CRISPR/Cas9-Mediated Resistance to Wheat Dwarf Virus in Hexaploid Wheat (Triticum aestivum L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus Propagation and Leafhopper Inoculation
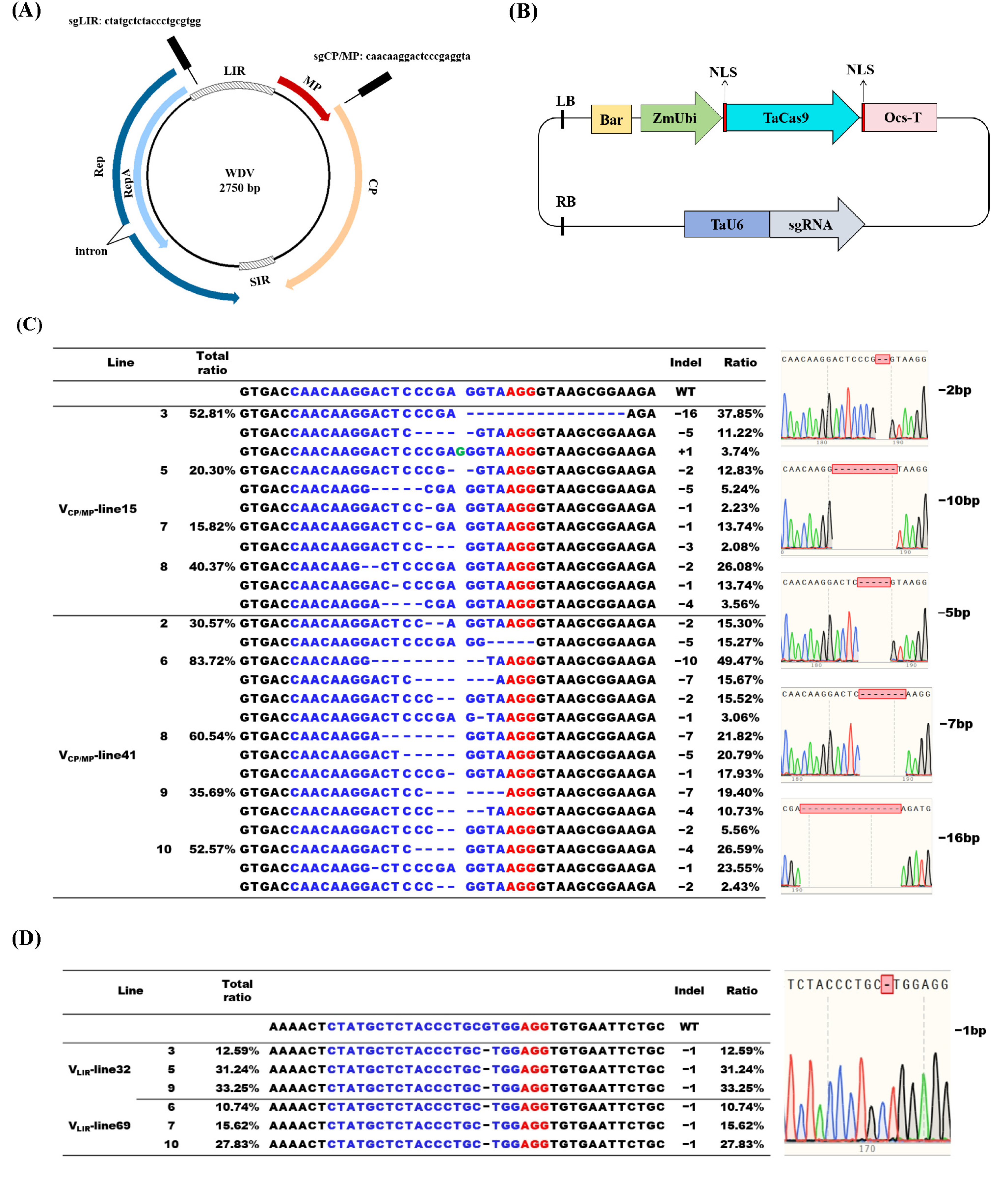
2.2. sgRNA Design and Construction of CRISPR/Cas9 Expression Vector
2.3. Agrobacterium-Mediated Wheat Transformation
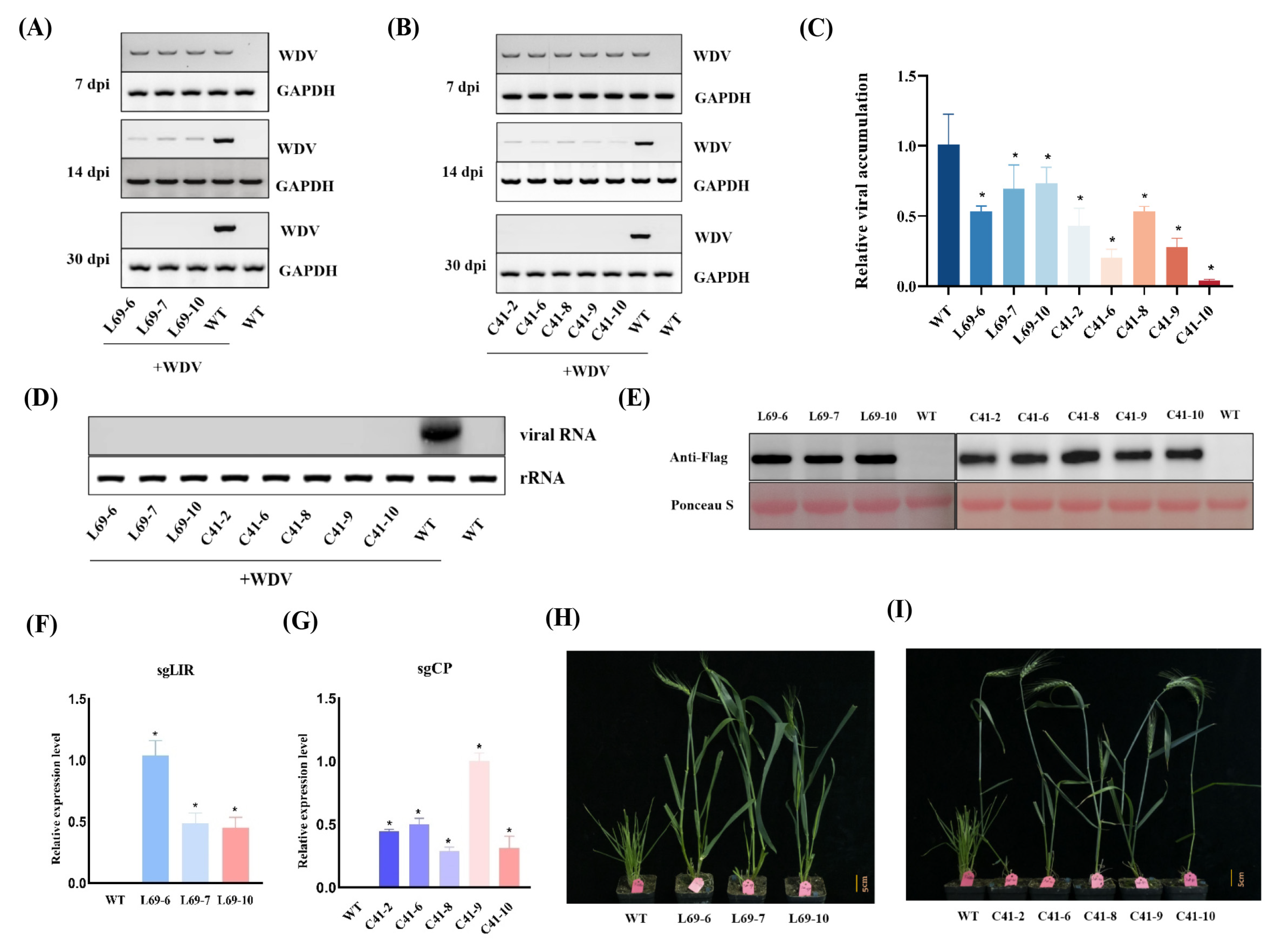
2.4. Transcriptional Analysis Using sqRT-PCR and RT-qPCR
2.5. Western Blot Assay
2.6. Northern Blot Analysis
2.7. Detection of Edited Mutations
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rotenberg, D.; Bockus, W.; Whitfield, A.; Hervey, K.; Baker, K.; Ou, Z.; Laney, A.; De Wolf, E.; Appel, J. Occurrence of viruses and associated grain yields of paired symptomatic and nonsymptomatic tillers in Kansas winter wheat fields. Phytopathology 2016, 106, 202–210. [Google Scholar] [CrossRef]
- Manurung, B.; Witsack, W.; Mehner, S.; Grüntzig, M.; Fuchs, E. The epidemiology of wheat dwarf virus in relation to occurrence of the leafhopper Psammotettix alienus in Middle-Germany. Virus Res. 2004, 100, 109–113. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, B.; Vida, G.; Cséplo-Károlyi, M.; Wu, B.; Wu, Y.; Wang, X. Genomic analysis of the natural population of wheat dwarf virus in wheat from China and Hungary. J. Integr. Agric. 2012, 11, 2020–2027. [Google Scholar] [CrossRef]
- Wang, Y.; Mao, Q.; Liu, W.; Mar, T.; Wei, T.; Liu, Y.; Wang, X. Localization and distribution of wheat dwarf virus in its vector leafhopper Psammotettix alienus. Phytopathology 2014, 104, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, C.; Ramirez-Parra, E.; Mar Castellano, M.; Sanz-Burgos, A.; Luque, A.; Missich, R. Geminivirus DNA replication and cell cycle interactions. Vet. Microbiol. 2004, 98, 111–119. [Google Scholar] [CrossRef]
- Liu, Y.; Jin, W.; Wang, L.; Wang, X. Replication-associated proteins encoded by wheat dwarf virus act as RNA silencing suppressors. Virus Res. 2014, 190, 34–39. [Google Scholar] [CrossRef]
- Nygren, J.; Shad, N.; Kvarnheden, A.; Westerbergh, A. Variation in susceptibility to wheat dwarf virus among wild and domesticated wheat. PLoS ONE 2015, 10, 4. [Google Scholar] [CrossRef]
- Robertson, G.; Burger, J.; Campa, M. CRISPR/Cas-based tools for the targeted control of plant viruses. Mol. Plant Pathol. 2022, 23, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Han, T.; Xie, K.; Zhao, J.; Song, J.; Liu, Y. Engineer complete resistance to Cotton Leaf Curl Multan virus by the CRISPR/Cas9 system in Nicotiana benthamiana. Phytopathol. Res. 2019, 1, 9. [Google Scholar] [CrossRef]
- Baltes, N.; Hummel, A.; Konecna, E.; Cegan, R.; Bruns, A.; Bisaro, D.; Voytas, A. Conferring resistance to geminiviruses with the CRISPR–Cas prokaryotic immune system. Nat. Plants 2015, 1, 15145. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Zhang, H.; Zhang, Y.; Wang, Y.; Gao, C. Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 2015, 1, 15144. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015, 16, 238. [Google Scholar] [CrossRef] [PubMed]
- Tashkandi, M.; Ali, Z.; Aljedaani, F.; Shami, A.; Mahfouz, M. Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal. Behav. 2018, 13, e1525996. [Google Scholar] [CrossRef]
- Khan, Z.; Khan, S.; Ahmad, A.; Aslam, S.; Mubarik, M.; Khan, S. CRISPR/dCas9-mediated inhibition of replication of Begomoviruses. Int. J. Agric. Biol. 2019, 21, 711–718. [Google Scholar]
- Gutierrez, C. DNA replication and cell cycle in plants: Learning from geminiviruses. EMBO J. 2000, 19, 792–799. [Google Scholar] [CrossRef]
- Kis, A.; Hamar, E.; Tholt, G.; Ban, R.; Havelda, Z. Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant Biotechnol. J. 2019, 17, 1004–1006. [Google Scholar] [CrossRef]
- Zhang, Z.; Hua, L.; Gupta, A.; Tricoli, D.; Edwards, K.; Yang, B.; Li, W. Development of an Agrobacterium-delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnol. J. 2019, 17, 1623–1635. [Google Scholar] [CrossRef]
- Ishida, Y.; Tsunashida, M.; Hiei, Y.; Komari, T. Wheat (Triticum aestivum L.) transformation using immature embryos. Methods Mol. Biol. 2015, 1223, 189–198. [Google Scholar]
- Wang, F. Semi-Quantitative RT-PCR: An effective method to explore the regulation of gene transcription level affected by environmental pollutants. Methods Mol. Biol. 2021, 2326, 95–103. [Google Scholar]
- Jarosová, J.; Kundu, J. Validation of reference genes as internal control for studying viral infections in cereals by quantitative real-time RT-PCR. BMC Plant Biol. 2010, 10, 146. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, C.; Jiao, X.; Zhang, H.; Song, L.; Li, Y.; Gao, C.; Wang, K. Hi-TOM: A platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 2019, 62, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lim, K.; Kim, J.; Bae, S. Cas-analyzer: An online tool for assessing genome editing results using NGS data. Bioinformatics 2017, 33, 286–288. [Google Scholar] [CrossRef] [PubMed]
- Bell, C.; Magor, G.; Gillinder, K.; Perkins, A. A high-throughput screening strategy for detecting CRISPR-Cas9 induced mutations using next-generation sequencing. BMC Genom. 2014, 15, 1002. [Google Scholar] [CrossRef]
- Lindsay, H.; Burger, A.; Biyong, B.; Felker, A.; Hess, C.; Zaugg, J.; Chiavacci, E.; Anders, C.; Jinek, M.; Mosimann, C.; et al. CrispR Variants charts the mutation spectrum of genome engineering experiments. Nat. Biotechnol. 2016, 34, 701–702. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, X.; Xu, K.; Yan, F.; Liu, Z.; Spetz, C.; Zhou, H.; Wang, X.; Jin, H.; Wang, X.; Liu, Y. CRISPR/Cas9-Mediated Resistance to Wheat Dwarf Virus in Hexaploid Wheat (Triticum aestivum L.). Viruses 2024, 16, 1382. https://doi.org/10.3390/v16091382
Yuan X, Xu K, Yan F, Liu Z, Spetz C, Zhou H, Wang X, Jin H, Wang X, Liu Y. CRISPR/Cas9-Mediated Resistance to Wheat Dwarf Virus in Hexaploid Wheat (Triticum aestivum L.). Viruses. 2024; 16(9):1382. https://doi.org/10.3390/v16091382
Chicago/Turabian StyleYuan, Xiaoyu, Keya Xu, Fang Yan, Zhiyuan Liu, Carl Spetz, Huanbin Zhou, Xiaojie Wang, Huaibing Jin, Xifeng Wang, and Yan Liu. 2024. "CRISPR/Cas9-Mediated Resistance to Wheat Dwarf Virus in Hexaploid Wheat (Triticum aestivum L.)" Viruses 16, no. 9: 1382. https://doi.org/10.3390/v16091382
APA StyleYuan, X., Xu, K., Yan, F., Liu, Z., Spetz, C., Zhou, H., Wang, X., Jin, H., Wang, X., & Liu, Y. (2024). CRISPR/Cas9-Mediated Resistance to Wheat Dwarf Virus in Hexaploid Wheat (Triticum aestivum L.). Viruses, 16(9), 1382. https://doi.org/10.3390/v16091382








