Humoral Responses Elicited after a Fifth Dose of SARS-CoV-2 mRNA Bivalent Vaccine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Human Subjects
2.3. Plasma Samples and Antibodies
2.4. Plasmids
2.5. Protein Expression and Purification
2.6. Cell Lines
2.7. Anti-Nucleocapsid (N) Assay
2.8. Anti-RBD ELISA and Avidity Assays
2.9. Cell-Surface Staining and Flow Cytometry Analysis
2.10. Antibody-Dependent Cellular Cytotoxicity Assay
2.11. Virus Neutralization Assay
2.12. Statistical Analysis
3. Results
3.1. Characteristics of the Cohort
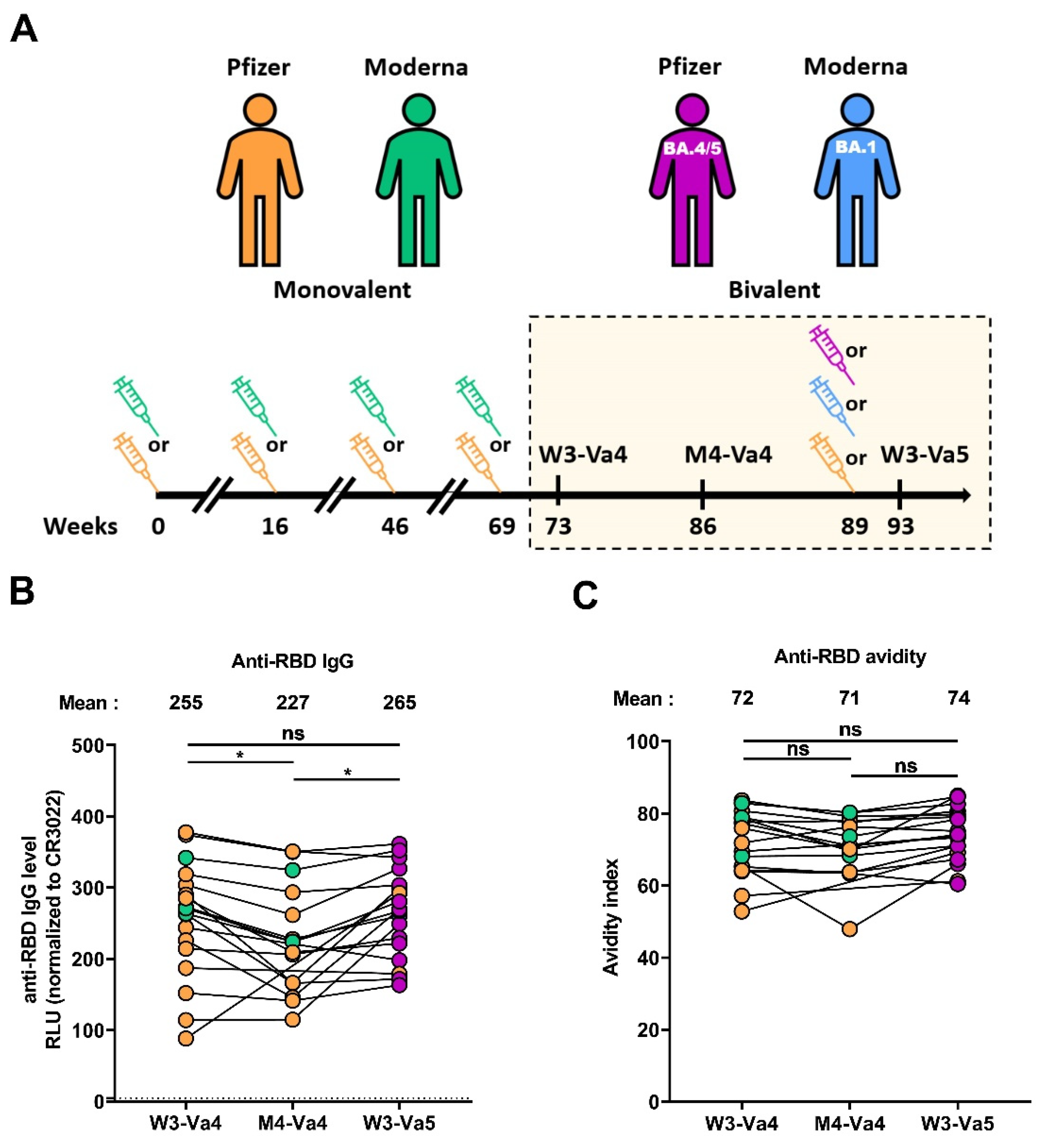
3.2. RBD-Specific IgG and Associated Avidity
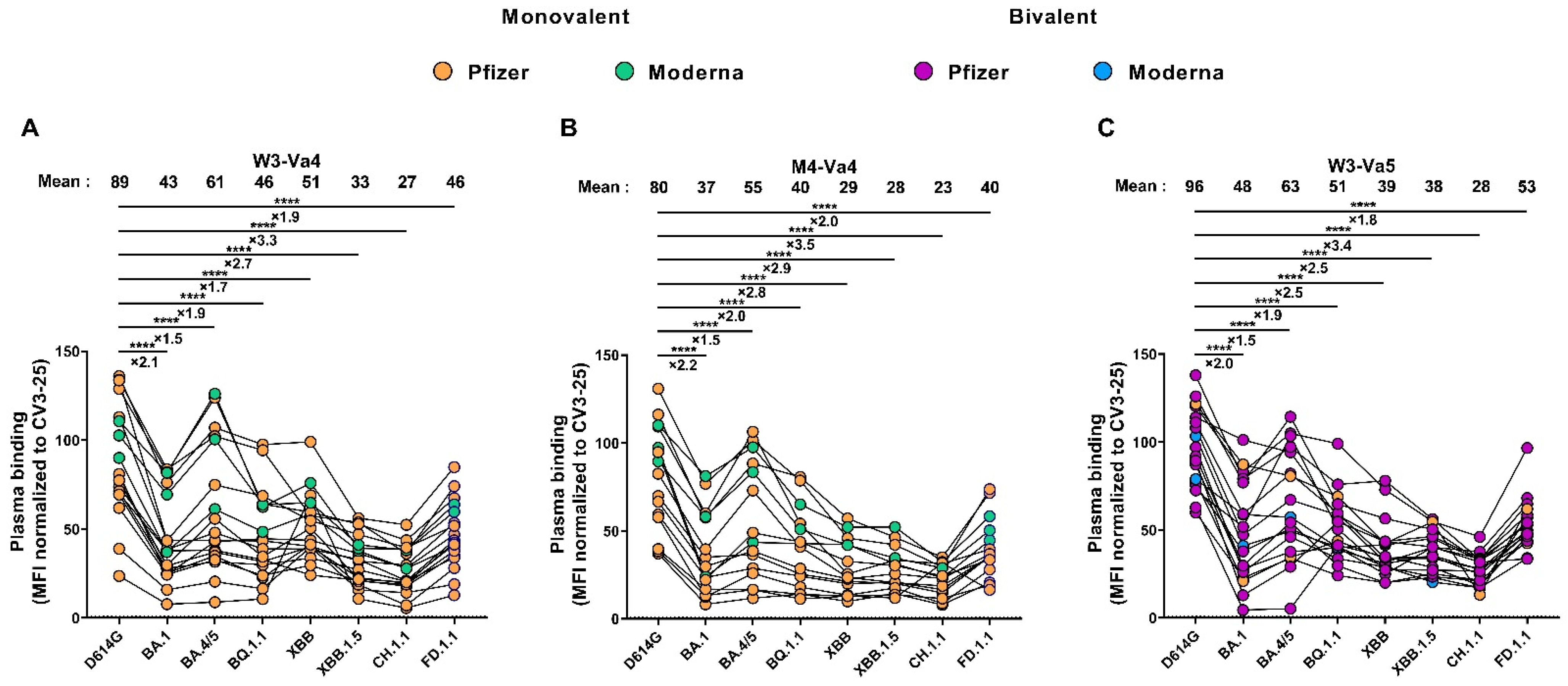
3.3. Recognition of SARS-CoV-2 Spike Variants by Plasma from Vaccinated Individuals
3.4. Neutralizing Activity against Different SARS-CoV-2 Spikes
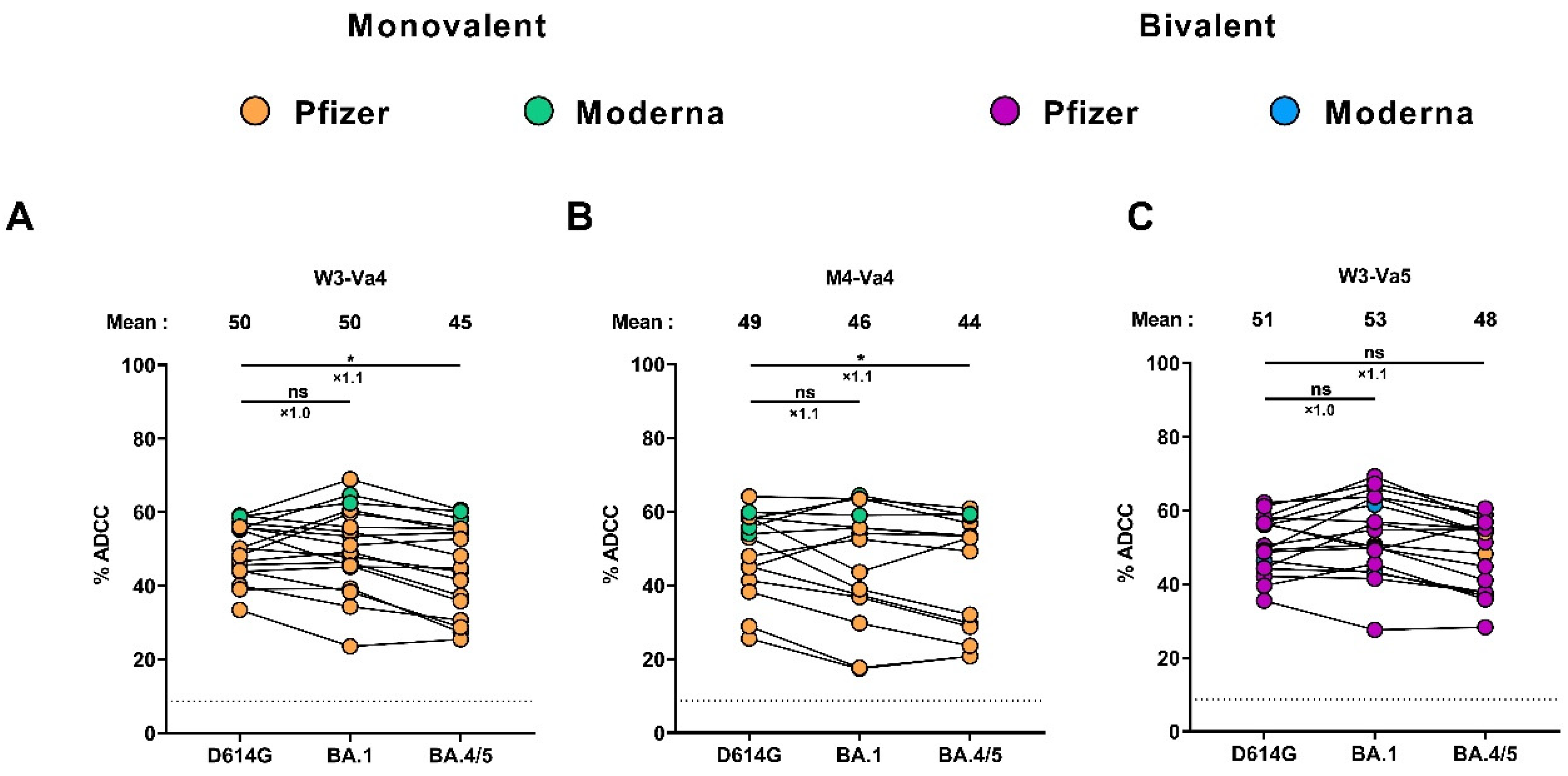
3.5. Fc-Effector Functions Elicited after the Fourth and the Fifth Doses of mRNA Vaccine
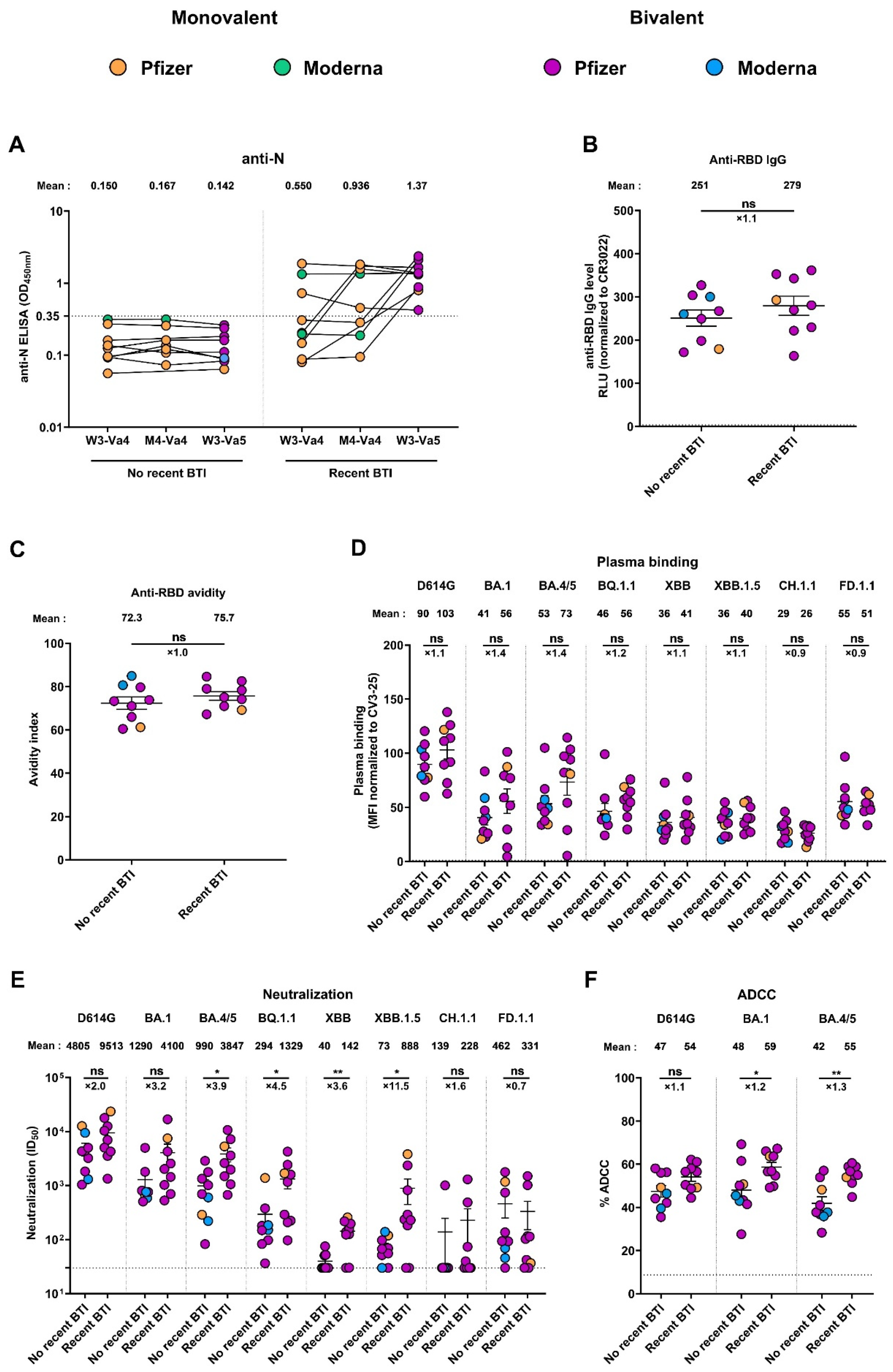
3.6. Recent Infection Leads to Antibodies with Better Functional Activities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the MRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 MRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Skowronski, D.; De Serres, G. Safety and Efficacy of the BNT162b2 MRNA COVID-19 Vaccine. N. Engl. J. Med. 2021, 384, NEJMc2036242. [Google Scholar] [CrossRef]
- Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. Available online: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (accessed on 4 July 2023).
- Andrews, N.; Stowe, J.; Kirsebom, F.; Toffa, S.; Rickeard, T.; Gallagher, E.; Gower, C.; Kall, M.; Groves, N.; O’Connell, A.-M.; et al. COVID-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N. Engl. J. Med. 2022, 386, 1532–1546. [Google Scholar] [CrossRef]
- Edara, V.-V.; Manning, K.E.; Ellis, M.; Lai, L.; Moore, K.M.; Foster, S.L.; Floyd, K.; Davis-Gardner, M.E.; Mantus, G.; Nyhoff, L.E.; et al. MRNA-1273 and BNT162b2 MRNA Vaccines Have Reduced Neutralizing Activity against the SARS-CoV-2 Omicron Variant. Cell Rep. Med. 2022, 3, 100529. [Google Scholar] [CrossRef]
- Viana, R.; Moyo, S.; Amoako, D.G.; Tegally, H.; Scheepers, C.; Althaus, C.L.; Anyaneji, U.J.; Bester, P.A.; Boni, M.F.; Chand, M.; et al. Rapid Epidemic Expansion of the SARS-CoV-2 Omicron Variant in Southern Africa. Nature 2022, 603, 679–686. [Google Scholar] [CrossRef]
- Tauzin, A.; Gong, S.Y.; Chatterjee, D.; Ding, S.; Painter, M.M.; Goel, R.R.; Beaudoin-Bussières, G.; Marchitto, L.; Boutin, M.; Laumaea, A.; et al. A Boost with SARS-CoV-2 BNT162b2 MRNA Vaccine Elicits Strong Humoral Responses Independently of the Interval between the First Two Doses. Cell Rep. 2022, 41, 111554. [Google Scholar] [CrossRef]
- Goel, R.R.; Painter, M.M.; Lundgreen, K.A.; Apostolidis, S.A.; Baxter, A.E.; Giles, J.R.; Mathew, D.; Pattekar, A.; Reynaldi, A.; Khoury, D.S.; et al. Efficient Recall of Omicron-Reactive B Cell Memory after a Third Dose of SARS-CoV-2 MRNA Vaccine. Cell 2022, 185, 1875–1887.e8. [Google Scholar] [CrossRef]
- Tauzin, A.; Nicolas, A.; Ding, S.; Benlarbi, M.; Medjahed, H.; Chatterjee, D.; Dionne, K.; Gong, S.Y.; Gendron-Lepage, G.; Bo, Y.; et al. Spike Recognition and Neutralization of SARS-CoV-2 Omicron Subvariants Elicited after the Third Dose of MRNA Vaccine. Cell Rep. 2023, 42, 111998. [Google Scholar] [CrossRef]
- Tegally, H.; Moir, M.; Everatt, J.; Giovanetti, M.; Scheepers, C.; Wilkinson, E.; Subramoney, K.; Makatini, Z.; Moyo, S.; Amoako, D.G.; et al. Emergence of SARS-CoV-2 Omicron Lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022, 28, 1785–1790. [Google Scholar] [CrossRef]
- Yu, J.; Collier, A.Y.; Rowe, M.; Mardas, F.; Ventura, J.D.; Wan, H.; Miller, J.; Powers, O.; Chung, B.; Siamatu, M.; et al. Neutralization of the SARS-CoV-2 Omicron BA.1 and BA.2 Variants. N. Engl. J. Med. 2022, 386, 1579–1580. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Kurhade, C.; Patel, S.; Kitchin, N.; Tompkins, K.; Cutler, M.; Cooper, D.; Yang, Q.; Cai, H.; Muik, A.; et al. Neutralization of BA.4–BA.5, BA.4.6, BA.2.75.2, BQ.1.1, and XBB.1 with Bivalent Vaccine. N. Engl. J. Med. 2023, 388, 854–857. [Google Scholar] [CrossRef] [PubMed]
- Les Variants du SRAS-CoV-2|INSPQ. Available online: https://www.inspq.qc.ca/covid-19/labo/variants (accessed on 5 July 2023).
- Miller, J.; Hachmann, N.P.; Collier, A.Y.; Lasrado, N.; Mazurek, C.R.; Patio, R.C.; Powers, O.; Surve, N.; Theiler, J.; Korber, B.; et al. Substantial Neutralization Escape by SARS-CoV-2 Omicron Variants BQ.1.1 and XBB.1. N. Engl. J. Med. 2023, 388, 662–664. [Google Scholar] [CrossRef] [PubMed]
- Qu, P.; Faraone, J.N.; Evans, J.P.; Zheng, Y.-M.; Carlin, C.; Anghelina, M.; Stevens, P.; Fernandez, S.; Jones, D.; Panchal, A.R.; et al. Enhanced Evasion of Neutralizing Antibody Response by Omicron XBB.1.5, CH.1.1, and CA.3.1 Variants. Cell Rep. 2023, 42, 112443. [Google Scholar] [CrossRef]
- Davis-Gardner, M.E.; Lai, L.; Wali, B.; Samaha, H.; Solis, D.; Lee, M.; Porter-Morrison, A.; Hentenaar, I.T.; Yamamoto, F.; Godbole, S.; et al. Neutralization against BA.2.75.2, BQ.1.1, and XBB from MRNA Bivalent Booster. N. Engl. J. Med. 2023, 388, 183–185. [Google Scholar] [CrossRef]
- Guidance on an Additional COVID-19 Booster Dose in the Spring of 2023 for Individuals at High Risk of Severe Illness Due to COVID-19. Available online: https://www.canada.ca/en/public-health/services/publications/vaccines-immunization/national-advisory-committee-immunization-guidance-additional-covid-19-booster-dose-spring-2023-individuals-high-risk-severe-illness-due-covid-19.html (accessed on 5 July 2023).
- How Boosters Produce Broad Protection against COVID-19. Available online: https://www.nih.gov/news-events/nih-research-matters/varied-boosters-produce-broad-protection-against-covid-19 (accessed on 5 July 2023).
- Tauzin, A.; Benlarbi, M.; Medjahed, H.; Grégoire, Y.; Perreault, J.; Gendron-Lepage, G.; Gokool, L.; Morrisseau, C.; Arlotto, P.; Tremblay, C.; et al. Humoral Responses against BQ.1.1 Elicited after Breakthrough Infection and SARS-CoV-2 MRNA Vaccination. Vaccines 2023, 11, 242. [Google Scholar] [CrossRef]
- Wang, Q.; Bowen, A.; Tam, A.R.; Valdez, R.; Stoneman, E.; Mellis, I.A.; Gordon, A.; Liu, L.; Ho, D.D. SARS-CoV-2 Neutralising Antibodies after Bivalent versus Monovalent Booster. Lancet Infect. Dis. 2023, 23, 527–528. [Google Scholar] [CrossRef]
- Kurhade, C.; Zou, J.; Xia, H.; Liu, M.; Chang, H.C.; Ren, P.; Xie, X.; Shi, P.-Y. Low Neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1, and XBB.1 by Parental MRNA Vaccine or a BA.5-Bivalent Booster. Nat. Med. 2022, 29, 344–347. [Google Scholar] [CrossRef]
- Gili, R.; Burioni, R. SARS-CoV-2 before and after Omicron: Two Different Viruses and Two Different Diseases? J. Transl. Med. 2023, 21, 251. [Google Scholar] [CrossRef]
- Tan, S.T.; Kwan, A.T.; Rodríguez-Barraquer, I.; Singer, B.J.; Park, H.J.; Lewnard, J.A.; Sears, D.; Lo, N.C. Infectiousness of SARS-CoV-2 Breakthrough Infections and Reinfections during the Omicron Wave. Nat. Med. 2023, 29, 358–365. [Google Scholar] [CrossRef]
- MN Department of Health. COVID-19 Vaccine Breakthrough Data COVID-19 Situation Update. Available online: https://www.health.state.mn.us/diseases/coronavirus/stats/vbt.html (accessed on 5 July 2023).
- Groenheit, R.; Bacchus, P.; Galanis, I.; Sondén, K.; Bujila, I.; Efimova, T.; Garli, F.; Lindsjö, O.K.; Mansjö, M.; Movert, E.; et al. High Prevalence of SARS-CoV-2 Omicron Infection Despite High Seroprevalence, Sweden, 2022. Emerg. Infect. Dis. J. 2023, 29, 1240. [Google Scholar] [CrossRef] [PubMed]
- Bivalent Boosters Provide Better Protection against Severe COVID-19. Available online: https://www.nih.gov/news-events/nih-research-matters/bivalent-boosters-provide-better-protection-against-severe-covid-19 (accessed on 5 July 2023).
- CDC. COVID Data Tracker Weekly Review. Available online: https://www.cdc.gov/coronavirus/2019-ncov/covid-data/covidview/index.html (accessed on 8 January 2023).
- Tenforde, M.W.; Weber, Z.A.; Natarajan, K.; Klein, N.P.; Kharbanda, A.B.; Stenehjem, E.; Embi, P.J.; Reese, S.E.; Naleway, A.L.; Grannis, S.J.; et al. Early Estimates of Bivalent MRNA Vaccine Effectiveness in Preventing COVID-19-Associated Emergency Department or Urgent Care Encounters and Hospitalizations Among Immunocompetent Adults—VISION Network, Nine States, September–November 2022. MMWR Morb. Mortal. Wkly. Rep. 2023, 71, 1637–1646. [Google Scholar] [CrossRef] [PubMed]
- ter Meulen, J.; van den Brink, E.N.; Poon, L.L.M.; Marissen, W.E.; Leung, C.S.W.; Cox, F.; Cheung, C.Y.; Bakker, A.Q.; Bogaards, J.A.; van Deventer, E.; et al. Human Monoclonal Antibody Combination against SARS Coronavirus: Synergy and Coverage of Escape Mutants. PLoS Med. 2006, 3, e237. [Google Scholar] [CrossRef] [PubMed]
- Prévost, J.; Gasser, R.; Beaudoin-Bussières, G.; Richard, J.; Duerr, R.; Laumaea, A.; Anand, S.P.; Goyette, G.; Benlarbi, M.; Ding, S.; et al. Cross-Sectional Evaluation of Humoral Responses against SARS-CoV-2 Spike. Cell Rep. Med. 2020, 1, 100126. [Google Scholar] [CrossRef] [PubMed]
- Jennewein, M.F.; MacCamy, A.J.; Akins, N.R.; Feng, J.; Homad, L.J.; Hurlburt, N.K.; Seydoux, E.; Wan, Y.-H.; Stuart, A.B.; Edara, V.V.; et al. Isolation and Characterization of Cross-Neutralizing Coronavirus Antibodies from COVID-19+ Subjects. Cell Rep. 2021, 36, 109353. [Google Scholar] [CrossRef]
- Chatterjee, D.; Tauzin, A.; Marchitto, L.; Gong, S.Y.; Boutin, M.; Bourassa, C.; Beaudoin-Bussières, G.; Bo, Y.; Ding, S.; Laumaea, A.; et al. SARS-CoV-2 Omicron Spike Recognition by Plasma from Individuals Receiving BNT162b2 MRNA Vaccination with a 16-Week Interval between Doses. Cell Rep. 2022, 38, 110429. [Google Scholar] [CrossRef]
- Anand, S.P.; Prévost, J.; Nayrac, M.; Beaudoin-Bussières, G.; Benlarbi, M.; Gasser, R.; Brassard, N.; Laumaea, A.; Gong, S.Y.; Bourassa, C.; et al. Longitudinal Analysis of Humoral Immunity against SARS-CoV-2 Spike in Convalescent Individuals up to Eight Months Post-Symptom Onset. Cell Rep. Med. 2021, 2, 100290. [Google Scholar] [CrossRef]
- Beaudoin-Bussières, G.; Richard, J.; Prévost, J.; Goyette, G.; Finzi, A. A New Flow Cytometry Assay to Measure Antibody-Dependent Cellular Cytotoxicity against SARS-CoV-2 Spike-Expressing Cells. STAR Protoc. 2021, 2, 100851. [Google Scholar] [CrossRef]
- Bazin, R.; Rochette, S.; Perreault, J.; Fournier, M.-J.; Grégoire, Y.; Boivin, A.; Lewin, A.; Germain, M.; Renaud, C. Evaluation of Anti-Nucleocapsid Level Variation to Assess SARS-CoV-2 Seroprevalence in a Vaccinated Population. Infect. Dis. Lond. Engl. 2023, 55, 425–430. [Google Scholar] [CrossRef]
- Tauzin, A.; Gendron-Lepage, G.; Nayrac, M.; Anand, S.P.; Bourassa, C.; Medjahed, H.; Goyette, G.; Dubé, M.; Bazin, R.; Kaufmann, D.E.; et al. Evolution of Anti-RBD IgG Avidity Following SARS-CoV-2 Infection. Viruses 2022, 14, 532. [Google Scholar] [CrossRef]
- Beaudoin-Bussières, G.; Tauzin, A.; Dionne, K.; Gendron-Lepage, G.; Medjahed, H.; Perreault, J.; Levade, I.; Alfadhli, L.; Bo, Y.; Bazin, R.; et al. A Recent SARS-CoV-2 Infection Enhances Antibody-Dependent Cellular Cytotoxicity against Several Omicron Subvariants Following a Fourth MRNA Vaccine Dose. Viruses 2023, 15, 1274. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin-Bussières, G.; Laumaea, A.; Anand, S.P.; Prévost, J.; Gasser, R.; Goyette, G.; Medjahed, H.; Perreault, J.; Tremblay, T.; Lewin, A.; et al. Decline of Humoral Responses against SARS-CoV-2 Spike in Convalescent Individuals. mBio 2020, 11, e02590-20. [Google Scholar] [CrossRef] [PubMed]
- Hägglöf, T.; Cipolla, M.; Loewe, M.; Chen, S.T.; Mesin, L.; Hartweger, H.; ElTanbouly, M.A.; Cho, A.; Gazumyan, A.; Ramos, V.; et al. Continuous Germinal Center Invasion Contributes to the Diversity of the Immune Response. Cell 2023, 186, 147–161.e15. [Google Scholar] [CrossRef]
- Kim, W.; Zhou, J.Q.; Sturtz, A.J.; Horvath, S.C.; Schmitz, A.J.; Lei, T.; Kalaidina, E.; Thapa, M.; Soussi, W.B.A.; Haile, A.; et al. Germinal Centre-Driven Maturation of B Cell Response to SARS-CoV-2 Vaccination. Nature 2022, 604, 141–145. [Google Scholar] [CrossRef]
- Tauzin, A.; Gong, S.Y.; Beaudoin-Bussières, G.; Vézina, D.; Gasser, R.; Nault, L.; Marchitto, L.; Benlarbi, M.; Chatterjee, D.; Nayrac, M.; et al. Strong Humoral Immune Responses against SARS-CoV-2 Spike after BNT162b2 MRNA Vaccination with a 16-Week Interval between Doses. Cell Host Microbe 2022, 30, 97–109.e5. [Google Scholar] [CrossRef] [PubMed]
- Ullah, I.; Prévost, J.; Ladinsky, M.S.; Stone, H.; Lu, M.; Anand, S.P.; Beaudoin-Bussières, G.; Symmes, K.; Benlarbi, M.; Ding, S.; et al. Live Imaging of SARS-CoV-2 Infection in Mice Reveals That Neutralizing Antibodies Require Fc Function for Optimal Efficacy. Immunity 2021, 54, 2143–2158.e15. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 Vaccine Breakthrough Infections Reported to CDC—United States, January 1–April 30, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 792. [CrossRef]
- Breakthrough Infections: Coronavirus after Vaccination. Available online: https://www.hopkinsmedicine.org/health/conditions-and-diseases/coronavirus/breakthrough-infections-coronavirus-after-vaccination (accessed on 27 June 2023).
- Hoffmann, M.; Arora, P.; Nehlmeier, I.; Kempf, A.; Cossmann, A.; Schulz, S.R.; Morillas Ramos, G.; Manthey, L.A.; Jäck, H.-M.; Behrens, G.M.N.; et al. Profound Neutralization Evasion and Augmented Host Cell Entry Are Hallmarks of the Fast-Spreading SARS-CoV-2 Lineage XBB.1.5. Cell. Mol. Immunol. 2023, 20, 419–422. [Google Scholar] [CrossRef]
- Wang, X.; Jiang, S.; Jiang, S.; Li, X.; Ai, J.; Lin, K.; Lv, S.; Zhang, S.; Li, M.; He, X.; et al. Neutralization of SARS-CoV-2 BQ.1.1 and XBB.1.5 by Breakthrough Infection Sera from Previous and Current Waves in China. bioRxiv 2023, 2023, 2023.02.07.527406. [Google Scholar]
- Statement on the Fifteenth Meeting of the International Health Regulations (2005) Emergency Committee Regarding the Coronavirus Disease (COVID-19) Pandemic. Available online: https://www.who.int/news/item/05-05-2023-statement-on-the-fifteenth-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-coronavirus-disease-(covid-19)-pandemic (accessed on 6 May 2023).

| Variants | Mutations in the Spike |
|---|---|
| D614G | D614G |
| BA.1 | A67V, Δ69–70, T95I, G142D/Δ143–145, Δ211/L212I, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F |
| BA.4/5 | T19I, LPPA24–27S, Δ69–70, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452R, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| BQ.1.1 | T19I, LPPA24–27S, Δ69–70, G142D, V213G, G339D, R346T, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, K444T, L452R, N460K, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| XBB | T19I, LPPA24–27S, V83A, G142D, Δ144, H146Q, Q183E, V213E, G339H, R346T, L368I, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, V445P, G446S, N460K, S477N, T478K, E484A, F486S, F490S, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| XBB.1.5 | T19I, LPPA24–27S, V83A, G142D, Δ144, H146Q, Q183E, V213E, G252V, G339H, R346T, L368I, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, V445P, G446S, N460K, S477N, T478K, E484A, F486P, F490S, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| CH.1.1 | T19I, LPPA24–27S, G142D, K147E, W152R, F157L, I210V, V213G, G257S, G339H, R346T, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, K444T, G446S, L452R, N460K, S477N, T478K, E484A, F486S, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| FD.1.1 | T19I, LPPA24–27S, V83A, G142D, Δ144, H146Q, Q183E, V213E, G252V, G339H, R346T, L368I, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, V445P, G446S, F456L, N460K, S477N, T478K, E484A, F486P, F490S, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K |
| Vaccinated Cohort | ||
|---|---|---|
| Number (n) a | 18 | |
| Age b | 59 (51–64) | |
| Sex a | Female (n) | 11 |
| Male (n) | 7 | |
| Days between the fourth and fifth doses b | 148 (129–156) | |
| Fifth dose (n) a | Pfizer monovalent | 2 |
| Moderna monovalent | 0 | |
| Pfizer BA.4/5 | 13 | |
| Moderna BA.1 | 3 | |
| Days between the fourth dose and W3-Va4 b | 29 (23–40) | |
| Days between the fourth dose and M4-Va4 b | 124 (116–133) | |
| Days between the fifth dose and W3-Va5 b | 22 (21–27) | |
| No Recent BTI | Recent BTI | ||
|---|---|---|---|
| Number (n) a | 9 | 9 | |
| Age b | 59 (50–67) | 59 (49–63) | |
| Sex a | Female (n) | 7 | 4 |
| Male (n) | 2 | 5 | |
| Days between the fourth and fifth doses b | 151 (118–154) | 147 (136–192) | |
| Fifth dose (n) a | Pfizer monovalent | 1 | 1 |
| Moderna monovalent | 0 | 0 | |
| Pfizer BA.4/5 | 6 | 7 | |
| Moderna BA.1 | 2 | 1 | |
| Days between the fourth dose and W3-Va4 b | 36 (24–43) | 27 (21–38) | |
| Days between the fourth dose and M4-Va4 b | 128 (118–148) | 124 (111–129) | |
| Days between the fifth dose and W3-Va5 b | 21 (21–25) | 26 (19–37) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tauzin, A.; Beaudoin-Bussières, G.; Benlarbi, M.; Nayrac, M.; Bo, Y.; Gendron-Lepage, G.; Medjahed, H.; Perreault, J.; Gokool, L.; Arlotto, P.; et al. Humoral Responses Elicited after a Fifth Dose of SARS-CoV-2 mRNA Bivalent Vaccine. Viruses 2023, 15, 1926. https://doi.org/10.3390/v15091926
Tauzin A, Beaudoin-Bussières G, Benlarbi M, Nayrac M, Bo Y, Gendron-Lepage G, Medjahed H, Perreault J, Gokool L, Arlotto P, et al. Humoral Responses Elicited after a Fifth Dose of SARS-CoV-2 mRNA Bivalent Vaccine. Viruses. 2023; 15(9):1926. https://doi.org/10.3390/v15091926
Chicago/Turabian StyleTauzin, Alexandra, Guillaume Beaudoin-Bussières, Mehdi Benlarbi, Manon Nayrac, Yuxia Bo, Gabrielle Gendron-Lepage, Halima Medjahed, Josée Perreault, Laurie Gokool, Pascale Arlotto, and et al. 2023. "Humoral Responses Elicited after a Fifth Dose of SARS-CoV-2 mRNA Bivalent Vaccine" Viruses 15, no. 9: 1926. https://doi.org/10.3390/v15091926
APA StyleTauzin, A., Beaudoin-Bussières, G., Benlarbi, M., Nayrac, M., Bo, Y., Gendron-Lepage, G., Medjahed, H., Perreault, J., Gokool, L., Arlotto, P., Morrisseau, C., Tremblay, C., Kaufmann, D. E., Martel-Laferrière, V., Levade, I., Côté, M., Bazin, R., & Finzi, A. (2023). Humoral Responses Elicited after a Fifth Dose of SARS-CoV-2 mRNA Bivalent Vaccine. Viruses, 15(9), 1926. https://doi.org/10.3390/v15091926








