DNA Virome in Cardiac Tissue from Green Sea Turtles (Chelonia mydas) with Myocarditis
Abstract
1. Introduction
2. Materials and Methods
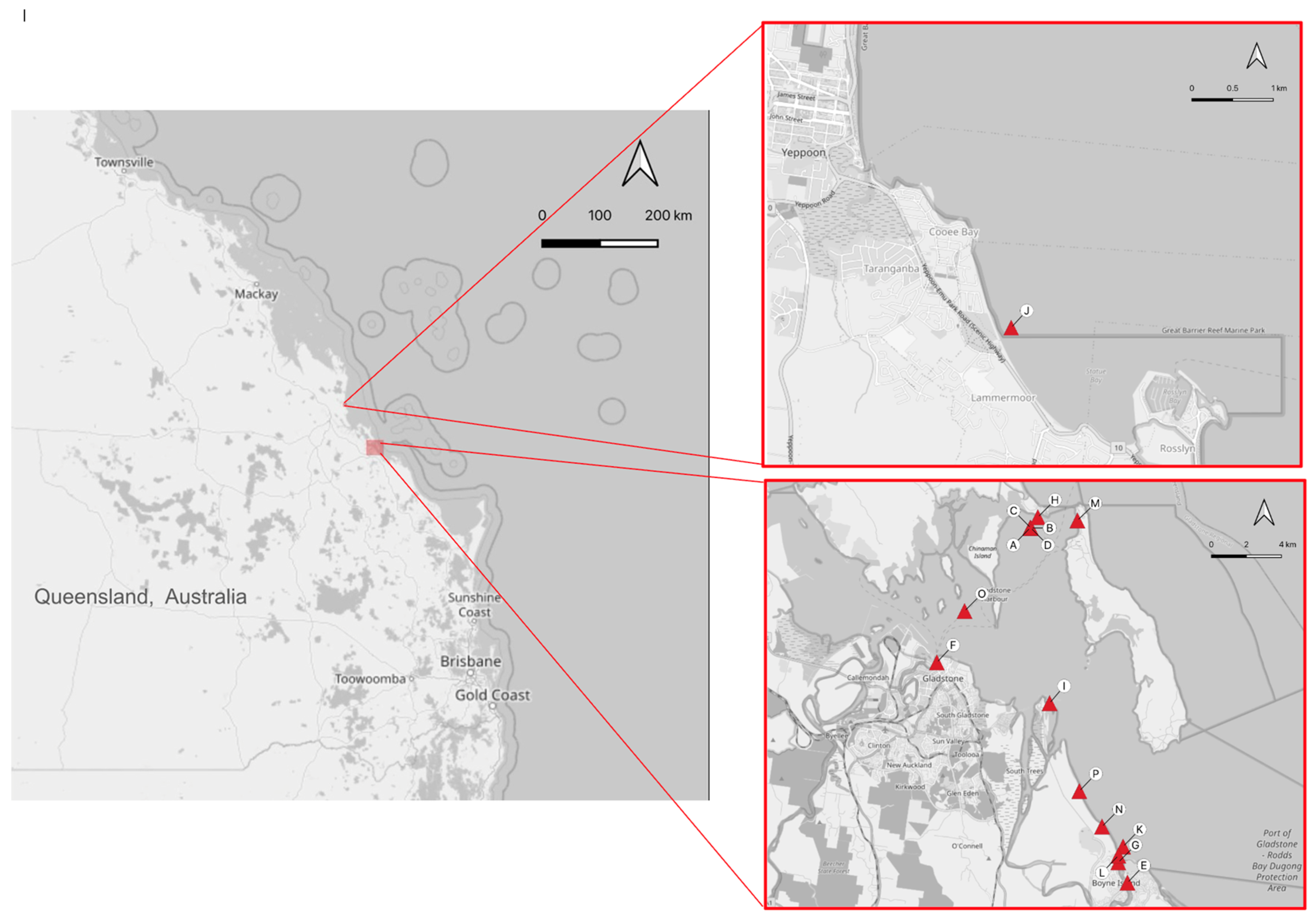
2.1. Animal Handling and Tissue Sampling
2.2. Histology
2.3. Nucleic Acid Recovery
2.4. Bioinformatics
2.5. Phylogenetic Analysis
2.6. Transmission Electron Microscopy (TEM)
3. Results
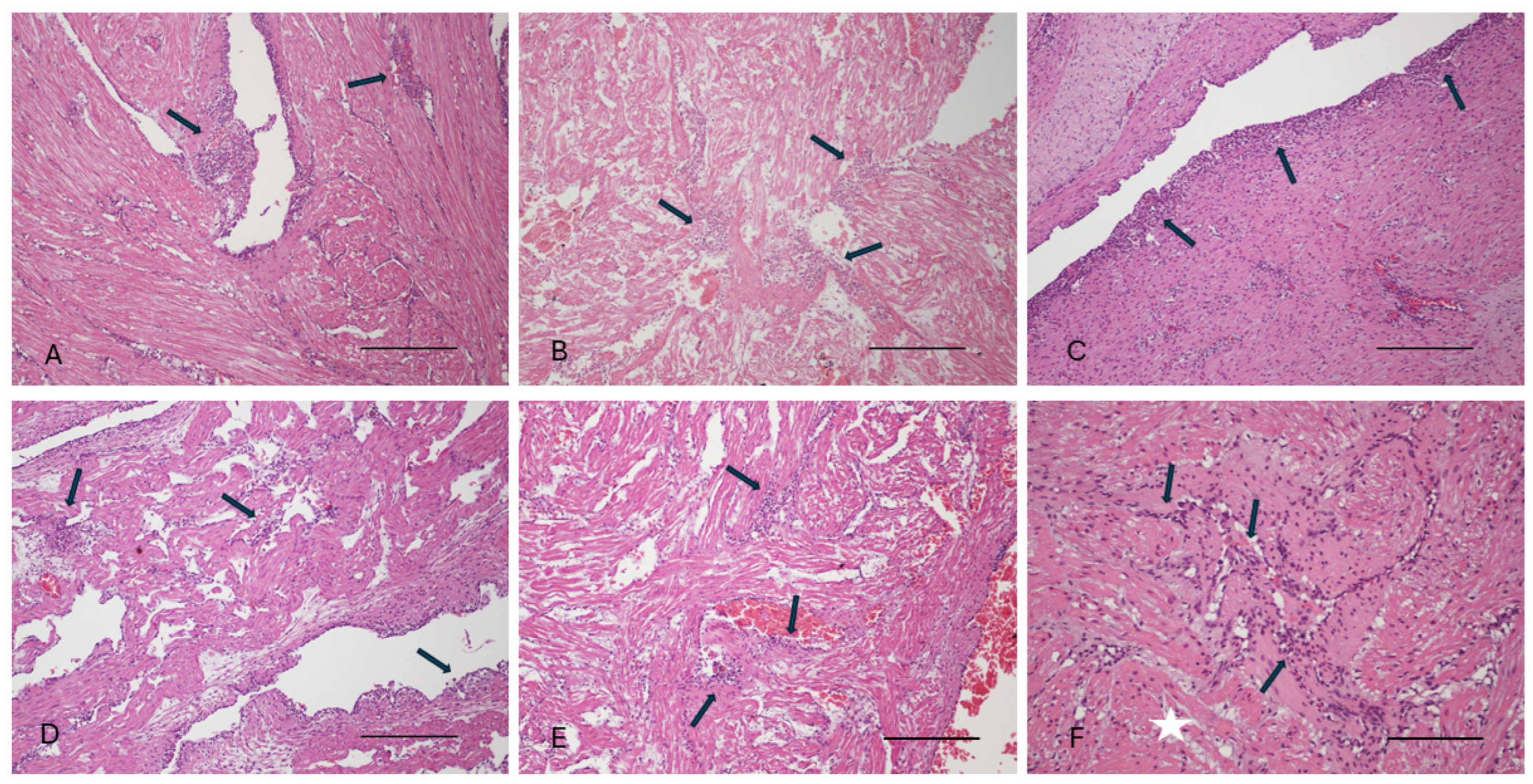
3.1. Gross Pathology and Histopathology Findings
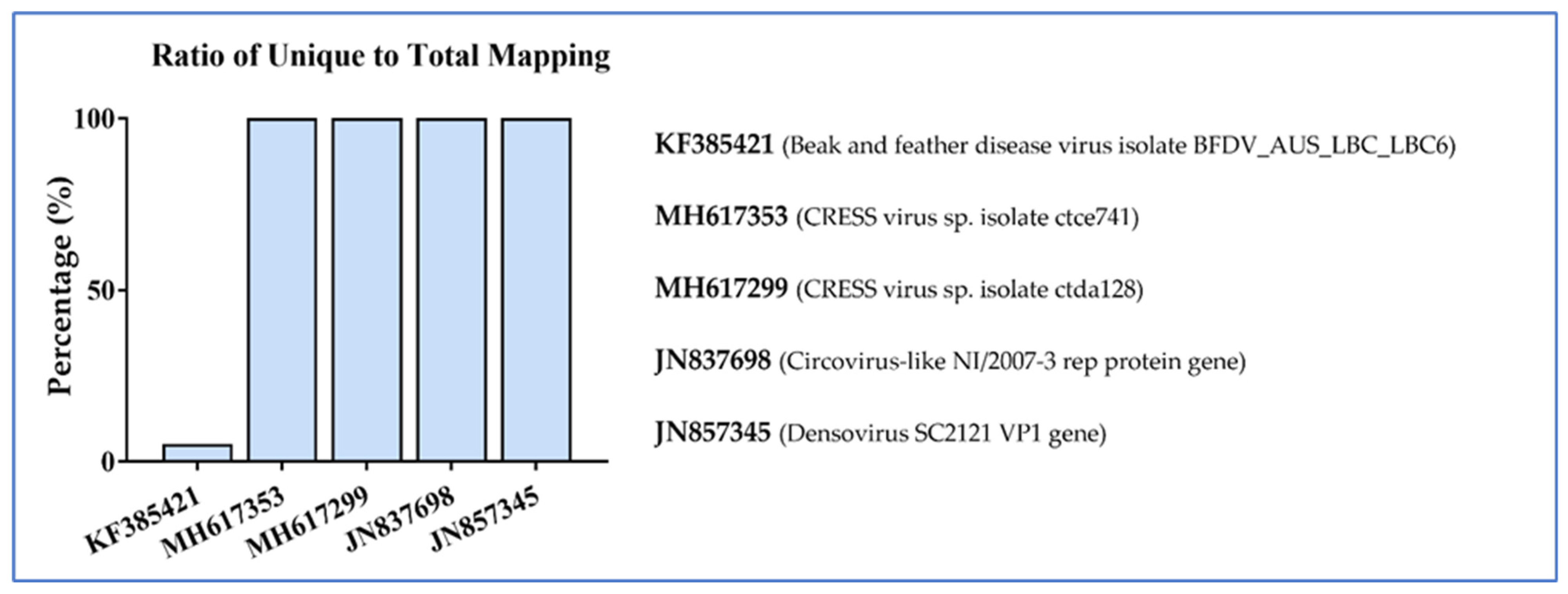
3.2. Nucleic Acid Recovery and Bioinformatics
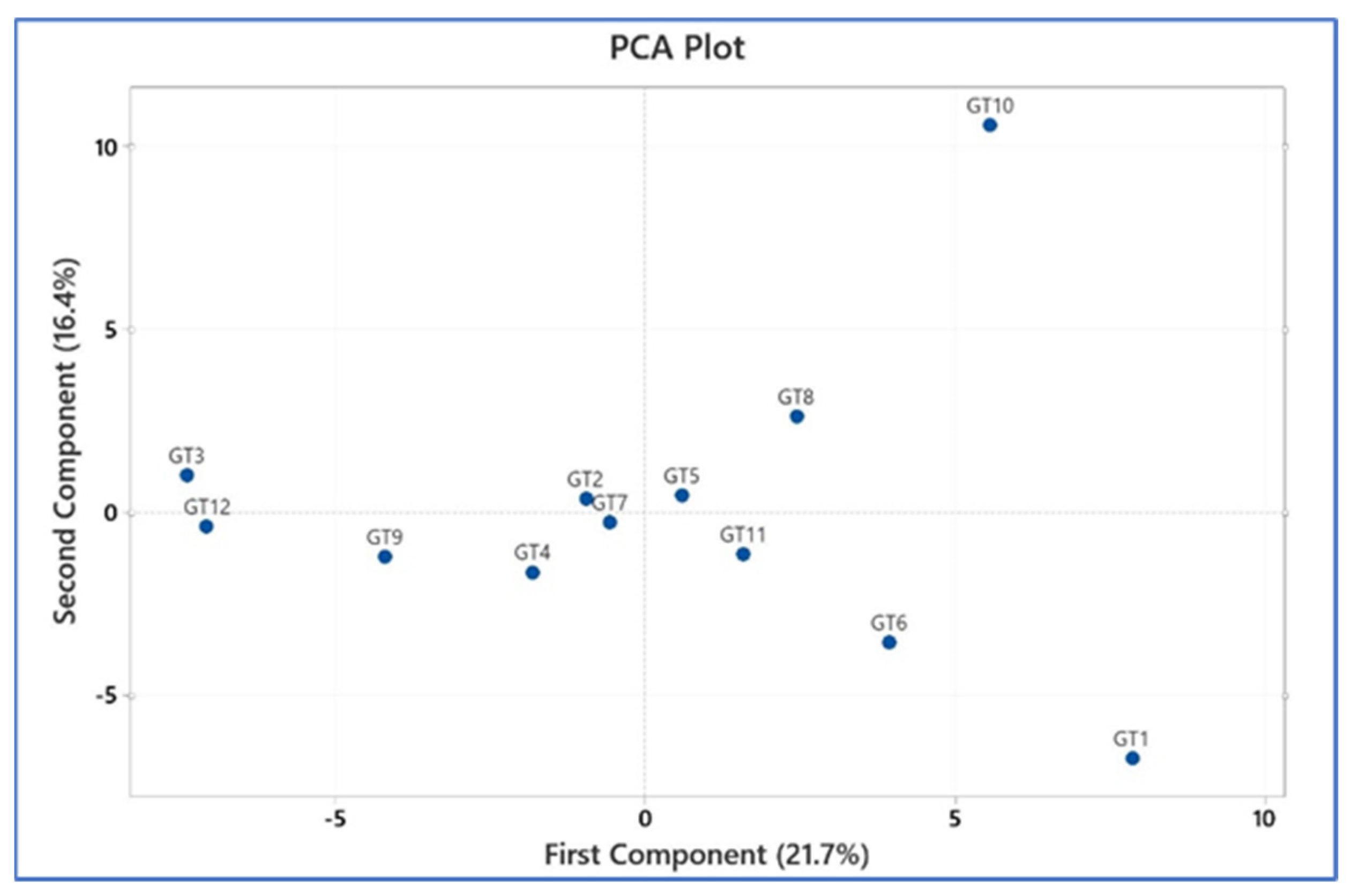
3.3. Phylogenetic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raidal, S.R.; Ohara, M.; Hobbs, R.P.; Prince, R.I. Gram-negative bacterial infections and cardiovascular parasitism in green sea turtles (Chelonia mydas). Aust. Vet. J. 1998, 76, 415–417. [Google Scholar] [CrossRef]
- Gordon, A.N.; Kelly, W.R.; Cribb, T.H. Lesions caused by cardiovascular flukes (Digenea: Spirorchidae) in stranded green turtles (Chelonia mydas). Vet. Pathol. 1998, 35, 21–30. [Google Scholar] [CrossRef]
- Cordero-Tapia, A.; Gardner, S.C.; Arellano-Blanco, J.; Inohuye-Rivera, R.B. Learedius learedi infection in black turtles (Chelonia mydas agassizii), Baja California Sur, Mexico. J. Parasitol. 2004, 90, 645–647. [Google Scholar] [CrossRef]
- Santoro, M.; Greiner, E.C.; Morales, J.A.; Rodríguez-Ortíz, B. A new pronocephalid, Pleurogonius tortugueroi n. sp. (Digenea), from the intestine of green sea turtles (Chelonia mydas) in Costa Rica. Parassitologia 2007, 49, 97–100. [Google Scholar]
- Chapman, P.A.; Cribb, T.H.; Flint, M.; Traub, R.J.; Blair, D.; Kyaw-Tanner, M.T.; Mills, P.C. Spirorchiidiasis in marine turtles: The current state of knowledge. Dis. Aquat. Org. 2019, 133, 217–245. [Google Scholar] [CrossRef] [PubMed]
- Homer, B.L.; Jacobson, E.R.; Schumacher, J.; Scherba, G. Chlamydiosis in mariculture-reared green sea turtles (Chelonia mydas). Vet. Pathol. 1994, 31, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Carossino, M.; Nevarez, J.G.; Sakaguchi, K.; Paulsen, D.B.; Langohr, I.M.; Strother, K.; Ferracone, J.; Roy, A.; Crossland, N.A.; Del Piero, F. An outbreak of systemic chlamydiosis in farmed American alligators (Alligator mississipiensis). Vet. Pathol. 2022, 59, 860–868. [Google Scholar] [CrossRef] [PubMed]
- Ammirati, E.; Veronese, G.; Bottiroli, M.; Wang, D.W.; Cipriani, M.; Garascia, A.; Pedrotti, P.; Adler, E.D.; Frigerio, M. Update on acute myocarditis. Trends Cardiovasc. Med. 2021, 31, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Zafeiri, M.; Knott, K.; Lampejo, T. Acute myocarditis: An overview of pathogenesis, diagnosis and management. Panminerva Med. 2024, 66, 174–187. [Google Scholar] [CrossRef]
- Mone, K.; Reddy, J. The knowns and unknowns of cardiac autoimmunity in viral myocarditis. Rev. Med. Virol. 2023, 33, e2478. [Google Scholar] [CrossRef]
- Flint, M.-S. A Veterinarian’s Guide for Sea Turtle Post Mortem Examination and Histological Investigation; School of Veterinary Science, University of Queensland: St. Lucia, Australia, 2009. [Google Scholar]
- QGIS.org. QGIS Geographic Information System. Open Source Geospatial Foundation Project. 2024. Available online: http://qgis.org (accessed on 2 June 2024).
- Sarker, S.; Isberg, S.R.; Moran, J.L.; Araujo, R.; Elliott, N.; Melville, L.; Beddoe, T.; Helbig, K.J. Crocodilepox virus evolutionary genomics supports observed poxvirus infection dynamics in saltwater crocodile (Crocodylus porosus). Viruses 2019, 11, 1116. [Google Scholar] [CrossRef] [PubMed]
- Sarker, S.; Hannon, C.; Athukurala, A.; Bielefeldt-Ohmann, H. Emergence of a novel pathogenic poxvirus infection in the endangered green sea turtle (Chelonia mydas) highlights a key threatening process. Viruses 2021, 13, 219. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- McCowan, C.; Crameri, S.; Kocak, A.; Shan, S.; Fegan, M.; Forshaw, D.; Rubbenstroth, D.; Chen, H.; Holmes, C.; Harper, J.; et al. A novel group A rotavirus associated with acute illness and hepatic necrosis in pigeons (Columba livia), in Australia. PLoS ONE 2018, 13, e0203853. [Google Scholar] [CrossRef] [PubMed]
- Brandes, N.; Linial, M. Giant viruses—Big surprises. Viruses 2019, 11, 404. [Google Scholar] [CrossRef] [PubMed]
- Bloom, D.C. Alphaherpesvirus latency: A dynamic state of transcription and reactivation. Adv. Virus Res. 2016, 94, 53–80. [Google Scholar] [CrossRef]
- Okoh, G.R.; Horwood, P.F.; Whitmore, D.; Ariel, E. Herpesviruses in reptiles. Front. Vet. Sci. 2021, 8, 642894. [Google Scholar] [CrossRef]
- Badrinath, A.; Bhatta, S.; Kloc, A. Persistent viral infections and their role in heart disease. Front. Microbiol. 2022, 13, 1030440. [Google Scholar] [CrossRef] [PubMed]
- Harding, E.F.; Russo, A.G.; Yan, G.J.H.; Mercer, L.K.; White, P.A. Revealing the uncharacterised diversity of amphibian and reptile viruses. ISME Commun. 2022, 2, 95. [Google Scholar] [CrossRef]
- Heidecker, B.; Williams, S.H.; Jain, K.; Oleynik, A.; Patriki, D.; Kottwitz, J.; Berg, J.; Garcia, J.A.; Baltensperger, N.; Lovrinovic, M.; et al. Virome sequencing in patients with myocarditis. Circ. Heart Fail. 2020, 13, e007103. [Google Scholar] [CrossRef]
- Takeuchi, S.; Kawada, J.; Okuno, Y.; Horiba, K.; Suzuki, T.; Torii, Y.; Yasuda, K.; Numaguchi, A.; Kato, T.; Takahashi, Y.; et al. Identification of potential pathogenic viruses in patients with acute myocarditis using next-generation sequencing. J. Med. Virol. 2018, 90, 1814–1821. [Google Scholar] [CrossRef] [PubMed]
- Rakibuzzaman, A.; Ramamoorthy, S. Comparative immunopathogenesis and biology of recently discovered porcine circoviruses. Transbound. Emerg. Dis. 2021, 68, 2957–2968. [Google Scholar] [CrossRef] [PubMed]
- Alomar, J.; Saporiti, V.; Pérez, M.; Gonçalvez, D.; Sibila, M.; Segalés, J. Multisystemic lymphoplasmacytic inflammation associated with PCV-3 in wasting pigs. Transbound. Emerg. Dis. 2021, 68, 2969–2974. [Google Scholar] [CrossRef] [PubMed]
- Shearer, P.L.; Bonne, N.; Clark, P.; Sharp, M.; Raidal, S.R. Development and applications of a monoclonal antibody to a recombinant beak and feather disease virus (BFDV) capsid protein. J. Virol. Methods 2008, 147, 206–212. [Google Scholar] [CrossRef]
- Sarker, S.; Athukorala, A.; Phalen, D.N. Genome sequence of a beak and feather disease virus from an unusual novel host, Australian boobook owl (Ninox boobook). Microbiol. Resour. Announc. 2022, 11, e0017222. [Google Scholar] [CrossRef]
- Bustard, R. Australian Sea Turtles. Their Natural History and Conservation; William Collins & Son Ltd.: London, UK, 1972. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hannon, C.; Sarker, S.; Suen, W.W.; Bielefeldt-Ohmann, H. DNA Virome in Cardiac Tissue from Green Sea Turtles (Chelonia mydas) with Myocarditis. Viruses 2024, 16, 1053. https://doi.org/10.3390/v16071053
Hannon C, Sarker S, Suen WW, Bielefeldt-Ohmann H. DNA Virome in Cardiac Tissue from Green Sea Turtles (Chelonia mydas) with Myocarditis. Viruses. 2024; 16(7):1053. https://doi.org/10.3390/v16071053
Chicago/Turabian StyleHannon, Christabel, Subir Sarker, Willy W. Suen, and Helle Bielefeldt-Ohmann. 2024. "DNA Virome in Cardiac Tissue from Green Sea Turtles (Chelonia mydas) with Myocarditis" Viruses 16, no. 7: 1053. https://doi.org/10.3390/v16071053
APA StyleHannon, C., Sarker, S., Suen, W. W., & Bielefeldt-Ohmann, H. (2024). DNA Virome in Cardiac Tissue from Green Sea Turtles (Chelonia mydas) with Myocarditis. Viruses, 16(7), 1053. https://doi.org/10.3390/v16071053







