Partial Alleviation of Homologous Superinfection Exclusion of SeMNPV Latently Infected Cells by G1 Phase Infection and G2/M Phase Arrest
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Virus Infection
2.2. Cell Cycle Synchronization
2.3. Cell Cycle Analysis by Flow Cytometry
2.4. Quantitative Real-Time PCR (qRT-PCR)
2.5. Viral Production Analysis in Infected Cells
2.6. Statistical Analysis
3. Results
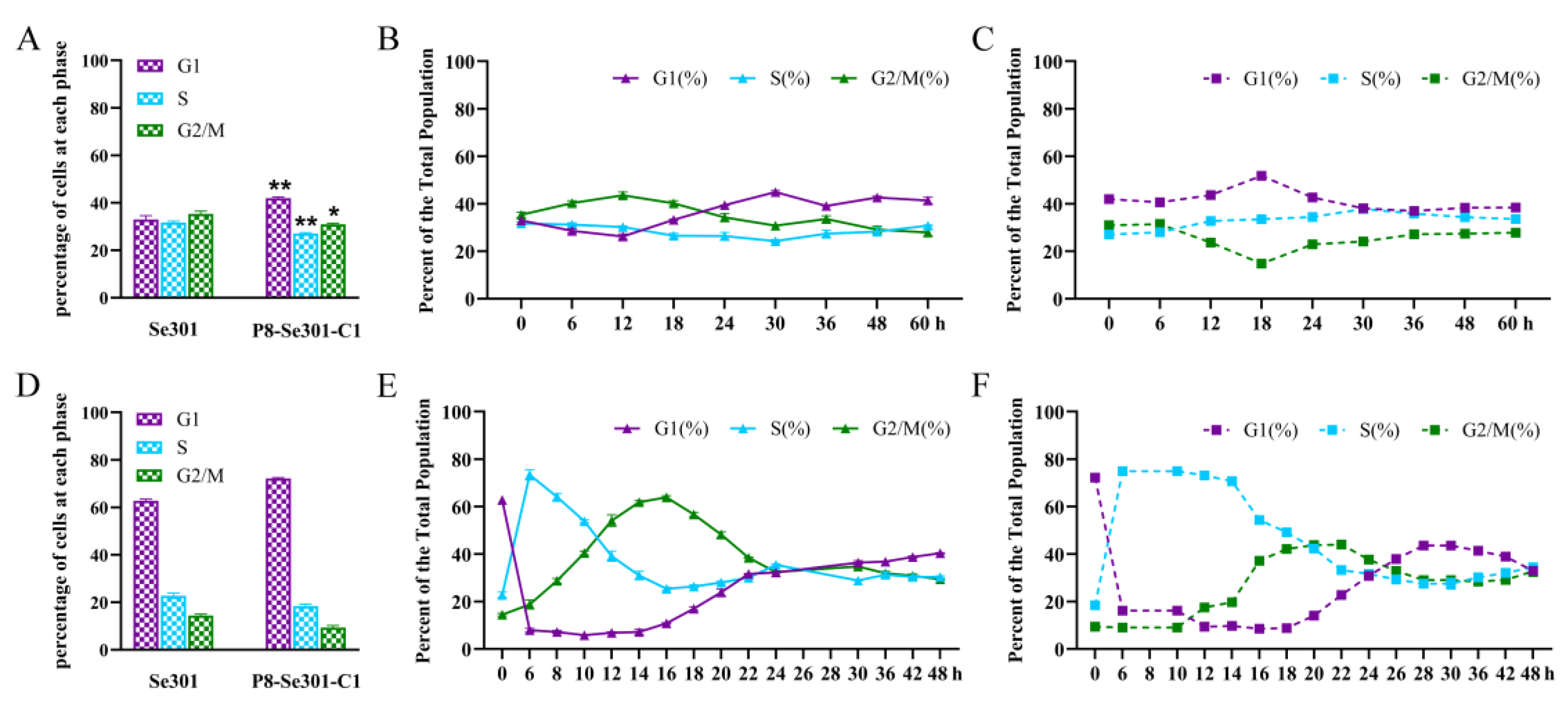
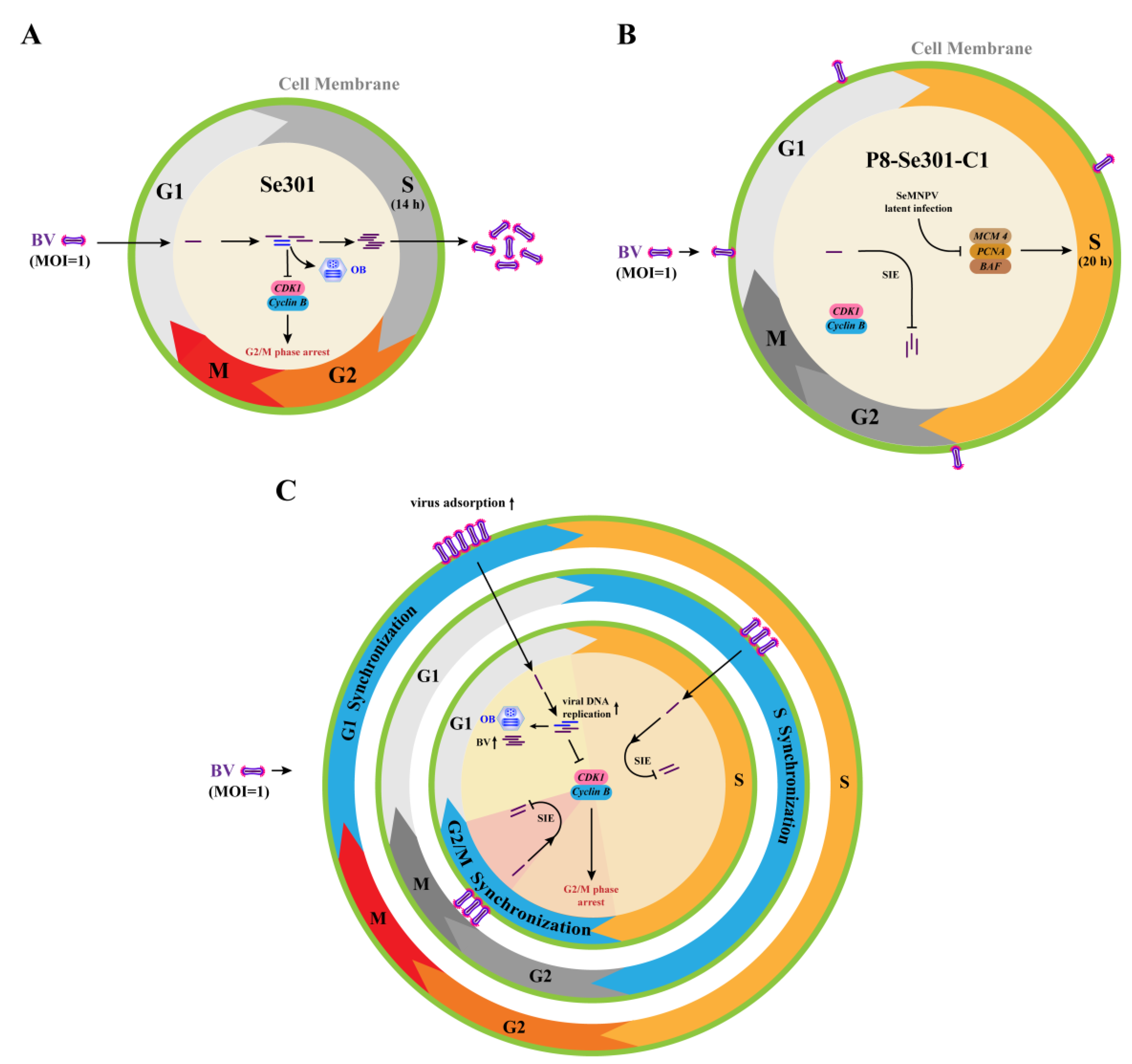
3.1. Cell Cycle Distribution and Differences between Se301 and P8-Se301-C1 Cells
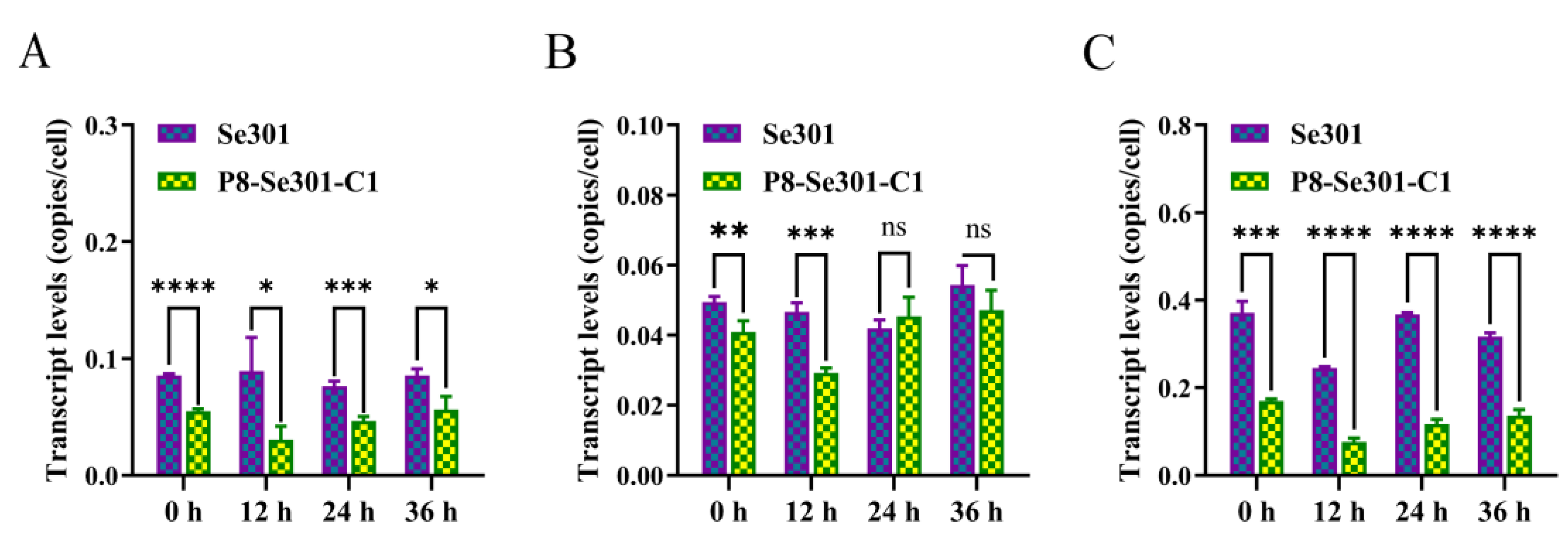
3.2. Transcription Analysis of DNA Replication-Related Genes in Se301 and P8-Se301-C1 Cells
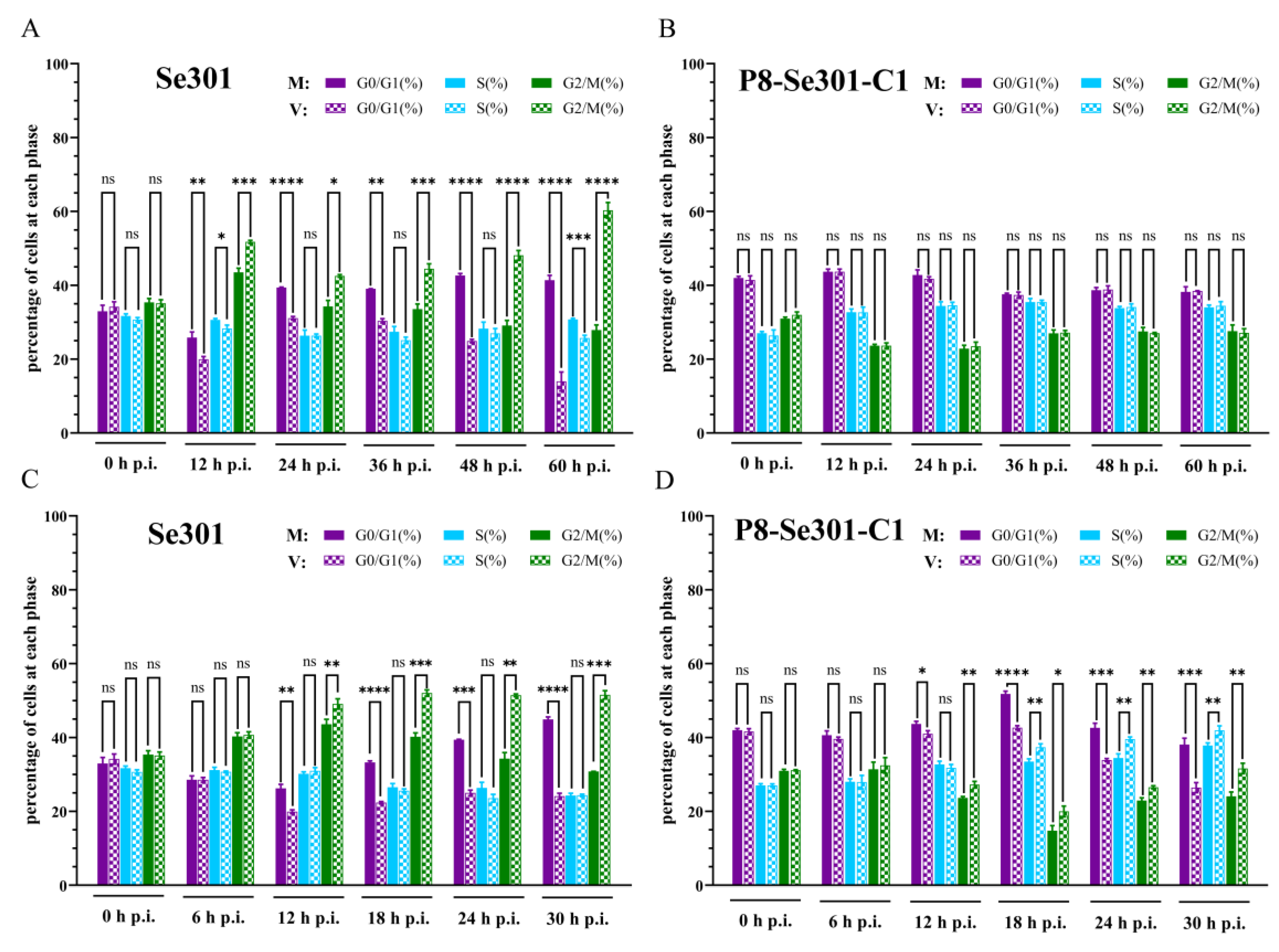
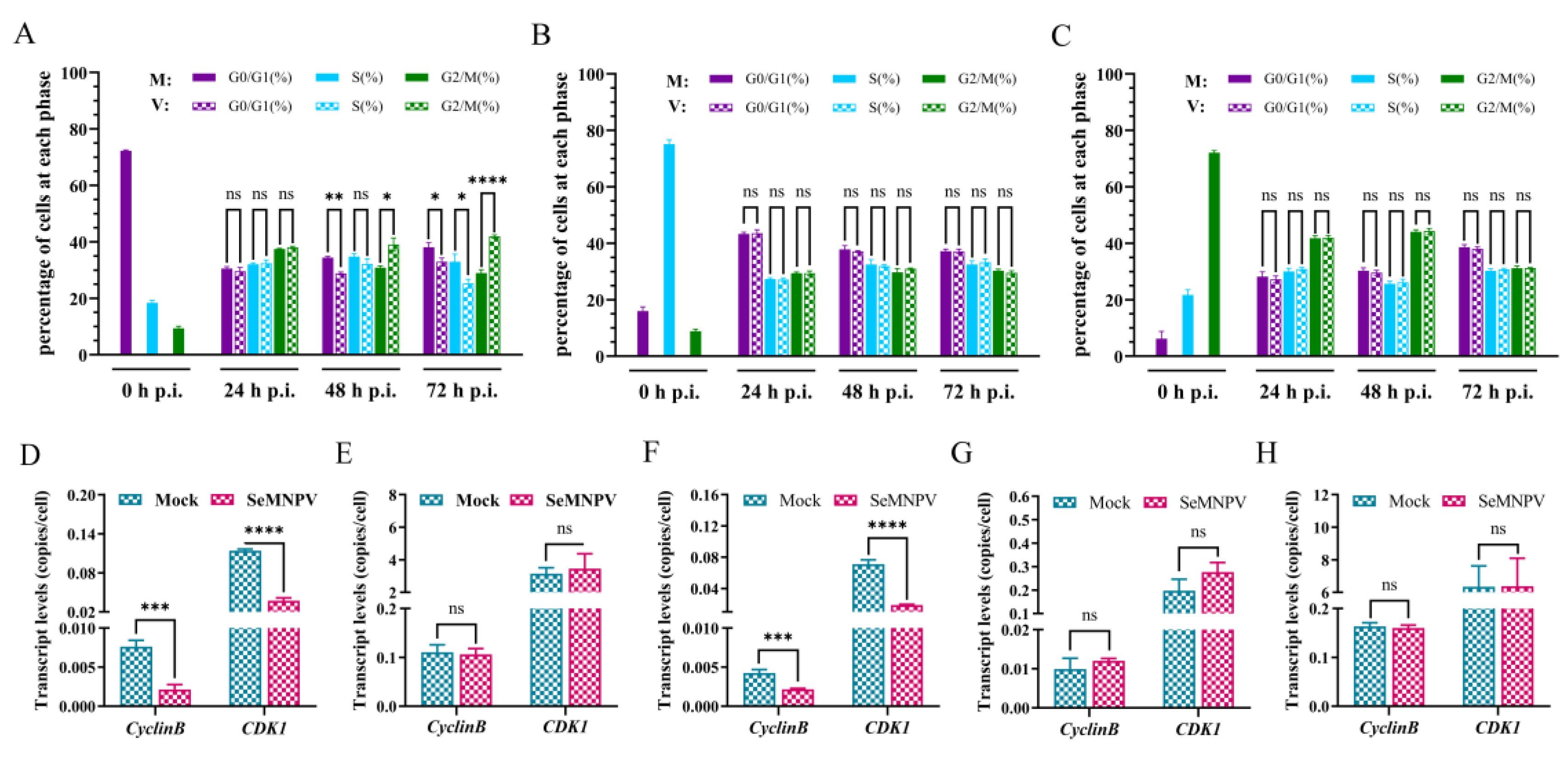
3.3. Effects of a Homologous Virus SeMNPV and Heterologous Virus AcMNPV Infection Affects the Cell Cycle of Se301 and P8-Se301-C1 Cells
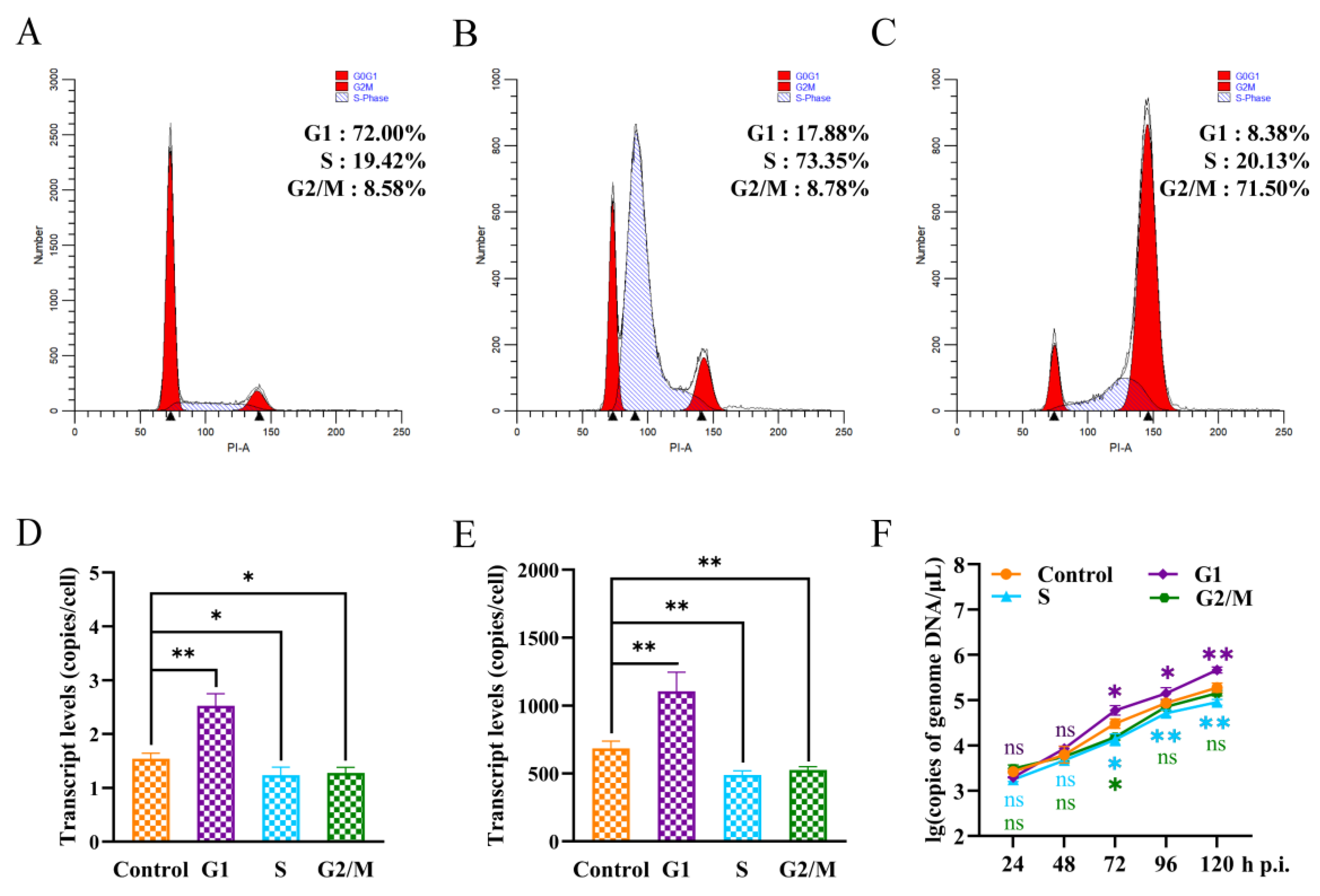
3.4. P8-Se301-C1 Cells in G1 Phase Were More Susceptible to SeMNPV Superinfection
3.5. SeMNPV Superinfection of G1 Phase P8-Se301-C1 Cells Induced G2/M Phase Arrest and Downregulated Cyclin B and CDK1 Expression
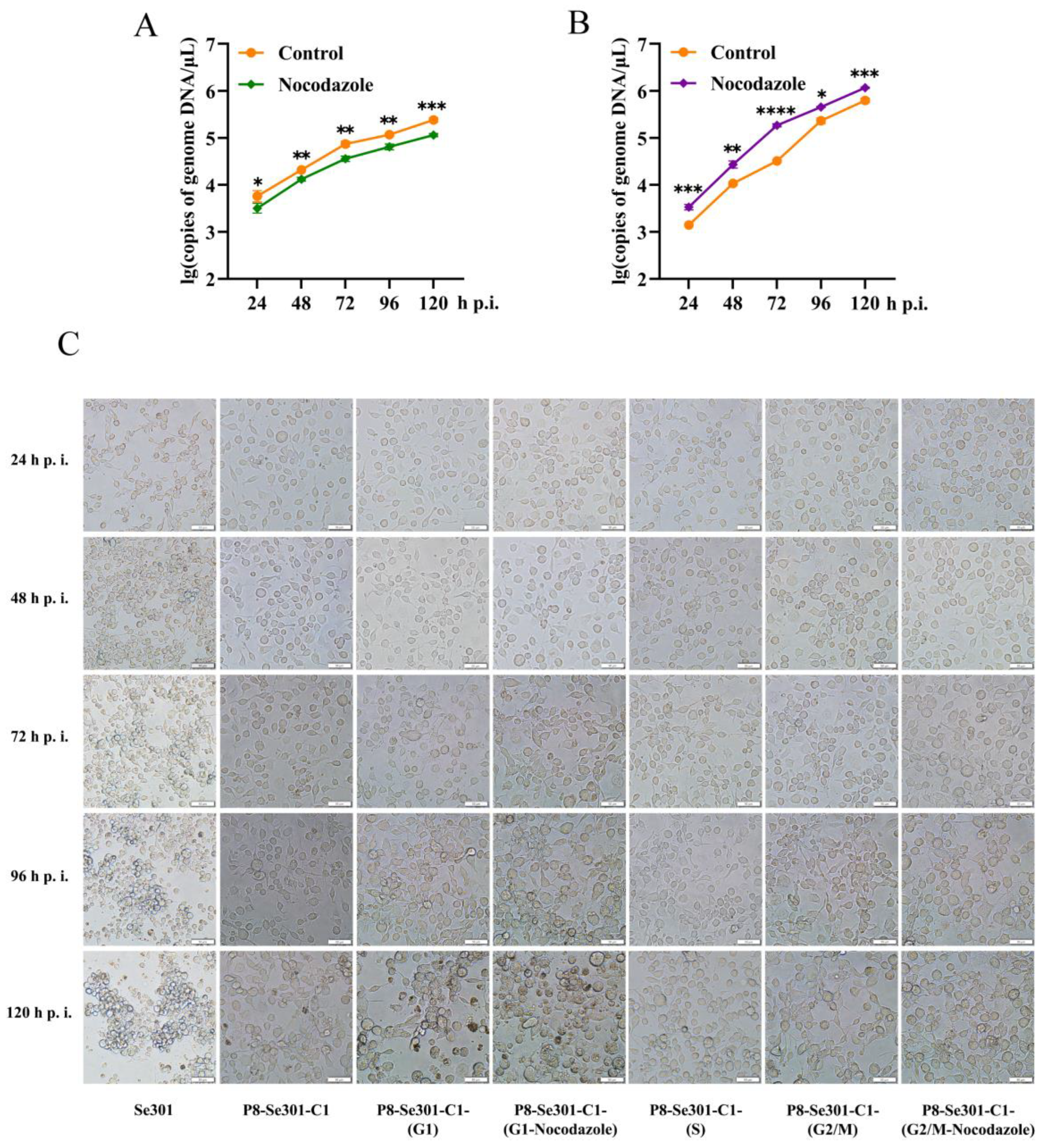
3.6. G2/M Phase Arrest Was Required for SeMNPV Replication in P8-Se301-C1 Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jakoby, M.; Schnittger, A. Cell cycle and differentiation. Curr. Opin. Plant Biol. 2004, 7, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Matthews, H.K.; Bertoli, C.; De Bruin, R.A.M. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2022, 23, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Adeyemi, R.O.; Pintel, D.J. Parvovirus-induced depletion of Cyclin B1 prevents mitotic entry of infected cells. PLoS Pathog. 2014, 10, e1003891. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Y.; Wang, S.; Meng, X.; Song, F.; Huo, W.; Zhang, S.; Chang, J.; Li, J.; Zheng, B.; et al. Coxsackievirus A6 induces cell cycle arrest in G0/G1 phase for viral production. Front. Cell. Infect. Microbiol. 2018, 8, 279. [Google Scholar] [CrossRef] [PubMed]
- Davy, C.; Doorbar, J. G2/M cell cycle arrest in the life cycle of viruses. Virology 2007, 368, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Caffarelli, N.; Fehr, A.R.; Yu, D. Cyclin A degradation by primate cytomegalovirus protein pUL21a counters its innate restriction of virus replication. PLoS Pathog. 2013, 9, e1003825. [Google Scholar] [CrossRef]
- Spector, D.H. Human cytomegalovirus riding the cell cycle. Med. Microbiol. Immunol. 2015, 204, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Shi, D.; Xing, X.; Qi, S.; Yang, D.; Zhang, J.; Han, Y.; Zhu, Q.; Sun, H.; Wang, X.; et al. Coronavirus porcine epidemic diarrhea virus nucleocapsid protein interacts with p53 To Induce cell cycle arrest in S-phase and promotes viral replication. J. Virol. 2021, 95, e0018721. [Google Scholar] [CrossRef] [PubMed]
- Bagga, S.; Bouchard, M.J. Cell cycle regulation during viral infection. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2014; Volume 1170, pp. 165–227. [Google Scholar] [CrossRef]
- Danquah, J.O.; Botchway, S.; Jeshtadi, A.; King, L.A. Direct interaction of baculovirus capsid proteins VP39 and EXON0 with kinesin-1 in insect cells determined by fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy. J. Virol. 2012, 86, 844–853. [Google Scholar] [CrossRef]
- Dong, Z.; Zhang, X.; Xiao, M.; Li, K.; Wang, J.; Chen, P.; Hu, Z.; Lu, C.; Pan, M. Baculovirus LEF-11 interacts with BmIMPI to induce cell cycle arrest in the G2/M phase for viral replication. Pestic. Biochem. Physiol. 2022, 188, 105231. [Google Scholar] [CrossRef]
- Bressy, C.; Droby, G.N.; Maldonado, B.D.; Steuerwald, N.; Grdzelishvili, V.Z. Cell cycle arrest in G2/M Phase enhances replication of interferon-sensitive cytoplasmic RNA viruses via inhibition of antiviral gene expression. J. Virol. 2019, 93, e01885-18. [Google Scholar] [CrossRef]
- Rohrmann, G.F. Baculovirus Molecular Biology, 4th ed.; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2019.
- Blissard, G.W.; Theilmann, D.A. Baculovirus entry and egress from insect cells. Annu. Rev. Virol. 2018, 5, 113–139. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Kobayashi, M. Cell-cycle perturbation in Sf9 cells infected with Autographa californica nucleopolyhedrovirus. Virology 1999, 258, 176–188. [Google Scholar] [CrossRef]
- Xiao, Q.; Dong, Z.Q.; Zhu, Y.; Zhang, Q.; Yang, X.; Xiao, M.; Chen, P.; Lu, C.; Pan, M.H. Bombyx mori nucleopolyhedrovirus (BmNPV) induces G2/M arrest to promote viral multiplication by depleting BmCDK1. Insects 2021, 12, 1098. [Google Scholar] [CrossRef] [PubMed]
- Prikhod’ko, E.A.; Miller, L.K. Role of baculovirus IE2 and its RING finger in cell cycle arrest. J. Virol. 1998, 72, 684–692. [Google Scholar] [CrossRef] [PubMed]
- Belyavskyi, M.; Braunagel, S.C.; Summers, M.D. The structural protein ODV-EC27 of Autographa californica nucleopolyhedrovirus is a multifunctional viral cyclin. Proc. Natl. Acad. Sci. USA 1998, 95, 11205–11210. [Google Scholar] [CrossRef]
- Saito, T.; Dojima, T.; Toriyama, M.; Park, E.Y. The effect of cell cycle on GFPuv gene expression in the baculovirus expression system. J. Biotechnol. 2002, 93, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.; Wei, W.; Xu, P.; Qin, Q.; Chen, J.; Chen, X.Z.; Chen, X.P.; Zhao, X.H. The cell cycle phase affects the potential of cells to replicate Autographa californica multiple nucleopolyhedrovirus. Acta Virol. 2012, 56, 133–137. [Google Scholar] [CrossRef]
- Lacey, L.A.; Grzywacz, D.; Shapiro-Ilan, D.I.; Frutos, R.; Brownbridge, M.; Goettel, M.S. Insect pathogens as biological control agents: Back to the future. J. Invertebr. Pathol. 2015, 132, 1–41. [Google Scholar] [CrossRef]
- Williams, T.; Virto, C.; Murillo, R.; Caballero, P. Covert infection of insects by baculoviruses. Front. Microbiol. 2017, 8, 1337. [Google Scholar] [CrossRef]
- Burden, J.P.; Nixon, C.P.; Hodgkinson, A.E.; Possee, R.D.; Sait, S.M.; King, L.A.; Hails, R.S. Covert infections as a mechanism for long-term persistence of baculoviruses. Ecol. Lett. 2003, 6, 524–531. [Google Scholar] [CrossRef]
- Vilaplana, L.; Wilson, K.; Redman, E.M.; Cory, J.S. Pathogen persistence in migratory insects: High levels of vertically-transmitted virus infection in field populations of the African armyworm. Evol. Ecol. 2010, 24, 147–160. [Google Scholar] [CrossRef]
- Cabodevilla, O.; Villar, E.; Virto, C.; Murillo, R.; Williams, T.; Caballero, P. Intra- and intergenerational persistence of an insect nucleopolyhedrovirus: Adverse effects of sublethal disease on host development, reproduction, and susceptibility to superinfection. Appl. Environ. Microbiol. 2011, 77, 2954–2960. [Google Scholar] [CrossRef]
- Weng, Q.; Yang, K.; Xiao, W.; Yuan, M.; Zhang, W.; Pang, Y. Establishment of an insect cell clone that harbours a partial baculoviral genome and is resistant to homologous virus infection. J. General. Virol. 2009, 90, 2871–2876. [Google Scholar] [CrossRef]
- Sun, Q.; Tsurimoto, T.; Juillard, F.; Li, L.; Li, S.; De León Vázquez, E.; Chen, S.; Kaye, K. Kaposi’s sarcoma-associated herpesvirus LANA recruits the DNA polymerase clamp loader to mediate efficient replication and virus persistence. Proc. Natl. Acad. Sci. USA 2014, 111, 11816–11821. [Google Scholar] [CrossRef]
- Dabral, P.; Uppal, T.; Rossetto, C.C.; Verma, S.C. Minichromosome maintenance proteins cooperate with LANA during the G1/S phase of the cell cycle to support viral DNA replication. J. Virol. 2019, 93, e02256-18. [Google Scholar] [CrossRef] [PubMed]
- Schang, L.M.; Hossain, A.; Jones, C. The latency-related gene of bovine herpesvirus 1 encodes a product which inhibits cell cycle progression. J. Virol. 1996, 70, 3807–3814. [Google Scholar] [CrossRef]
- An, F.Q.; Compitello, N.; Horwitz, E.; Sramkoski, M.; Knudsen, E.S.; Renne, R. The latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus modulates cellular gene expression and protects lymphoid cells from p16 INK4A-induced cell cycle arrest. J. Biol. Chem. 2005, 280, 3862–3874. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Qu, J.; Peng, Q.; Gan, R. Molecular mechanisms of EBV-driven cell cycle progression and oncogenesis. Med. Microbiol. Immunol. 2019, 208, 573–583. [Google Scholar] [CrossRef]
- Kang, M.S.; Kieff, E. Epstein-Barr virus latent genes. Exp. Mol. Med. 2015, 47, e131. [Google Scholar] [CrossRef]
- Laliberte, J.P.; Moss, B. A Novel Mode of Poxvirus Superinfection Exclusion That Prevents Fusion of the Lipid Bilayers of Viral and Cellular Membranes. J. Virol. 2014, 88, 9751–9768. [Google Scholar] [CrossRef] [PubMed]
- Biryukov, J.; Meyers, C. Superinfection exclusion between two high-risk human papillomavirus types during a coinfection. J. Virol. 2018, 92, e01993-17. [Google Scholar] [CrossRef]
- Zhang, X.F.; Sun, R.; Guo, Q.; Zhang, S.; Meulia, T.; Halfmann, R.; Li, D.; Qu, F. A self-perpetuating repressive state of a viral replication protein blocks superinfection by the same virus. PLoS Pathog. 2017, 13, e1006253. [Google Scholar] [CrossRef]
- Berngruber, T.W.; Weissing, F.J.; Gandon, S. Inhibition of superinfection and the evolution of viral latency. J. Virol. 2010, 84, 10200–10208. [Google Scholar] [CrossRef]
- Beperet, I.; Irons, S.L.; Simón, O.; King, L.A.; Williams, T.; Possee, R.D.; López-Ferber, M.; Caballero, P. Superinfection exclusion in alphabaculovirus infections is concomitant with actin reorganization. J. Virol. 2014, 88, 3548–3556. [Google Scholar] [CrossRef]
- Fang, Z.; Shao, J.; Weng, Q. De novo transcriptome analysis of Spodoptera exigua multiple nucleopolyhedrovirus (SeMNPV) genes in latently infected Se301 cells. Virol. Sin. 2016, 31, 425–436. [Google Scholar] [CrossRef]
- Arrizubieta, M.; Williams, T.; Caballero, P.; Simón, O. Selection of a nucleopolyhedrovirus isolate from Helicoverpa armigera as the basis for a biological insecticide. Pest Manag. Sci. 2014, 70, 967–976. [Google Scholar] [CrossRef]
- Wu, W.; Lin, T.; Pan, L.; Yu, M.; Li, Z.; Pang, Y.; Yang, K. Autographa californica multiple nucleopolyhedrovirus nucleocapsid assembly is interrupted upon deletion of the 38K gene. J. Virol. 2006, 80, 11475–11485. [Google Scholar] [CrossRef] [PubMed]
- Braunagel, S.C.; Parr, R.; Belyavskyi, M.; Summers, M.D. Autographa californica nucleopolyhedrovirus infection results in Sf9 cell cycle arrest at G2/M phase. Virology 1998, 244, 195–211. [Google Scholar] [CrossRef]
- Zhou, R.; Yu, Z.H.; Li, X.Q.; Jia, F.; Wu, J.H.; Chen, X. Heliocoverpa armigera single nucleocapsid nucleopolyhedrovirus induces Hz-AM1 cell cycle arrest at the G2 phase with accumulation of Cyclin B1. Virus Res. 2004, 105, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, M.L.; Sherlock, G.; Saldanha, A.J.; Murray, J.I.; Ball, C.A.; Alexander, K.E.; Matese, J.C.; Perou, C.M.; Hurt, M.M.; Brown, P.O.; et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 2002, 13, 1977–2000. [Google Scholar] [CrossRef] [PubMed]
- Spencer, S.L.; Cappell, S.D.; Tsai, F.C.; Overton, K.W.; Wang, C.L.; Meyer, T. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell 2013, 155, 369–383. [Google Scholar] [CrossRef]
- Yano, S.; Miwa, S.; Mii, S.; Hiroshima, Y.; Uehara, F.; Kishimoto, H.; Tazawa, H.; Zhao, M.; Bouvet, M.; Fujiwara, T.; et al. Cancer cells mimic in vivo spatial-temporal cell-cycle phase distribution and chemosensitivity in 3-dimensional Gelfoam® histoculture but not 2-dimensional culture as visualized with real-time FUCCI imaging. Cell Cycle 2015, 14, 808–819. [Google Scholar] [CrossRef] [PubMed]
- Limas, J.C.; Cook, J.G. Preparation for DNA replication: The key to a successful S phase. FEBS Lett. 2019, 593, 2853–2867. [Google Scholar] [CrossRef]
- Lin, Q.; Yu, B.; Wang, X.; Zhu, S.; Zhao, G.; Jia, M.; Huang, F.; Xu, N.; Ren, H.; Jiang, Q.; et al. K6-linked SUMOylation of BAF regulates nuclear integrity and DNA replication in mammalian cells. Proc. Natl. Acad. Sci. USA 2020, 117, 10378–10387. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.; Cao, T.V.; Winter, S.L.; Alexandrow, M.G. Mammalian MCM loading in late-G1 coincides with Rb hyperphosphorylation and the transition to post-transcriptional control of progression into S-phase. PLoS ONE 2009, 4, e5462. [Google Scholar] [CrossRef]
- Iwahori, S.; Ikeda, M.; Kobayashi, M. Association of Sf9 cell proliferating cell nuclear antigen with the DNA replication site of Autographa californica multicapsid nucleopolyhedrovirus. J. Gen. Virol. 2004, 85, 2857–2862. [Google Scholar] [CrossRef]
- Xin, X.; Wang, H.; Han, L.; Wang, M.; Fang, H.; Hao, Y.; Li, J.; Zhang, H.; Zheng, C.; Shen, C. Single-Cell analysis of the impact of host cell heterogeneity on infection with Foot-and-Mouth disease virus. J. Virol. 2018, 92, e00179-18. [Google Scholar] [CrossRef]
- Icard, P.; Simula, L. Metabolic oscillations during cell-cycle progression. Trends Endocrinol. Metab. 2022, 33, 447–450. [Google Scholar] [CrossRef]
- Boward, B.; Wu, T.; Dalton, S. Concise review: Control of cell fate through cell cycle and pluripotency networks. Stem Cells 2016, 34, 1427–1436. [Google Scholar] [CrossRef]
- Frye, K.; Renda, F.; Fomicheva, M.; Zhu, X.; Gong, L.; Khodjakov, A.; Kaverina, I. Cell cycle-dependent dynamics of the golgi-centrosome association in motile Cells. Cells 2020, 9, 1069. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Enden, G.; Wei, W.; Zhou, F.; Chen, J.; Merchuk, J.C. Baculovirus transit through insect cell membranes: A mechanistic approach. Chem. Eng. Sci. 2020, 223, 115727. [Google Scholar] [CrossRef]
- Chaurushiya, M.S.; Weitzman, M.D. Viral manipulation of DNA repair and cell cycle checkpoints. DNA Repair. 2009, 8, 1166–1176. [Google Scholar] [CrossRef]
- Flemington, E.K. Herpesvirus lytic replication and the cell cycle: Arresting new developments. J. Virol. 2001, 75, 4475–4481. [Google Scholar] [CrossRef] [PubMed]
- Ohi, R.; Gould, K.L. Regulating the onset of mitosis. Curr. Opin. Cell Biol. 1999, 11, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, H.; Kang, J.; Xu, L.; Zhang, K.; Li, X.; Hou, W.; Wang, Z.; Wang, T. The severe fever with thrombocytopenia syndrome virus NSs protein interacts with CDK1 to induce G2 cell cycle arrest and positively regulate viral replication. J. Virol. 2020, 94, e01575-19. [Google Scholar] [CrossRef]
- Koutsoudakis, G.; Herrmann, E.; Kallis, S.; Bartenschlager, R.; Pietschmann, T. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J. Virol. 2007, 81, 588–598. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Q.-M.; Fang, Z.; Ren, L.; Wu, Q.-S.; Zhang, J.-B.; Liu, Q.-P.; Tan, L.-T.; Weng, Q.-B. Partial Alleviation of Homologous Superinfection Exclusion of SeMNPV Latently Infected Cells by G1 Phase Infection and G2/M Phase Arrest. Viruses 2024, 16, 736. https://doi.org/10.3390/v16050736
Fu Q-M, Fang Z, Ren L, Wu Q-S, Zhang J-B, Liu Q-P, Tan L-T, Weng Q-B. Partial Alleviation of Homologous Superinfection Exclusion of SeMNPV Latently Infected Cells by G1 Phase Infection and G2/M Phase Arrest. Viruses. 2024; 16(5):736. https://doi.org/10.3390/v16050736
Chicago/Turabian StyleFu, Qi-Ming, Zheng Fang, Lou Ren, Qing-Shan Wu, Jun-Bo Zhang, Qiu-Ping Liu, Lei-Tao Tan, and Qing-Bei Weng. 2024. "Partial Alleviation of Homologous Superinfection Exclusion of SeMNPV Latently Infected Cells by G1 Phase Infection and G2/M Phase Arrest" Viruses 16, no. 5: 736. https://doi.org/10.3390/v16050736
APA StyleFu, Q.-M., Fang, Z., Ren, L., Wu, Q.-S., Zhang, J.-B., Liu, Q.-P., Tan, L.-T., & Weng, Q.-B. (2024). Partial Alleviation of Homologous Superinfection Exclusion of SeMNPV Latently Infected Cells by G1 Phase Infection and G2/M Phase Arrest. Viruses, 16(5), 736. https://doi.org/10.3390/v16050736






