Galectin-3-ITGB1 Signaling Mediates Interleukin 10 Production of Hepatic Conventional Natural Killer Cells in Hepatitis B Virus Transgenic Mice and Correlates with Hepatocellular Carcinoma Progression in Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Reagents
2.3. Mononuclear Cell (MNC) Preparation
2.4. Purification of cNK Cells
2.5. Flow Cytometry Analysis
2.6. Electron Microscopic Morphology of NK Cells
2.7. Serum Transaminase Activity Assays
2.8. Galectin-3 Binding Analysis
2.9. Galectin-3 Stimulation In Vitro
2.10. Correlation Analysis
2.11. Statistical Analysis
3. Results
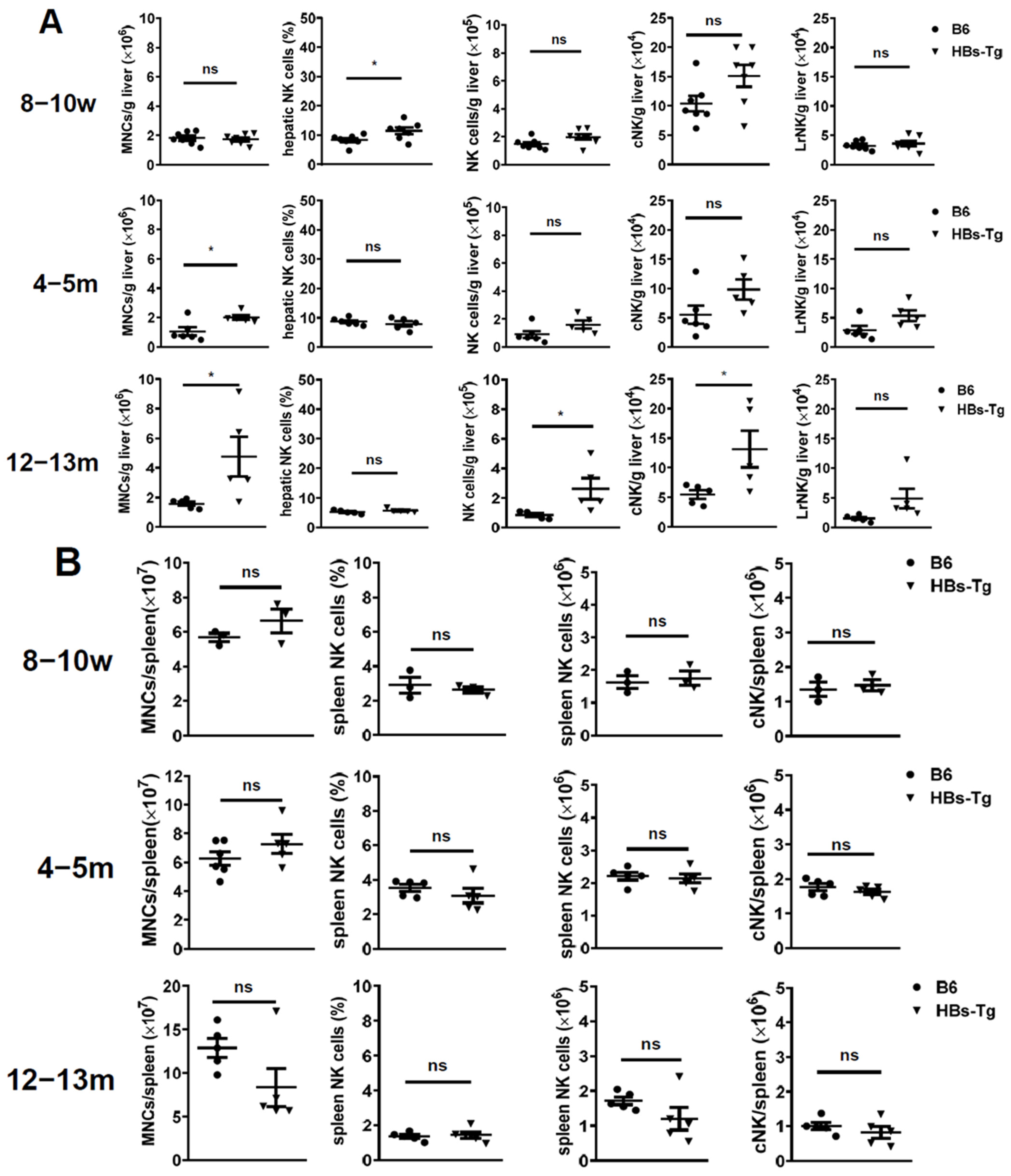
3.1. Increased Hepatic cNK Cells Were Skewed to Produce IL-10 and Exhibited Immune Inhibitory Features with Ageing in HBs-Tg Mice
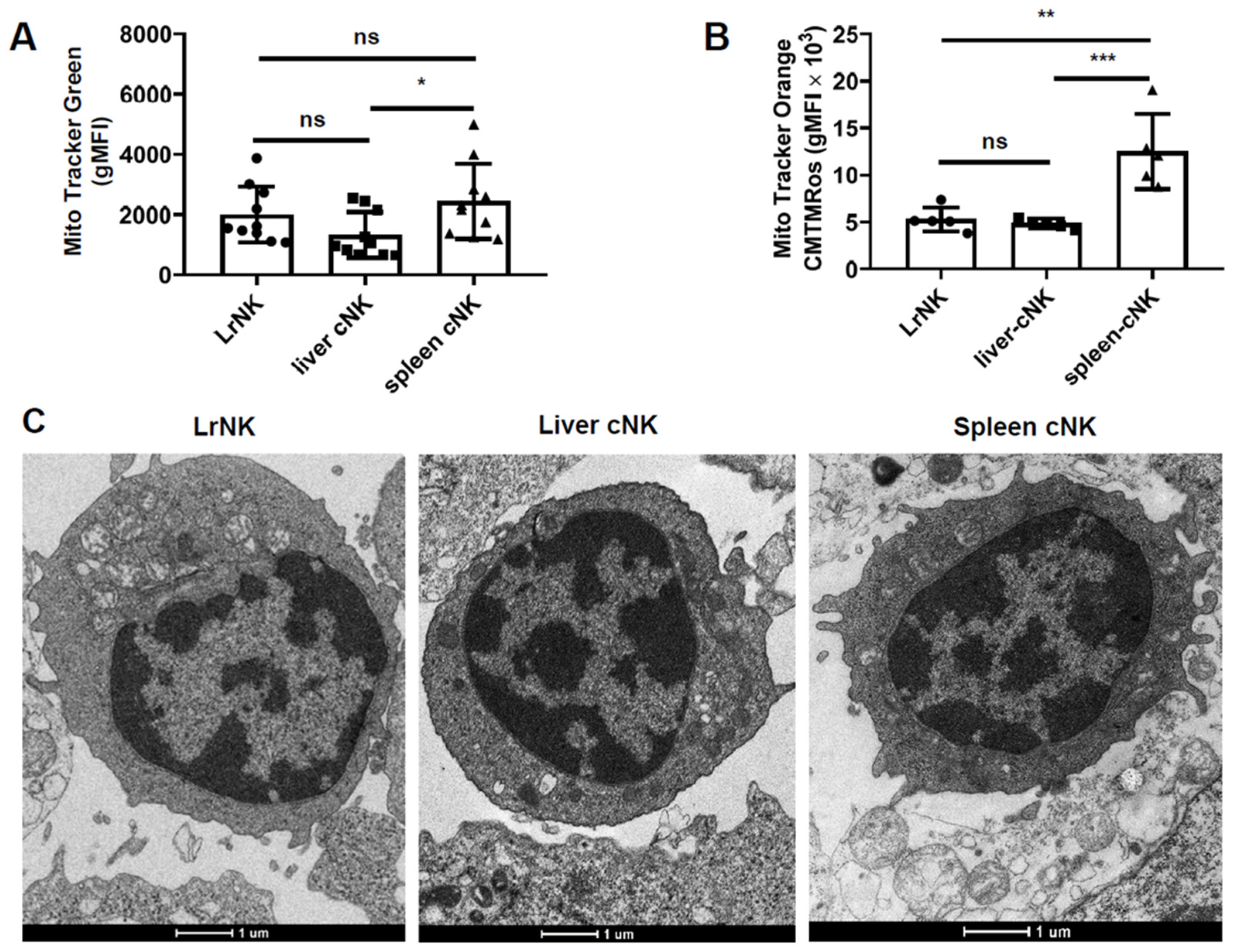
3.2. Hepatic cNK Cells Showed a Distinct Mitochondrial Signature in HBs-Tg Mice
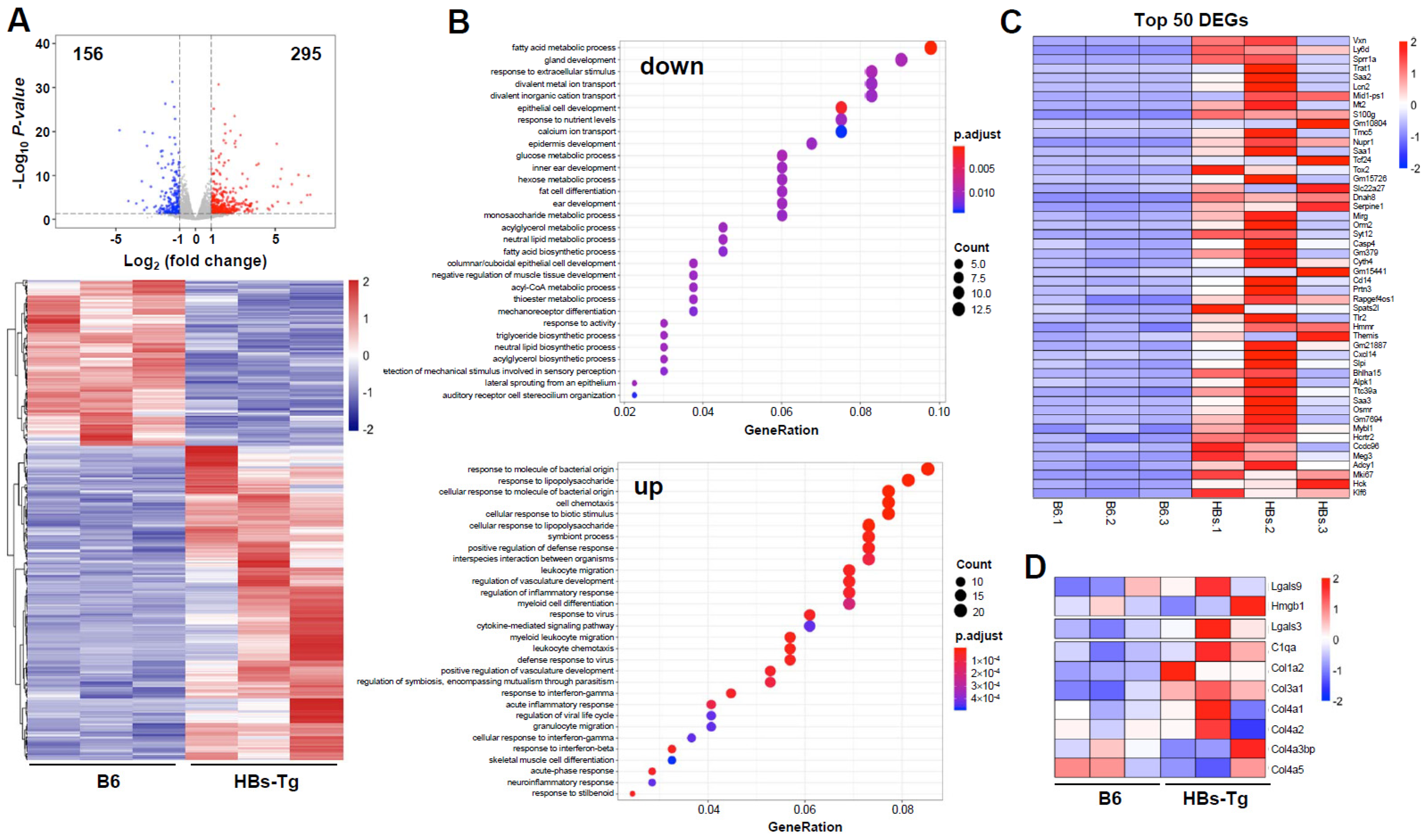
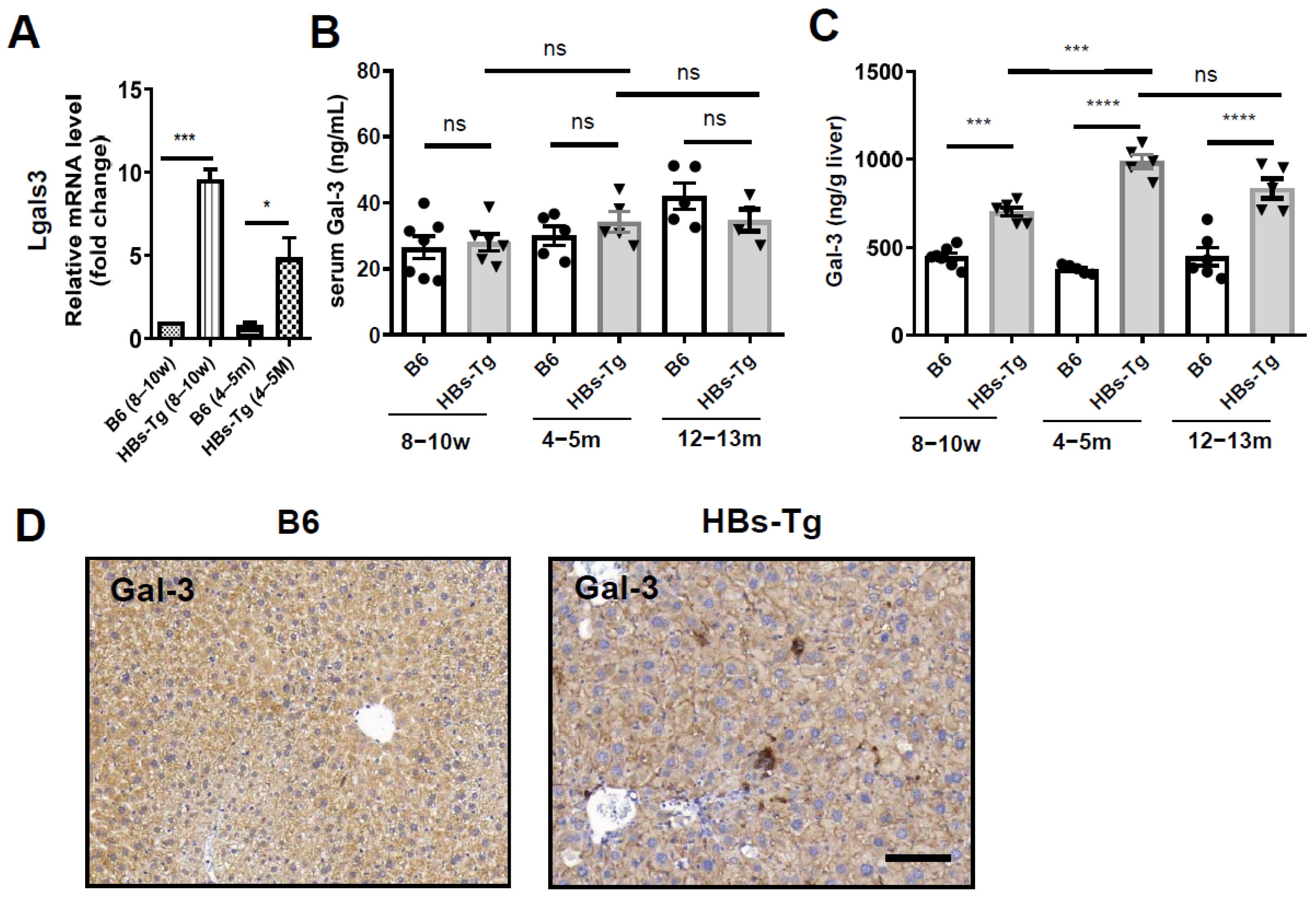
3.3. HBsAg+ Hepatocytes Enhanced Galectin-3 Expression in HBs-Tg Mice
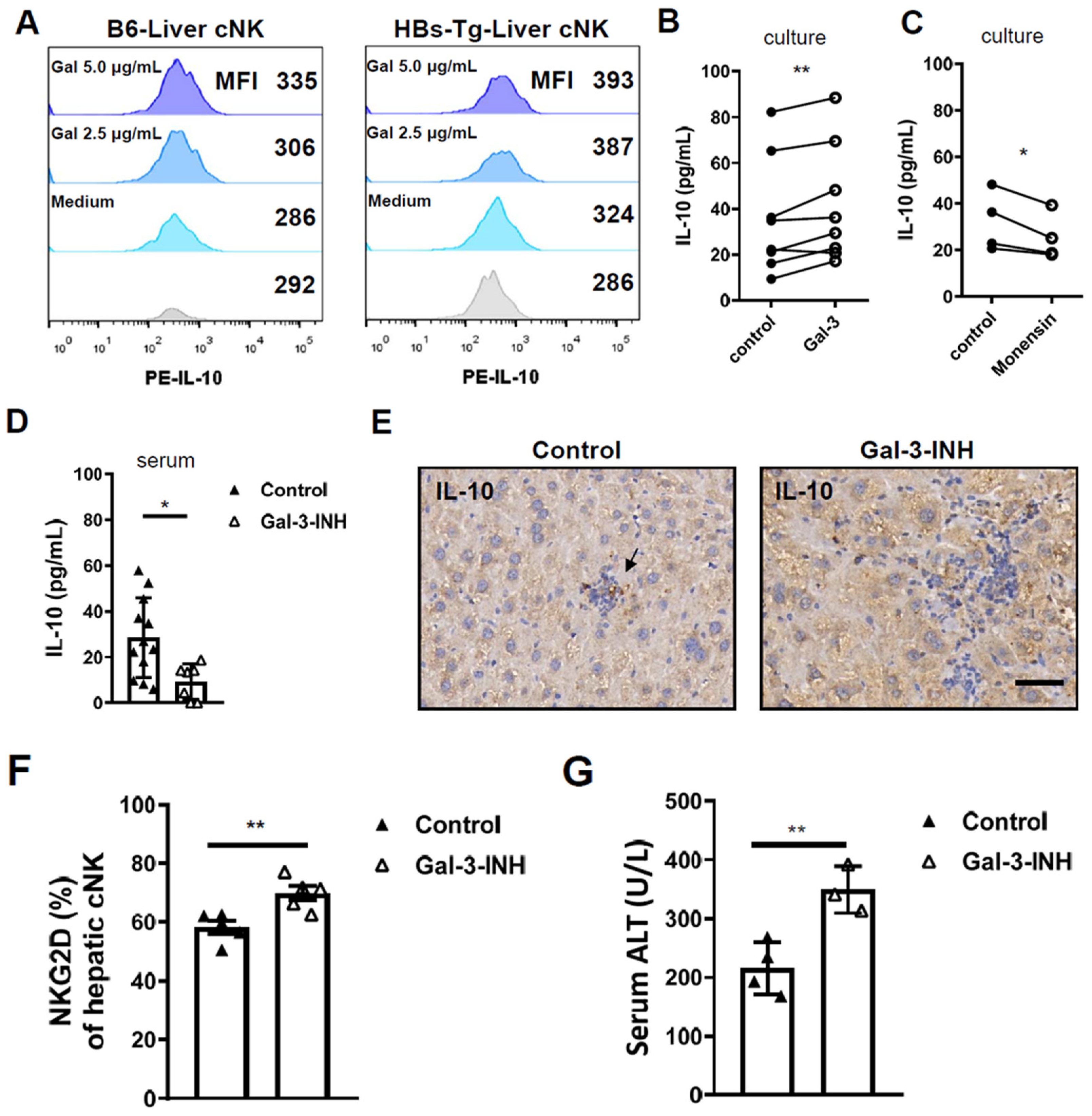
3.4. Galectin-3 Accounted for IL-10 Production in Hepatic cNK Cells and Prevented Liver Injury in HBs-Tg Mice
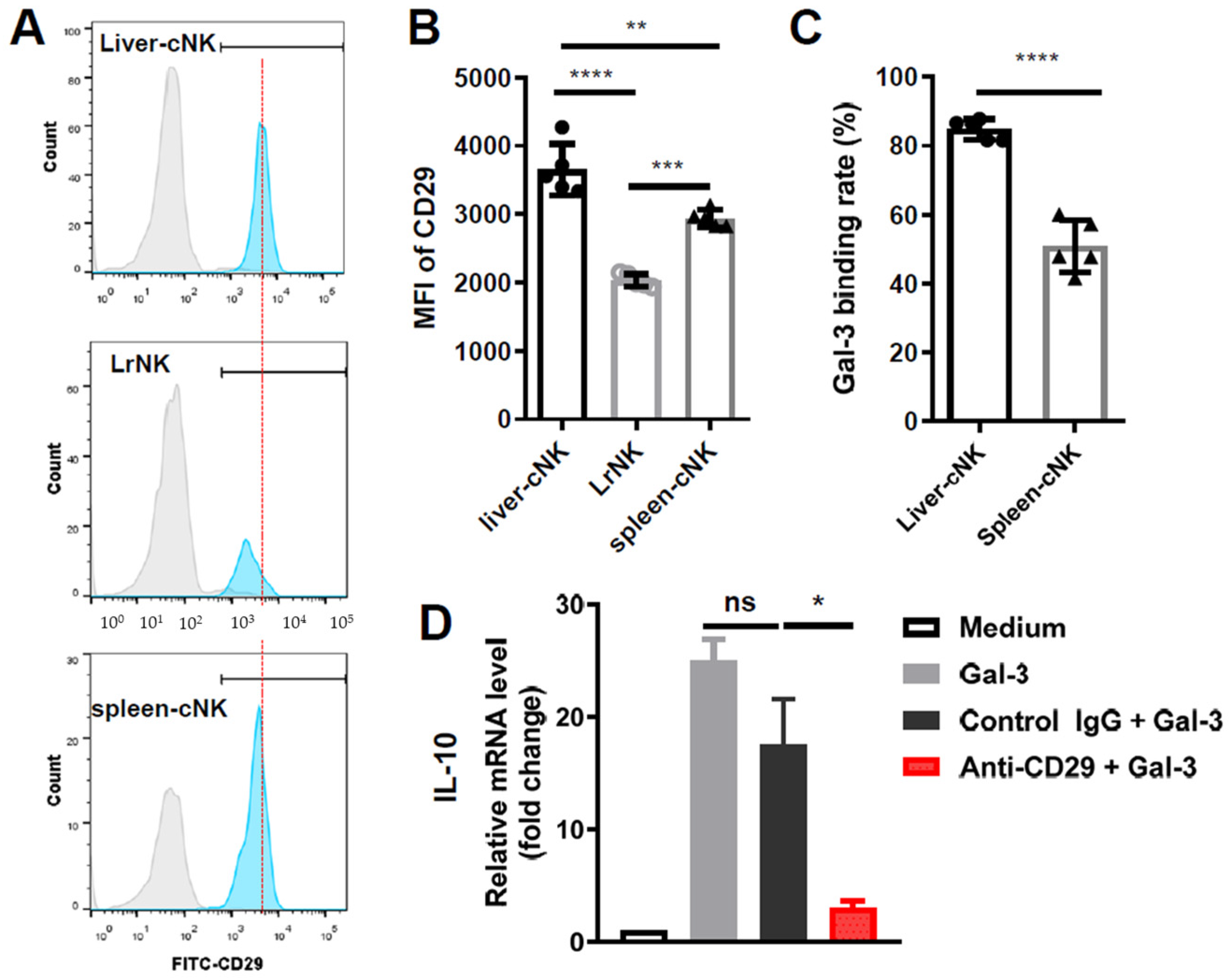
3.5. Galectin-3 Induced IL-10 Transcription via ITGB1 Signaling in Hepatic cNK Cells
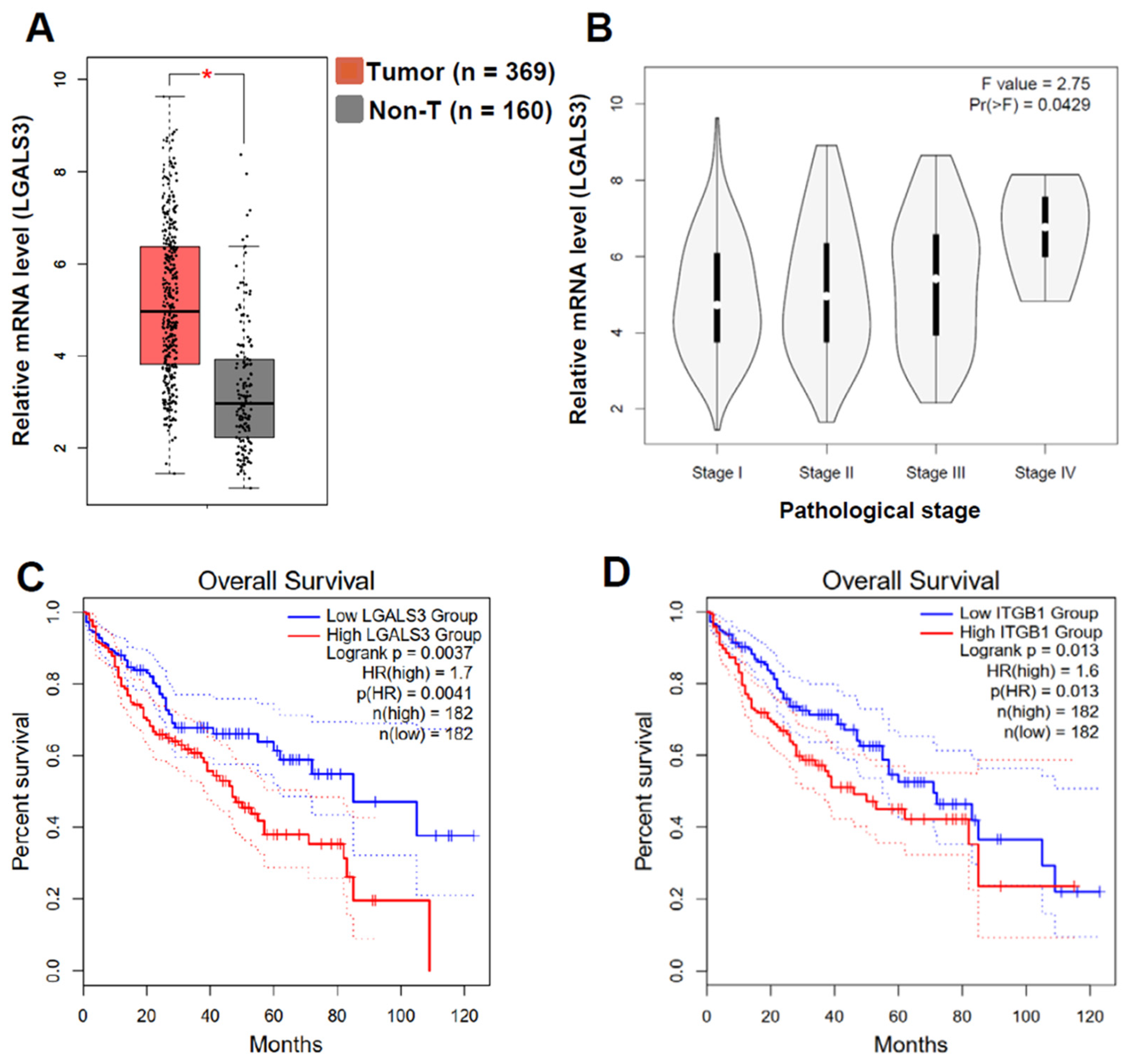
3.6. LGALS3 and ITGB1 Expression Negatively Correlated with the Poor Progression of HCC
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NK | natural killer cell |
| cNK | conventional NK cell |
| LrNK | liver-resident NK cell |
| NKT | natural killer T |
| HBs-Tg | HBV transgenic mice |
| HBV | hepatitis B virus |
| HBsAg | hepatitis B virus surface antigen |
| HCC | hepatocellular carcinoma |
| ALT | alanine aminotransferase |
| AST | aspartate aminotransferase |
| MNC | mononuclear cell |
| PBS | phosphate-buffered saline |
| mAb | monoclonal antibody |
| FITC | fluorescein isothiocyanate |
| IFN-γ | interferon-γ |
| IL | interleukin |
| NKG2D | natural killer cell group 2D |
| ITGB1 | integrin β1 |
References
- Petruzziello, A. Epidemiology of Hepatitis B Virus (HBV) and Hepatitis C Virus (HCV) Related Hepatocellular Carcinoma. Open Virol. J. 2018, 12, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Men, P.; Xiao, Y.; Gao, P.; Lv, M.; Yuan, Q.; Chen, W.; Bai, S.; Wu, J. Hepatitis B infection in the general population of China: A systematic review and meta-analysis. BMC Infect. Dis. 2019, 19, 811. [Google Scholar] [CrossRef] [PubMed]
- Kubes, P.; Jenne, C. Immune Responses in the Liver. Annu. Rev. Immunol. 2018, 36, 247–277. [Google Scholar] [CrossRef] [PubMed]
- Khanam, A.; Chua, J.V.; Kottilil, S. Immunopathology of Chronic Hepatitis B Infection: Role of Innate and Adaptive Immune Response in Disease Progression. Int. J. Mol. Sci. 2021, 22, 5497. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tian, Z. HBV-Induced Immune Imbalance in the Development of HCC. Front. Immunol. 2019, 10, 2048. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Jeong, W.I.; Tian, Z. Liver: An organ with predominant innate immunity. Hepatology 2008, 47, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.D.; Ljunggren, H.G.; La Cava, A.; Van Kaer, L. Organ-specific features of natural killer cells. Nat. Rev. Immunol. 2011, 11, 658–671. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wei, H.; Sun, R.; Dong, Z.; Zhang, J.; Tian, Z. Increased susceptibility to liver injury in hepatitis B virus transgenic mice involves NKG2D-ligand interaction and natural killer cells. Hepatology 2007, 46, 706–715. [Google Scholar] [CrossRef] [PubMed]
- Vilarinho, S.; Ogasawara, K.; Nishimura, S.; Lanier, L.L.; Baron, J.L. Blockade of NKG2D on NKT cells prevents hepatitis and the acute immune response to hepatitis B virus. Proc. Natl. Acad. Sci. USA 2007, 104, 18187–18192. [Google Scholar] [CrossRef]
- Chen, Y.; Hao, X.; Sun, R.; Wei, H.; Tian, Z. Natural Killer Cell-Derived Interferon-Gamma Promotes Hepatocellular Carcinoma Through the Epithelial Cell Adhesion Molecule-Epithelial-to-Mesenchymal Transition Axis in Hepatitis B Virus Transgenic Mice. Hepatology 2019, 69, 1735–1750. [Google Scholar] [CrossRef]
- Sun, C.; Sun, H.; Zhang, C.; Tian, Z. NK cell receptor imbalance and NK cell dysfunction in HBV infection and hepatocellular carcinoma. Cell. Mol. Immunol. 2015, 12, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Maini, M.K.; Peppa, D. NK cells: A double-edged sword in chronic hepatitis B virus infection. Front. Immunol. 2013, 4, 57. [Google Scholar] [CrossRef]
- Peppa, D.; Micco, L.; Javaid, A.; Kennedy, P.T.F.; Schurich, A.; Dunn, C.; Pallant, C.; Ellis, G.; Khanna, P.; Dusheiko, G.; et al. Blockade of immunosuppressive cytokines restores NK cell antiviral function in chronic hepatitis B virus infection. PLoS Pathog. 2010, 6, e1001227. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Vienne, M.; Tang, L.; Kerdiles, Y.; Etiennot, M.; Escalière, B.; Galluso, J.; Wei, H.; Sun, R.; Vivier, E.; et al. Liver type 1 innate lymphoid cells develop locally via an interferon-gamma-dependent loop. Science 2021, 371, eaba4177. [Google Scholar] [CrossRef]
- Peng, H.; Jiang, X.; Chen, Y.; Sojka, D.K.; Wei, H.; Gao, X.; Sun, R.; Yokoyama, W.M.; Tian, Z. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J. Clin. Investig. 2013, 123, 1444–1456. [Google Scholar] [CrossRef]
- Vivier, E.; Artis, D.; Colonna, M.; Diefenbach, A.; Di Santo, J.P.; Eberl, G.; Koyasu, S.; Locksley, R.M.; McKenzie, A.N.J.; Mebius, R.E.; et al. Innate Lymphoid Cells: 10 Years On. Cell 2018, 174, 1054–1066. [Google Scholar] [CrossRef]
- Daussy, C.; Faure, F.; Mayol, K.; Viel, S.; Gasteiger, G.; Charrier, E.; Bienvenu, J.; Henry, T.; Debien, E.; Hasan, U.A.; et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med. 2014, 211, 563–577. [Google Scholar] [CrossRef]
- Sojka, D.K.; Plougastel-Douglas, B.; Yang, L.; Pak-Wittel, M.A.; Artyomov, M.N.; Ivanova, Y.; Zhong, C.; Chase, J.M.; Rothman, P.B.; Yu, J.; et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife 2014, 3, e01659. [Google Scholar] [CrossRef] [PubMed]
- Mackay, L.K.; Minnich, M.; Kragten, N.A.M.; Liao, Y.; Nota, B.; Seillet, C.; Zaid, A.; Man, K.; Preston, S.; Freestone, D.; et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 2016, 352, 459–463. [Google Scholar] [CrossRef]
- Pikovskaya, O.; Chaix, J.; Rothman, N.J.; Collins, A.; Chen, Y.-H.; Scipioni, A.M.; Vivier, E.; Reiner, S.L. Cutting Edge: Eomesodermin Is Sufficient To Direct Type 1 Innate Lymphocyte Development into the Conventional NK Lineage. J. Immunol. 2016, 196, 1449–1454. [Google Scholar] [CrossRef]
- Zheng, M.; Sun, R.; Wei, H.; Tian, Z. NK Cells Help Induce Anti-Hepatitis B Virus CD8+ T Cell Immunity in Mice. J. Immunol. 2016, 196, 4122–4131. [Google Scholar] [CrossRef]
- Yang, Z.; Tang, T.; Wei, X.; Yang, S.; Tian, Z. Type 1 innate lymphoid cells contribute to the pathogenesis of chronic hepatitis B. Innate Immun. 2015, 21, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Tian, Z. NK cells in liver homeostasis and viral hepatitis. Sci. China Life Sci. 2018, 61, 1477–1485. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Pan, Q.; Yang, J.; He, J.; Zeng, J.; Cheng, S.; Huang, Y.; Zhou, Z.Q.; Zhu, Q.; Yang, C.; et al. Galectin-3 favours tumour metastasis via the activation of beta-catenin signalling in hepatocellular carcinoma. Br. J. Cancer 2020, 123, 1521–1534. [Google Scholar] [CrossRef]
- An, Y.; Xu, S.; Liu, Y.; Xu, X.; Philips, C.A.; Chen, J.; Méndez-Sánchez, N.; Guo, X.; Qi, X. Role of Galectins in the Liver Diseases: A Systematic Review and Meta-Analysis. Front. Med. 2021, 8, 744518. [Google Scholar] [CrossRef]
- Wang, W.; Guo, H.; Geng, J.; Zheng, X.; Wei, H.; Sun, R.; Tian, Z. Tumor-released Galectin-3, a soluble inhibitory ligand of human NKp30, plays an important role in tumor escape from NK cell attack. J. Biol. Chem. 2014, 289, 33311–33319. [Google Scholar] [CrossRef] [PubMed]
- Hsu, D.K.; Dowling, C.A.; Jeng, K.C.; Chen, J.T.; Yang, R.Y.; Liu, F.T. Galectin-3 expression is induced in cirrhotic liver and hepatocellular carcinoma. Int. J. Cancer 1999, 81, 519–526. [Google Scholar] [CrossRef]
- Ulu, M.; Alacacioglu, A.; Yuksel, E.; Pamukk, B.; Bozkaya, G.; Ari, A.; Yuksel, A.; Sop, G.; Alacacioglu, I. Prognostic significance of serum galectin-3 levels in patients with hepatocellular cancer and chronic viral hepatitis. Saudi J. Gastroenterol. 2015, 21, 47–50. [Google Scholar] [CrossRef]
- Güneş, A.; Uluca, Ü.; Şen, V.; Ece, A.; Tan, I.; Karabel, D.; Aktar, F.; Karabel, M.; Balık, H. Serum galectin-3 levels in children with chronic hepatitis B infection and inactive hepatitis B carriers. Med. Sci. Monit. 2015, 21, 1376–1380. [Google Scholar] [CrossRef]
- Chisari, F.V.; Klopchin, K.; Moriyama, T.; Pasquinelli, C.; Dunsford, H.A.; Sell, S.; Pinkert, C.A.; Brinster, R.L.; Palmiter, R.D. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell 1989, 59, 1145–1156. [Google Scholar] [CrossRef]
- Faure-Dupuy, S.; Delphin, M.; Aillot, L.; Dimier, L.; Lebossé, F.; Fresquet, J.; Parent, R.; Matter, M.S.; Rivoire, M.; Bendriss-Vermare, N.; et al. Hepatitis B virus-induced modulation of liver macrophage function promotes hepatocyte infection. J. Hepatol. 2019, 71, 1086–1098. [Google Scholar] [CrossRef] [PubMed]
- De Pasquale, C.; Campana, S.; Barberi, C.; Migliore, G.S.; Oliveri, D.; Lanza, M.; Musolino, C.; Raimondo, G.; Ferrone, S.; Pollicino, T.; et al. Human Hepatitis B Virus Negatively Impacts the Protective Immune Crosstalk Between Natural Killer and Dendritic Cells. Hepatology 2021, 74, 550–565. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhai, N.; Wang, Z.; Song, H.; Yang, Y.; Cui, A.; Li, T.; Wang, G.; Niu, J.; Crispe, I.N.; et al. Regulatory NK cells mediated between immunosuppressive monocytes and dysfunctional T cells in chronic HBV infection. Gut 2018, 67, 2035–2044. [Google Scholar] [CrossRef]
- Gu, Y.; Huang, Z.; Li, X.; Chen, Y.; Liao, C.; Bi, Y.; Huang, Y. Serum HBV pregenomic RNA exhibited opposite associations with NKdim and NKbright cell immunity in treatment-naive chronic hepatitis B patients. Biosci. Rep. 2021, 41, BSR20210600. [Google Scholar] [CrossRef] [PubMed]
- Zecca, A.; Barili, V.; Boni, C.; Fisicaro, P.; Vecchi, A.; Rossi, M.; Reverberi, V.; Montali, A.; Pedrazzi, G.; Ferrari, C.; et al. High CD49a+ NK cell infiltrate is associated with poor clinical outcomes in Hepatocellular Carcinoma. Heliyon 2023, 9, e22680. [Google Scholar] [CrossRef] [PubMed]
- Wijaya, R.S.; A Read, S.; Truong, N.R.; Han, S.; Chen, D.; Shahidipour, H.; Fewings, N.L.; Schibeci, S.; Azardaryany, M.K.; Parnell, G.P.; et al. HBV vaccination and HBV infection induces HBV-specific natural killer cell memory. Gut 2021, 70, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, A.D.; Gude, R.P.; Kalraiya, R.D.; Chiplunkar, S.V. Endogenous galectin-3 expression levels modulate immune responses in galectin-3 transgenic mice. Mol. Immunol. 2015, 68 Pt A, 300–311. [Google Scholar] [CrossRef]
- Radosavljevic, G.; Jovanovic, I.; Majstorovic, I.; Mitrovic, M.; Lisnic, V.J.; Arsenijevic, N.; Jonjic, S.; Lukic, M.L. Deletion of galectin-3 in the host attenuates metastasis of murine melanoma by modulating tumor adhesion and NK cell activity. Clin. Exp. Metastasis 2011, 28, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Hollyoake, M.; Campbell, R.D.; Aguado, B. NKp30 (NCR3) is a pseudogene in 12 inbred and wild mouse strains, but an expressed gene in Mus caroli. Mol. Biol. Evol. 2005, 22, 1661–1672. [Google Scholar] [CrossRef]
- Tsuboi, S.; Sutoh, M.; Hatakeyama, S.; Hiraoka, N.; Habuchi, T.; Horikawa, Y.; Hashimoto, Y.; Yoneyama, T.; Mori, K.; Koie, T.; et al. A novel strategy for evasion of NK cell immunity by tumours expressing core2 O-glycans. EMBO J. 2011, 30, 3173–3185. [Google Scholar] [CrossRef]
- Zhao, W.; Ajani, J.A.; Sushovan, G.; Ochi, N.; Hwang, R.; Hafley, M.; Johnson, R.L.; Bresalier, R.S.; Logsdon, C.D.; Zhang, Z.; et al. Galectin-3 Mediates Tumor Cell-Stroma Interactions by Activating Pancreatic Stellate Cells to Produce Cytokines via Integrin Signaling. Gastroenterology 2018, 154, 1524–1537.e6. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.-S.; Weng, D.-S.; Wang, Q.-J.; Pan, K.; Zhang, Y.-J.; Li, Y.-Q.; Li, J.-J.; Zhao, J.-J.; He, J.; Lv, L.; et al. Galectin-3 is associated with a poor prognosis in primary hepatocellular carcinoma. J. Transl. Med. 2014, 12, 273. [Google Scholar] [CrossRef] [PubMed]
- Setayesh, T.; Colquhoun, S.D.; Wan, Y.Y. Overexpression of Galectin-1 and Galectin-3 in hepatocellular carcinoma. Liver Res. 2020, 4, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Zhang, R.; Xia, Y.; Jiang, X.; Zhou, K.; Li, J.; Guo, M.; Cao, X.; Zhang, S. The necroptosis related gene LGALS3 can be used as a biomarker for the adverse progression from chronic HBV infection to HCC. Front. Immunol. 2023, 14, 1142319. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, Y.; Tsuneyama, K.; Nomoto, K.; Fujimoto, M.; Salunga, T.L.; Nakajima, T.; Miwa, S.; Murai, Y.; Hayashi, S.; Kato, I.; et al. Nonalcoholic steatohepatitis and hepatocellular carcinoma in galectin-3 knockout mice. Hepatol. Res. 2008, 38, 1241–1251. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Tian, F.; Ying, W.; Qian, X. Quantitative proteomics reveal the anti-tumour mechanism of the carbohydrate recognition domain of Galectin-3 in Hepatocellular carcinoma. Sci. Rep. 2017, 7, 5189. [Google Scholar] [CrossRef]
- Jaruga, B.; Hong, F.; Kim, W.H.; Gao, B. IFN-gamma/STAT1 acts as a proinflammatory signal in T cell-mediated hepatitis via induction of multiple chemokines and adhesion molecules: A critical role of IRF-1. Am. J. Physiol. Gastrointest. Liver Physiol. 2004, 287, G1044–G1052. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Zhang, W.; Cheng, M.; Hao, X.; Wei, H.; Sun, R.; Tian, Z. Galectin-3-ITGB1 Signaling Mediates Interleukin 10 Production of Hepatic Conventional Natural Killer Cells in Hepatitis B Virus Transgenic Mice and Correlates with Hepatocellular Carcinoma Progression in Patients. Viruses 2024, 16, 737. https://doi.org/10.3390/v16050737
Chen Y, Zhang W, Cheng M, Hao X, Wei H, Sun R, Tian Z. Galectin-3-ITGB1 Signaling Mediates Interleukin 10 Production of Hepatic Conventional Natural Killer Cells in Hepatitis B Virus Transgenic Mice and Correlates with Hepatocellular Carcinoma Progression in Patients. Viruses. 2024; 16(5):737. https://doi.org/10.3390/v16050737
Chicago/Turabian StyleChen, Yongyan, Wendi Zhang, Min Cheng, Xiaolei Hao, Haiming Wei, Rui Sun, and Zhigang Tian. 2024. "Galectin-3-ITGB1 Signaling Mediates Interleukin 10 Production of Hepatic Conventional Natural Killer Cells in Hepatitis B Virus Transgenic Mice and Correlates with Hepatocellular Carcinoma Progression in Patients" Viruses 16, no. 5: 737. https://doi.org/10.3390/v16050737
APA StyleChen, Y., Zhang, W., Cheng, M., Hao, X., Wei, H., Sun, R., & Tian, Z. (2024). Galectin-3-ITGB1 Signaling Mediates Interleukin 10 Production of Hepatic Conventional Natural Killer Cells in Hepatitis B Virus Transgenic Mice and Correlates with Hepatocellular Carcinoma Progression in Patients. Viruses, 16(5), 737. https://doi.org/10.3390/v16050737




