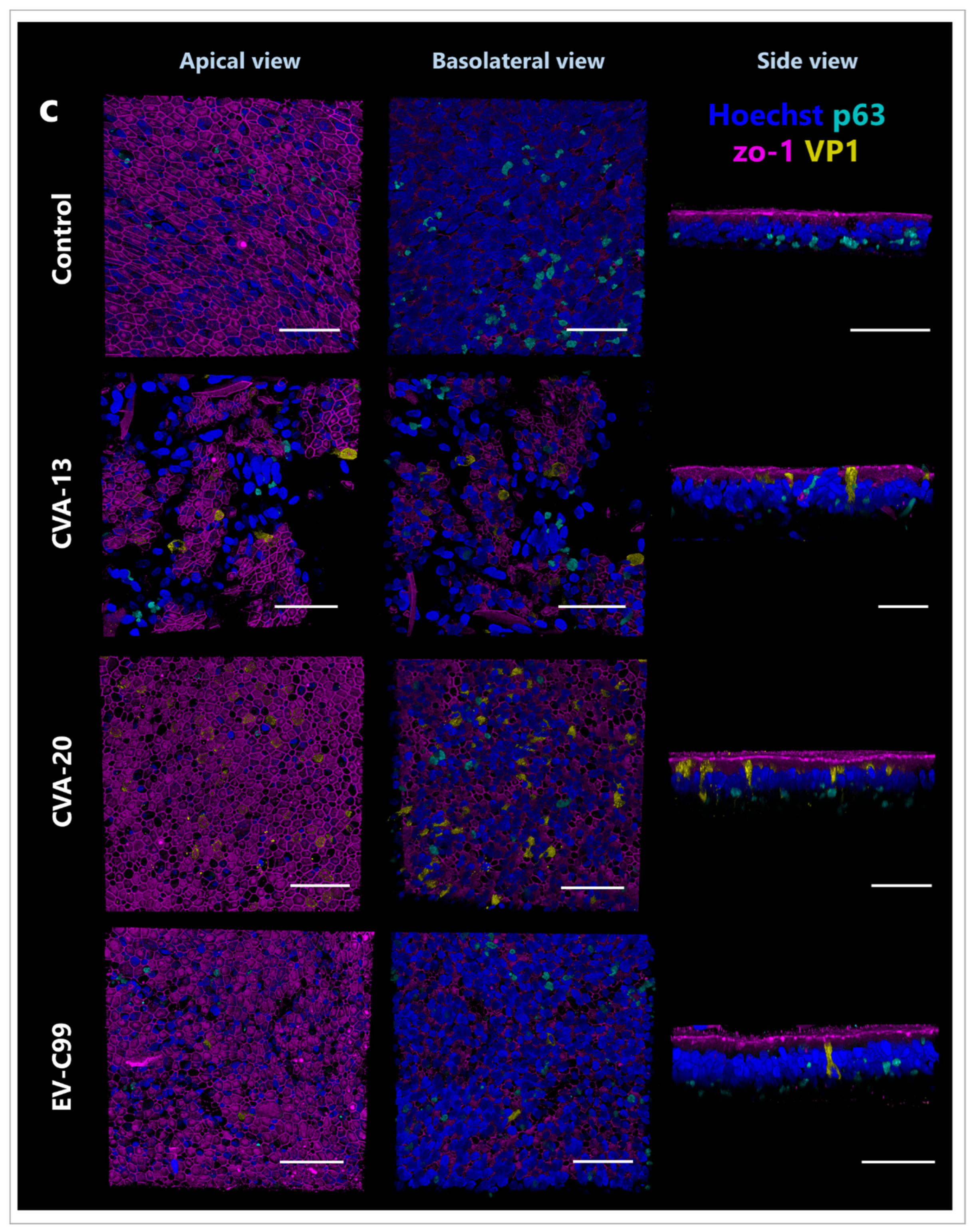
3.3. CVA-13, CVA-20, and EV-C99 Infect Ciliated Cells in the Airway Epithelium
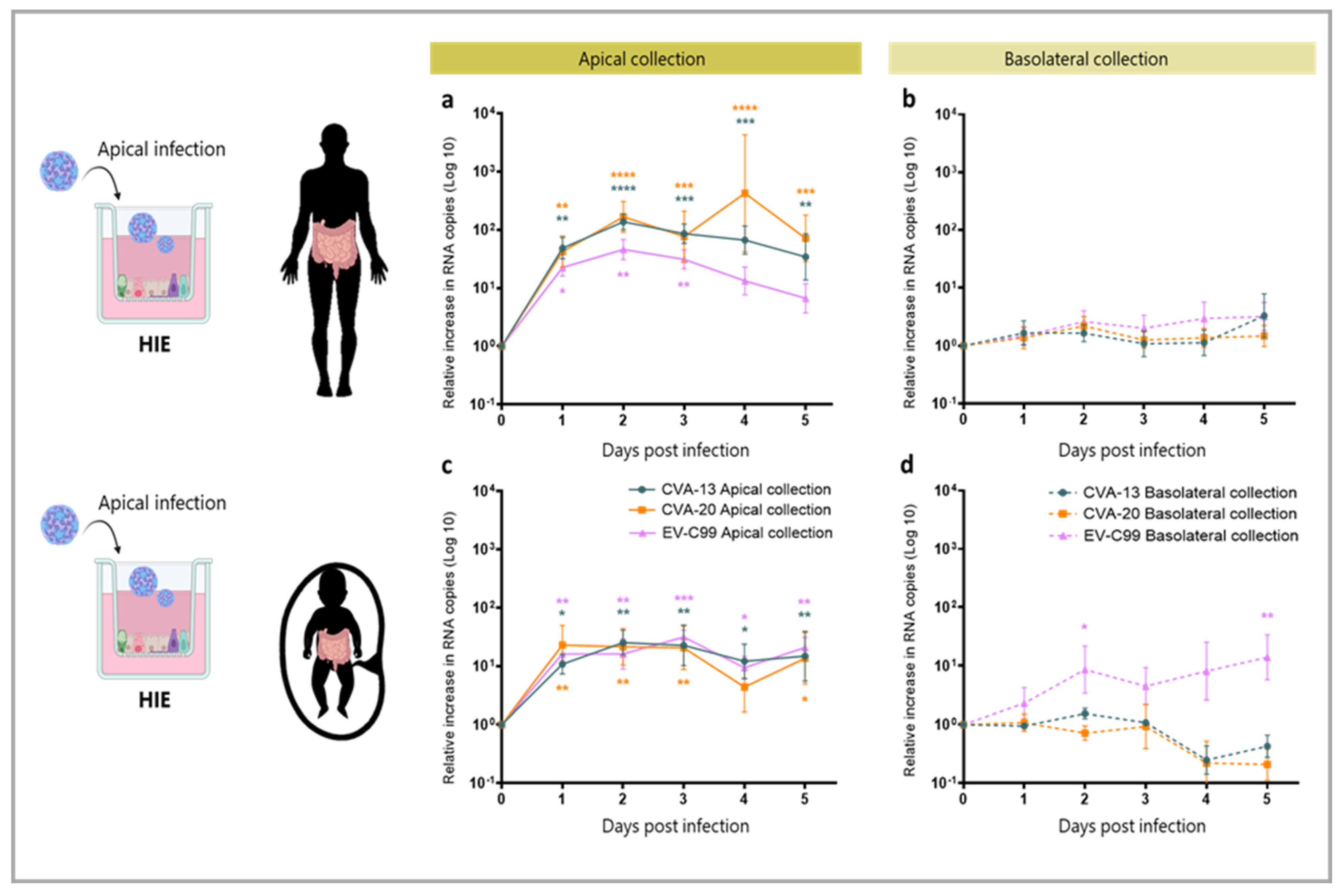
3.4. CVA-13, CVA-20 and EV-C99 Replicate in the Human Intestinal Epithelial (HIE) Model
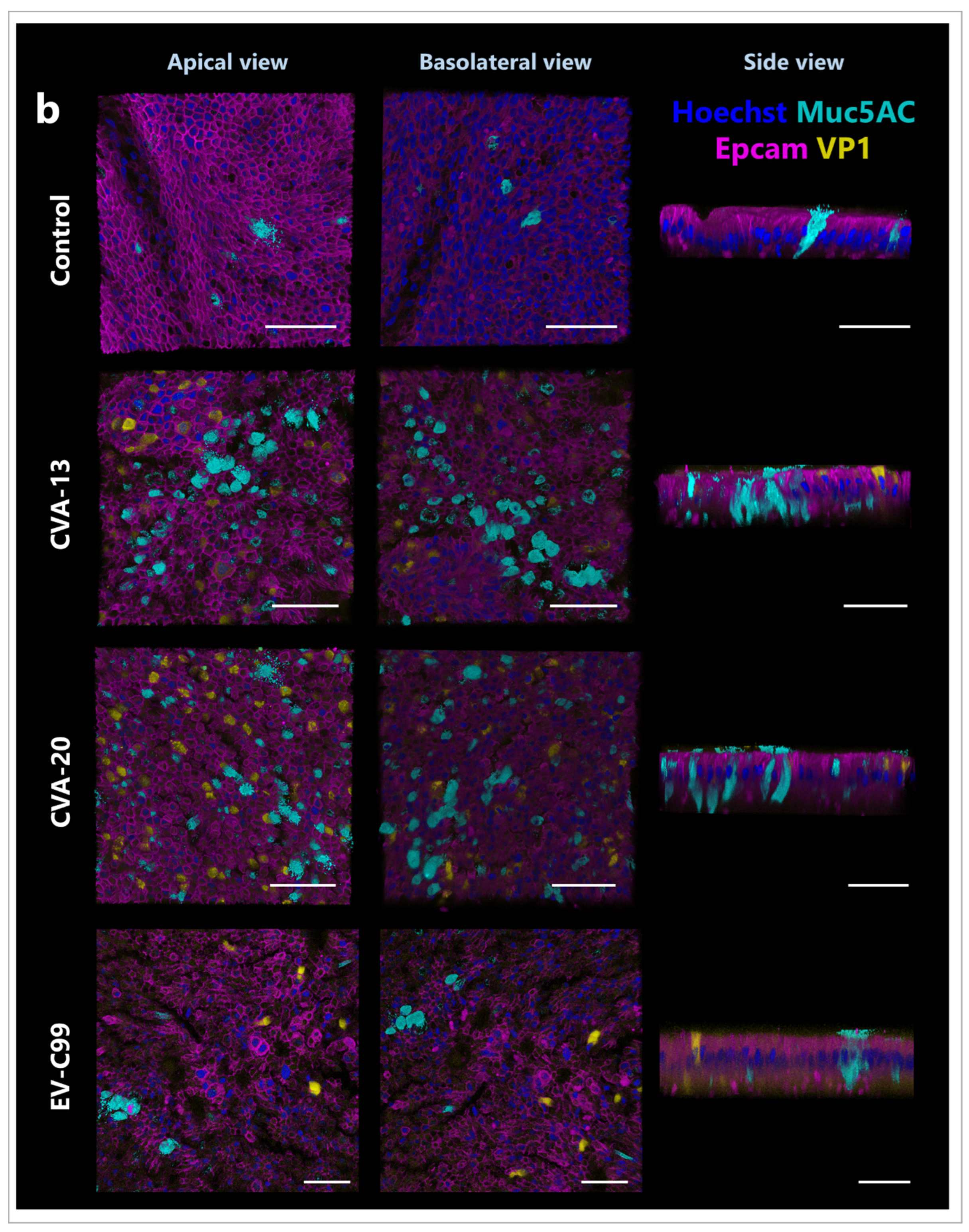
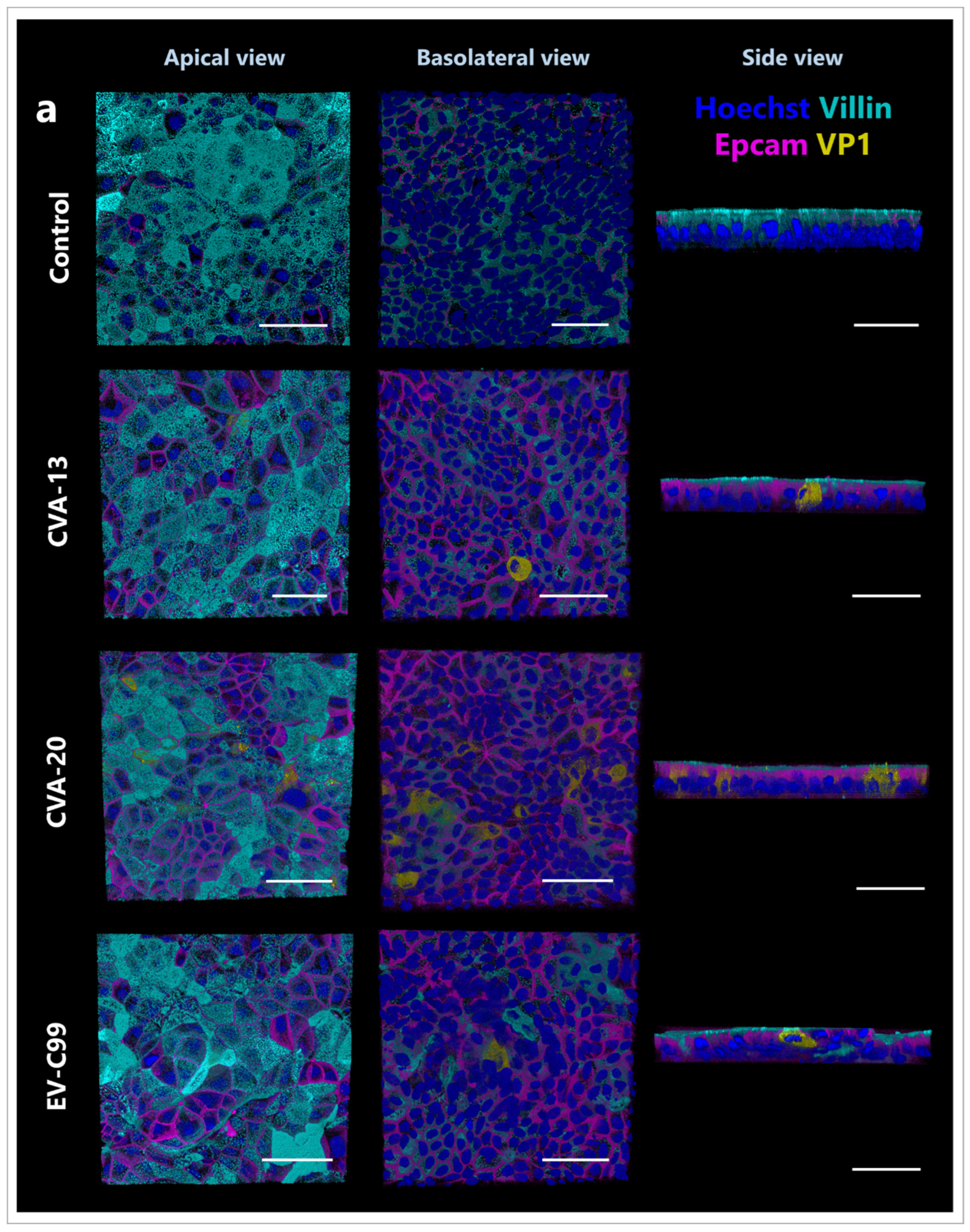
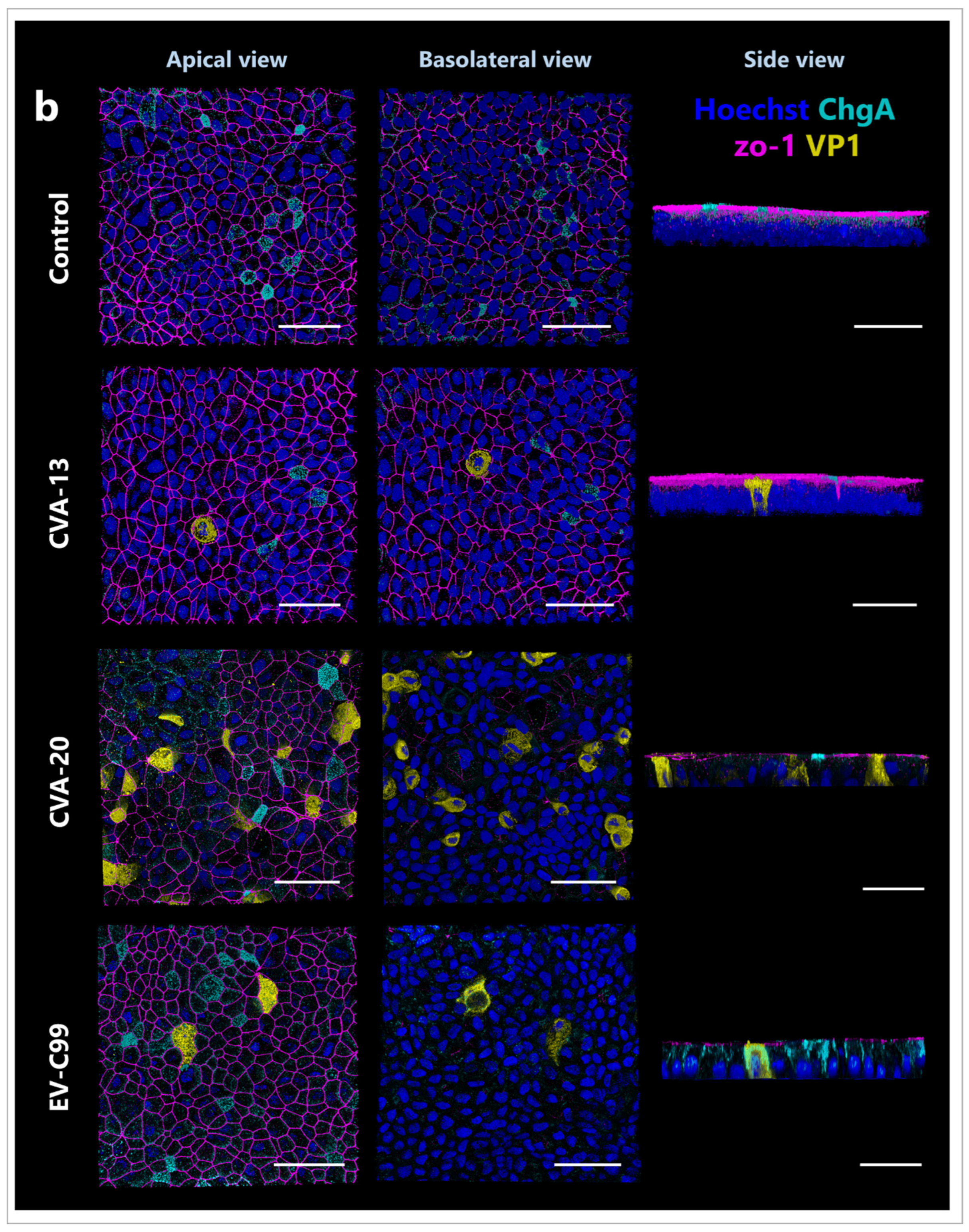
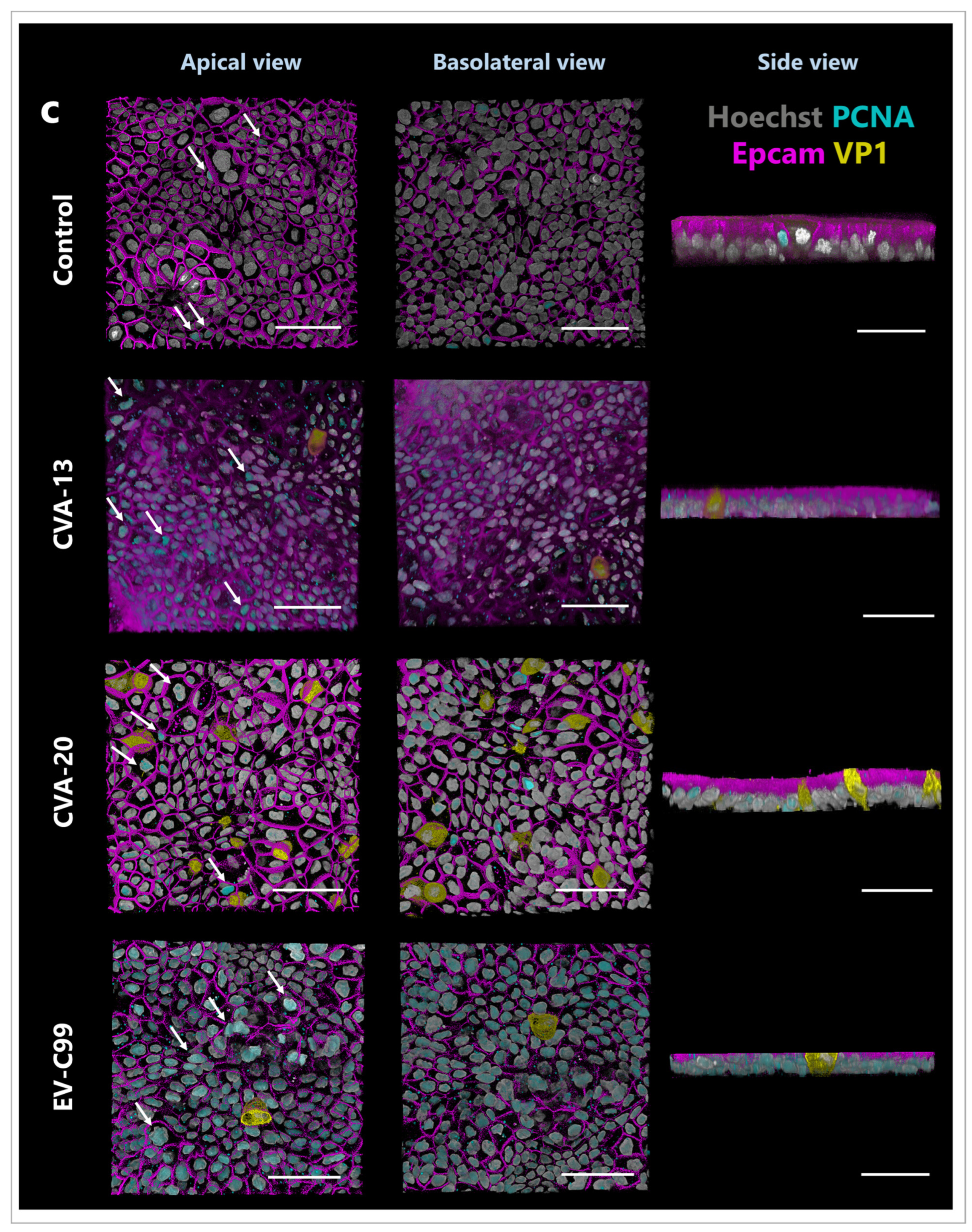
3.5. CVA-13 and CVA-20 Infect Enterocytes While EV-C99 Infects Both Enterocytes and Enteroendocrine Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Available online: https://www.picornaviridae.com/ (accessed on 6 February 2023).
- Royston, L.; Tapparel, C. Rhinoviruses and Respiratory Enteroviruses: Not as Simple as ABC. Viruses 2016, 8, 16. [Google Scholar] [PubMed]
- Chen, B.S.; Lee, H.C.; Lee, K.M.; Gong, Y.N.; Shih, S.R. Enterovirus and Encephalitis. Front. Microbiol. 2020, 11, 261. [Google Scholar] [PubMed]
- Sawyer, M.H. Enterovirus infections: Diagnosis and treatment. Semin. Pediatr. Infect. Dis. 2002, 13, 40–47. [Google Scholar] [PubMed]
- Rhoades, R.E.; Tabor-Godwin, J.M.; Tsueng, G.; Feuer, R. Enterovirus infections of the central nervous system. Virology 2011, 411, 288–305. [Google Scholar]
- Tapparel, C.; Siegrist, F.; Petty, T.J.; Kaiser, L. Picornavirus and enterovirus diversity with associated human diseases. Infect. Genet. Evol. 2013, 14, 282–293. [Google Scholar] [PubMed]
- de Crom, S.C.; Rossen, J.W.; van Furth, A.M.; Obihara, C.C. Enterovirus and parechovirus infection in children: A brief overview. Eur. J. Pediatr. 2016, 175, 1023–1029. [Google Scholar]
- Suresh, S.; Rawlinson, W.D.; Andrews, P.I.; Stelzer-Braid, S. Global epidemiology of nonpolio enteroviruses causing severe neurological complications: A systematic review and meta-analysis. Rev. Med. Virol. 2020, 30, e2082. [Google Scholar]
- Worobey, M.; Holmes, E.C. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 1999, 80 Pt 10, 2535–2543. [Google Scholar]
- Lowry, K.; Woodman, A.; Cook, J.; Evans, D.J. Recombination in enteroviruses is a biphasic replicative process involving the generation of greater-than genome length ‘imprecise’ intermediates. PLoS Pathog. 2014, 10, e1004191. [Google Scholar]
- Muslin, C.; Joffret, M.L.; Pelletier, I.; Blondel, B.; Delpeyroux, F. Evolution and Emergence of Enteroviruses through Intra- and Inter-species Recombination: Plasticity and Phenotypic Impact of Modular Genetic Exchanges in the 5′ Untranslated Region. PLoS Pathog. 2015, 11, e1005266. [Google Scholar]
- Bessaud, M.; Joffret, M.L.; Blondel, B.; Delpeyroux, F. Exchanges of genomic domains between poliovirus and other cocirculating species C enteroviruses reveal a high degree of plasticity. Sci. Rep. 2016, 6, 38831. [Google Scholar]
- Jegouic, S.; Joffret, M.L.; Blanchard, C.; Riquet, F.B.; Perret, C.; Pelletier, I.; Colbere-Garapin, F.; Rakoto-Andrianarivelo, M.; Delpeyroux, F. Recombination between polioviruses and co-circulating Coxsackie A viruses: Role in the emergence of pathogenic vaccine-derived polioviruses. PLoS Pathog. 2009, 5, e1000412. [Google Scholar]
- Li, K.S.; Guan, Y.; Wang, J.; Smith, G.J.; Xu, K.M.; Duan, L.; Rahardjo, A.P.; Puthavathana, P.; Buranathai, C.; Nguyen, T.D.; et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 2004, 430, 209–213. [Google Scholar]
- Smith, G.J.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S.; et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009, 459, 1122–1125. [Google Scholar]
- Combelas, N.; Holmblat, B.; Joffret, M.L.; Colbère-Garapin, F.; Delpeyroux, F. Recombination between poliovirus and coxsackie A viruses of species C: A model of viral genetic plasticity and emergence. Viruses 2011, 3, 1460–1484. [Google Scholar] [PubMed]
- Chansaenroj, J.; Vongpunsawad, S.; Puenpa, J.; Theamboonlers, A.; Vuthitanachot, V.; Chattakul, P.; Areechokchai, D.; Poovorawan, Y. Epidemic outbreak of acute haemorrhagic conjunctivitis caused by coxsackievirus A24 in Thailand, 2014. Epidemiol. Infect. 2015, 143, 3087–3093. [Google Scholar]
- Brouwer, L.; Moreni, G.; Wolthers, K.C.; Pajkrt, D. World-Wide Prevalence and Genotype Distribution of Enteroviruses. Viruses 2021, 13, 434. [Google Scholar]
- Brouwer, L.; van der Sanden, S.M.G.; Calis, J.C.J.; Bruning, A.H.L.; Wang, S.; Wildenbeest, J.G.; Rebers, S.P.H.; Phiri, K.S.; Westerhuis, B.M.; van Hensbroek, M.B.; et al. High frequency of Polio-like Enterovirus C strains with differential clustering of CVA-13 and EV-C99 subgenotypes in a cohort of Malawian children. Arch. Virol. 2018, 163, 2645–2653. [Google Scholar] [PubMed]
- Racaniello, V.R. One hundred years of poliovirus pathogenesis. Virology 2006, 344, 9–16. [Google Scholar] [PubMed]
- Moreni, G.; Aknouch, I.; Ras, M.; Brouwer, L.; Spijker, R.; Calitz, C.; Stittelaar, K.J.; Sridhar, A.; Wolthers, K.C.; Pajkrt, D. Bridging the gap between emerging models and humans by learning from polio animal studies: A systematic review. Clin. Transl. Discov. 2022, 2, e42. [Google Scholar]
- Pfeiffer, J.K. Innate host barriers to viral trafficking and population diversity: Lessons learned from poliovirus. Adv. Virus. Res. 2010, 77, 85–118. [Google Scholar]
- Wells, A.I.; Coyne, C.B. Enteroviruses: A Gut-Wrenching Game of Entry, Detection, and Evasion. Viruses 2019, 11, 460. [Google Scholar]
- Baggen, J.; Thibaut, H.J.; Strating, J.; van Kuppeveld, F.J.M. The life cycle of non-polio enteroviruses and how to target it. Nat Rev. Microbiol. 2018, 16, 368–381. [Google Scholar]
- Brouwer, L.; Benschop, K.S.M.; Nguyen, D.; Kamau, E.; Pajkrt, D.; Simmonds, P.; Wolthers, K.C. Recombination Analysis of Non-Poliovirus Members of the Enterovirus C Species; Restriction of Recombination Events to Members of the Same 3DPol Cluster. Viruses 2020, 12, 706. [Google Scholar]
- Tokarz, R.; Haq, S.; Sameroff, S.; Howie, S.R.C.; Lipkin, W.I. Genomic analysis of coxsackieviruses A1, A19, A22, enteroviruses 113 and 104: Viruses representing two clades with distinct tropism within enterovirus C. J. Gen. Virol. 2013, 94, 1995–2004. [Google Scholar]
- Donbraye, E.; Olasunkanmi, O.I.; Opabode, B.A.; Ishola, T.R.; Faleye, T.O.C.; Adewumi, O.M.; Adeniji, J.A. Abundance of enterovirus C in RD-L20B cell culture-negative stool samples from acute flaccid paralysis cases in Nigeria is geographically defined. J Med. Microbiol. 2018, 67, 854–865. [Google Scholar] [PubMed]
- Adeniji, J.A.; Oragwa, A.O.; George, U.E.; Ibok, U.I.; Faleye, T.O.C.; Adewumi, M.O. Preponderance of enterovirus C in RD-L20B-cell-culture-negative stool samples from children diagnosed with acute flaccid paralysis in Nigeria. Arch. Virol. 2017, 162, 3089–3101. [Google Scholar]
- Rakoto-Andrianarivelo, M.; Rousset, D.; Razafindratsimandresy, R.; Chevaliez, S.; Guillot, S.; Balanant, J.; Delpeyroux, F. High frequency of human enterovirus species C circulation in Madagascar. J. Clin. Microbiol. 2005, 43, 242–249. [Google Scholar] [PubMed]
- Zhang, Y.; Sun, Q.; Cui, H.; Yan, D.; Fan, Q.; Song, Y.; Zhu, S.; Li, X.; Huang, G.; Ji, T.; et al. Circulation of multiple serotypes of highly divergent enterovirus C in the Xinjiang Uighur Autonomous Region of China. Sci. Rep. 2016, 6, 33595. [Google Scholar] [PubMed]
- Van Leer-Buter, C.C.; Poelman, R.; Borger, R.; Niesters, H.G. Newly Identified Enterovirus C Genotypes, Identified in the Netherlands through Routine Sequencing of All Enteroviruses Detected in Clinical Materials from 2008 to 2015. J. Clin. Microbiol. 2016, 54, 2306–2314. [Google Scholar]
- Smura, T.; Blomqvist, S.; Vuorinen, T.; Ivanova, O.; Samoilovich, E.; Al-Hello, H.; Savolainen-Kopra, C.; Hovi, T.; Roivainen, M. Recombination in the Evolution of Enterovirus C Species Sub-Group that Contains Types CVA-21, CVA-24, EV-C95, EV-C96 and EV-C99. PLoS ONE 2014, 9, e94579. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, A.; Simmini, S.; Ribeiro, C.M.S.; Tapparel, C.; Evers, M.M.; Pajkrt, D.; Wolthers, K. A Perspective on Organoids for Virology Research. Viruses 2020, 12, 1314. [Google Scholar] [CrossRef] [PubMed]
- Karelehto, E.; Cristella, C.; Yu, X.; Sridhar, A.; Hulsdouw, R.; de Haan, K.; van Eijk, H.; Koekkoek, S.; Pajkrt, D.; de Jong, M.D.; et al. Polarized Entry of Human Parechoviruses in the Airway Epithelium. Front. Cell. Infect. Microbiol. 2018, 8, 294. [Google Scholar] [CrossRef]
- Sridhar, A.; Depla, J.A.; Mulder, L.A.; Karelehto, E.; Brouwer, L.; Kruiswijk, L.; Vieira de Sá, R.; Meijer, A.; Evers, M.M.; van Kuppeveld, F.J.M.; et al. Enterovirus D68 Infection in Human Primary Airway and Brain Organoids: No Additional Role for Heparan Sulfate Binding for Neurotropism. Microbiol. Spectr. 2022, 10, e0169422. [Google Scholar] [CrossRef]
- García-Rodríguez, I.; Sridhar, A.; Pajkrt, D.; Wolthers, K.C. Put Some Guts into It: Intestinal Organoid Models to Study Viral Infection. Viruses 2020, 12, 1288. [Google Scholar] [CrossRef]
- Aknouch, I.; García-Rodríguez, I.; Giugliano, F.P.; Calitz, C.; Koen, G.; van Eijk, H.; Johannessson, N.; Rebers, S.; Brouwer, L.; Muncan, V.; et al. Amino acid variation at VP1-145 of enterovirus A71 determines the viral infectivity and receptor usage in a primary human intestinal model. Front. Microbiol. 2023, 14, 1045587. [Google Scholar] [CrossRef]
- Aknouch, I.; Sridhar, A.; Freeze, E.; Giugliano, F.P.; van Keulen, B.J.; Romijn, M.; Calitz, C.; García-Rodríguez, I.; Mulder, L.; Wildenberg, M.E.; et al. Human milk inhibits some enveloped virus infections, including SARS-CoV-2, in an intestinal model. Life Sci. Alliance 2022, 5, e202201432. [Google Scholar] [CrossRef] [PubMed]
- Roodsant, T.; Navis, M.; Aknouch, I.; Renes, I.B.; van Elburg, R.M.; Pajkrt, D.; Wolthers, K.C.; Schultsz, C.; van der Ark, K.C.H.; Sridhar, A.; et al. A Human 2D Primary Organoid-Derived Epithelial Monolayer Model to Study Host-Pathogen Interaction in the Small Intestine. Front. Cell. Infect. Microbiol. 2020, 10, 272. [Google Scholar] [CrossRef] [PubMed]
- Benschop, K.S.M.; Broberg, E.K.; Hodcroft, E.; Schmitz, D.; Albert, J.; Baicus, A.; Bailly, J.L.; Baldvinsdottir, G.; Berginc, N.; Blomqvist, S.; et al. Molecular Epidemiology and Evolutionary Trajectory of Emerging Echovirus 30, Europe. Emerg. Infect. Dis. 2021, 27, 1616–1626. [Google Scholar] [CrossRef]
- Fulcher, M.L.; Gabriel, S.; Burns, K.A.; Yankaskas, J.R.; Randell, S.H. Well-differentiated human airway epithelial cell cultures. Methods Mol. Med. 2005, 107, 183–206. [Google Scholar]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Benschop, K.; Minnaar, R.; Koen, G.; van Eijk, H.; Dijkman, K.; Westerhuis, B.; Molenkamp, R.; Wolthers, K. Detection of human enterovirus and human parechovirus (HPeV) genotypes from clinical stool samples: Polymerase chain reaction and direct molecular typing, culture characteristics, and serotyping. Diagn. Microbiol. Infect. Dis. 2010, 68, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A SIMPLE METHOD OF ESTIMATING FIFTY PER CENT ENDPOINTS12. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Jiang, P.; Liu, Y.; Ma, H.C.; Paul, A.V.; Wimmer, E. Picornavirus morphogenesis. Microbiol. Mol. Biol. Rev. 2014, 78, 418–437. [Google Scholar] [CrossRef] [PubMed]
- Pozzetto, B.; Gaudin, O.G. Coxsackieviruses (Picornaviridae). Encycl. Virol. 1999, 305–311. [Google Scholar]
- Wenner, H.A.; Lenahan, M.F. Propagation of group A Coxsackie viruses in tissue cultures. II. Some interactions between virus and mammalian cells. Yale. J. Biol. Med. 1961, 34, 421–438. [Google Scholar]
- Lipson, S.M.; Walderman, R.; Costello, P.; Szabo, K. Sensitivity of rhabdomyosarcoma and guinea pig embryo cell cultures to field isolates of difficult-to-cultivate group A coxsackieviruses. J. Clin. Microbiol. 1988, 26, 1298–1303. [Google Scholar] [CrossRef]
- García-Rodríguez, I.; van Eijk, H.; Koen, G.; Pajkrt, D.; Sridhar, A.; Wolthers, K.C. Parechovirus A Infection of the Intestinal Epithelium: Differences Between Genotypes A1 and A3. Front. Cell. Infect. Microbiol. 2021, 11, 740662. [Google Scholar] [CrossRef]
- Chang, C.S.; Liao, C.C.; Liou, A.T.; Chou, Y.C.; Yu, Y.Y.; Lin, C.Y.; Lin, J.S.; Suen, C.S.; Hwang, M.J.; Shih, C. Novel Naturally Occurring Mutations of Enterovirus 71 Associated With Disease Severity. Front. Microbiol. 2020, 11, 610568. [Google Scholar] [CrossRef]
- Bochkov, Y.A.; Watters, K.; Basnet, S.; Sijapati, S.; Hill, M.; Palmenberg, A.C.; Gern, J.E. Mutations in VP1 and 3A proteins improve binding and replication of rhinovirus C15 in HeLa-E8 cells. Virology 2016, 499, 350–360. [Google Scholar] [CrossRef] [PubMed]
- Cagno, V.; Tseligka, E.D.; Jones, S.T.; Tapparel, C. Heparan Sulfate Proteoglycans and Viral Attachment: True Receptors or Adaptation Bias? Viruses 2019, 11, 596. [Google Scholar] [CrossRef]
- Patton, J.S.; Byron, P.R. Inhaling medicines: Delivering drugs to the body through the lungs. Nat. Rev. Drug Discov. 2007, 6, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Filipe, I.C.; Tee, H.K.; Prados, J.; Piuz, I.; Constant, S.; Huang, S.; Tapparel, C. Comparison of tissue tropism and host response to enteric and respiratory enteroviruses. PLoS Pathog. 2022, 18, e1010632. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, C.; Liu, X.; Chiu, M.C.; Wang, D.; Wei, Y.; Chu, H.; Cai, J.P.; Hau-Yee Chan, I.; Kak-Yuen Wong, K.; et al. Human Intestinal Organoids Recapitulate Enteric Infections of Enterovirus and Coronavirus. Stem Cell Rep. 2021, 16, 493–504. [Google Scholar] [CrossRef]
- Drummond, C.G.; Bolock, A.M.; Ma, C.; Luke, C.J.; Good, M.; Coyne, C.B. Enteroviruses infect human enteroids and induce antiviral signaling in a cell lineage-specific manner. Proc. Natl. Acad. Sci. USA 2017, 114, 1672–1677. [Google Scholar] [CrossRef]
- Ohka, S.; Igarashi, H.; Nagata, N.; Sakai, M.; Koike, S.; Nochi, T.; Kiyono, H.; Nomoto, A. Establishment of a poliovirus oral infection system in human poliovirus receptor-expressing transgenic mice that are deficient in alpha/beta interferon receptor. J. Virol. 2007, 81, 7902–7912. [Google Scholar] [CrossRef]
- Suresh, S.; Forgie, S.; Robinson, J. Non-polio Enterovirus detection with acute flaccid paralysis: A systematic review. J. Med. Virol. 2018, 90, 3–7. [Google Scholar] [CrossRef]
- Oberste, M.S.; Pallansch, M.A. Coxsackieviruses. In Encyclopedia of Virology, 3rd ed.; Mahy, B.W.J., Van Regenmortel, M.H.V., Eds.; Academic Press: Oxford, UK, 2008; pp. 580–587. [Google Scholar]
- Magré, L.; Verstegen, M.M.A.; Buschow, S.; van der Laan, L.J.W.; Peppelenbosch, M.; Desai, J. Emerging organoid-immune co-culture models for cancer research: From oncoimmunology to personalized immunotherapies. J. Immunother. Cancer 2023, 11, e006290. [Google Scholar] [CrossRef] [PubMed]
- Sittipo, P.; Lee, Y.K. The application of intestinal organoids and their co-culture systems in the study of gastrointestinal diseases. Organoid 2022, 2, e3. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreni, G.; van Eijk, H.; Koen, G.; Johannesson, N.; Calitz, C.; Benschop, K.; Cremer, J.; Pajkrt, D.; Sridhar, A.; Wolthers, K. Non-Polio Enterovirus C Replicate in Both Airway and Intestine Organotypic Cultures. Viruses 2023, 15, 1823. https://doi.org/10.3390/v15091823
Moreni G, van Eijk H, Koen G, Johannesson N, Calitz C, Benschop K, Cremer J, Pajkrt D, Sridhar A, Wolthers K. Non-Polio Enterovirus C Replicate in Both Airway and Intestine Organotypic Cultures. Viruses. 2023; 15(9):1823. https://doi.org/10.3390/v15091823
Chicago/Turabian StyleMoreni, Giulia, Hetty van Eijk, Gerrit Koen, Nina Johannesson, Carlemi Calitz, Kimberley Benschop, Jeroen Cremer, Dasja Pajkrt, Adithya Sridhar, and Katja Wolthers. 2023. "Non-Polio Enterovirus C Replicate in Both Airway and Intestine Organotypic Cultures" Viruses 15, no. 9: 1823. https://doi.org/10.3390/v15091823
APA StyleMoreni, G., van Eijk, H., Koen, G., Johannesson, N., Calitz, C., Benschop, K., Cremer, J., Pajkrt, D., Sridhar, A., & Wolthers, K. (2023). Non-Polio Enterovirus C Replicate in Both Airway and Intestine Organotypic Cultures. Viruses, 15(9), 1823. https://doi.org/10.3390/v15091823





