Attaining the Promise of Geminivirus-Based Vectors in Plant Genome Editing
Abstract
1. Introduction
2. Harnessing Virus Engineering for Advanced Crop Breeding
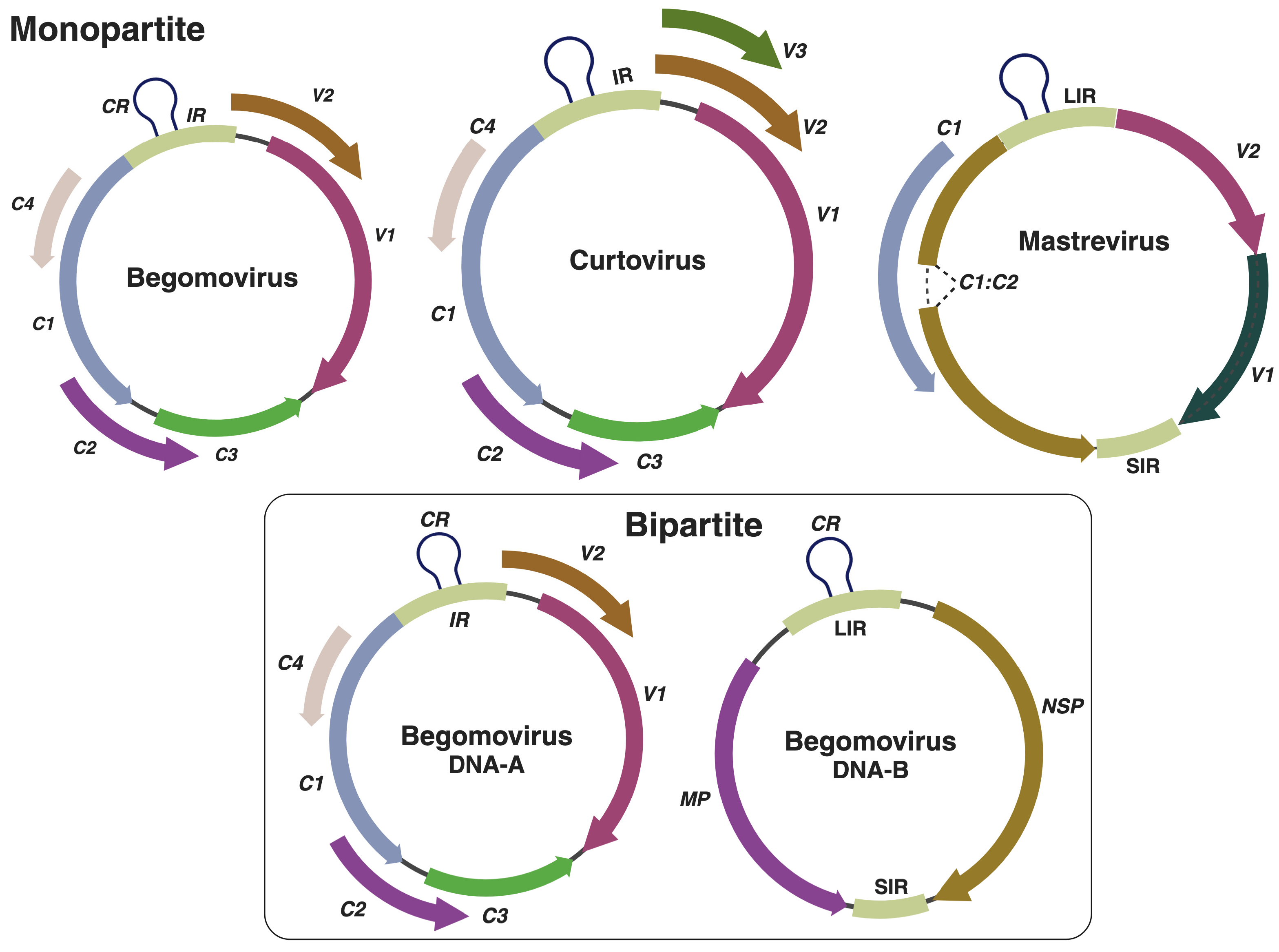
3. Geminiviruses and Their Unique Characteristics
4. Geminiviruses: Master of the Host Genome’s Manipulator
Geminiviruses Lifecycle: Hijacking Host Nuclear Machinery
5. Geminiviruses: How They Can Help in Plant Genome Editing
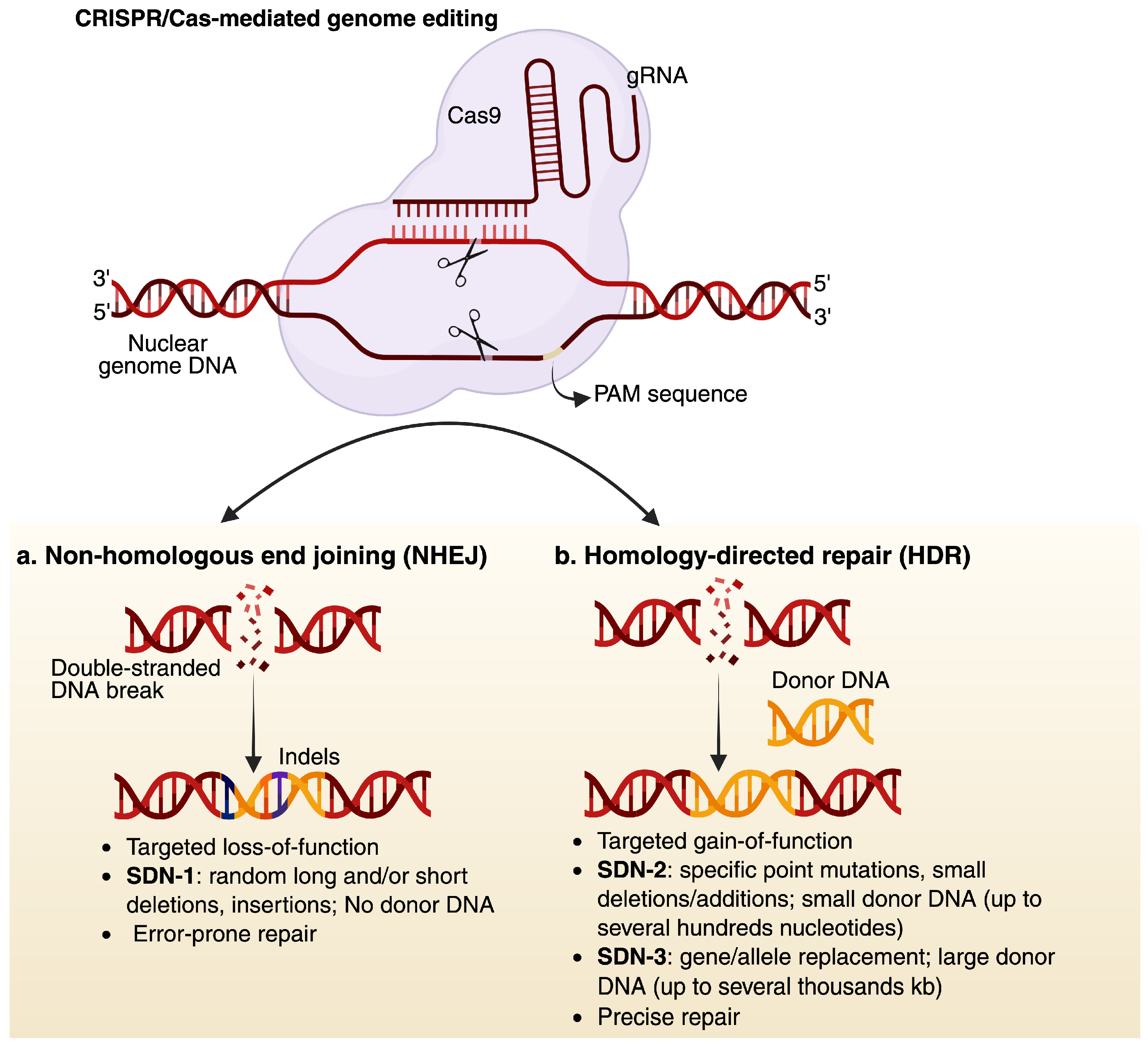
5.1. Overview of CRISPR-Cas System
5.2. Geminivirus-Based Vectors: A Stalwart Approach for Genome Editing
5.3. Full and Deconstructed Virus Vector Strategy
5.4. BeYDV: A Successful and Pioneer Geminivirus-Based Vector
5.5. Geminivirus-Based Homology-Directed Repair: Case Studies
| Viruses | Genus | GE Platform | gRNA Type | Plant Species | Targeted Gene * | Heritability | Reference |
|---|---|---|---|---|---|---|---|
| BeYDV | Mastrevirus | CRISPR-Cas9 | AtU6-gRNA | Solanum tuberosum | StALS1 | Yes | [61] |
| TALEN and CRISPR-Cas9 | AtU6-gRNA | S. lycopersicum | ALS1 | No | [51] | ||
| ZFN, TALEN and CRISPR-Cas9 | AtU6-gRNA | N. tabacum | ALS and P-GUS: NPTII | No | [15] | ||
| TALEN and CRISPR-Cas9 | AtU6-gRNAs | Tomato cv. MicroTom | ANT1 | Yes | [14] | ||
| CRISPR-Cas9 | AtU6-gRNAs | Lycopersicon esculentum | CRTISO and PSY1 | Yes | [62] | ||
| WDV | CRISPR-Cas9 | TaU6-gRNA | Triticum aestivum | Ubi, MLO, and GFP | No | [13] | |
| Cas9 expressing rice | OsU6-gRNAs | Oryza sativa | ACT1 and GST | No | [41] | ||
| CaLCuV | Begomovirus | Cas9 expressing tobacco | AtU6-gRNA | N. benthamiana | NbPDS3 and NbIspH | No | [50] |
| SPLCV | LwaCas13, LbCas12a and Cas9 | AtU6-gRNA | N. benthamiana | NbPDS1 and mGFP5 | No | [56] | |
| BCTV | Curtovirus | CRISPR/Cas12a | - | N. benthamiana | NbGFP | Yes | [55] |
6. Pros and Cons of Geminivirus-Based Vectors
7. Outlook of Viral Vector-Based Crop Design
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GT | Gene targeting. |
| HR | Homologous recombination. |
| IR | Intergenic region. |
| NSP | Nuclear shuttle protein. |
| MP | Movement protein. |
| CR | Common region. |
| ORFs | Open reading frames. |
| CP | Coat protein. |
| PD | Plasmodesmata. |
| ZFNs | Zinc-finger nucleases. |
| SDNs | Site-directed nucleases. |
| TALENs | Transcription activator-like effector nucleases. |
| CRISPR/Cas | Clustered, regularly interspaced short palindromic repeats-CRISPR/CRISPR-associated proteins. |
| DSBs | Double-stranded breaks. |
| NHEJ | Nonhomologous end joining. |
| HDR | Homology-directed repair. |
| SNV | Single-nucleotide variant. |
| KI | Knock-in. |
| PEG | Polyethylene glycol. |
| LIR | Long intergenic region. |
| GOI | Gene of interest. |
| SIR | Small intergenic region. |
| GB | GoldenBraid. |
| VIGE | Virus-induced genome editing. |
| PE | Prime editing. |
| VIGS | Virus-induced gene silencing. |
| BaMV | Bamboo mosaic virus. |
| BNYVV | Beet necrotic yellow vein virus. |
| BYSMV | Barley yellow striate mosaic virus. |
| PEBV | Pea early browning virus. |
| BMV | Brome mosaic virus. |
| CMV | Cucumber mosaic virus. |
| RTBV | Rice tungro bacilliform virus. |
| CymMV | Cymbidium mosaic virus. |
| WSMV | Wheat streak mosaic virus. |
| FoMV | Foxtail mosaic virus. |
| TYLCV | Tomato yellow leaf curl virus. |
| CMD | Cassava mosaic disease. |
| CMGs | Cassava mosaic geminiviruses. |
| CLCuD | Cotton leaf curl disease. |
| CLCuVs | Cotton leaf curl viruses. |
| BeYDV | Bean yellow dwarf virus. |
| WDV | Wheat dwarf virus. |
| ToLCV | Tomato leaf curl virus. |
| BCTV | Beet curly top virus. |
| CaMV | Cauliflower mosaic virus. |
| SPLCV | Sweet potato leaf curl virus. |
| SYNV | Sonchus yellow net rhabdovirus. |
| SCSMV | Sugarcane streak mosaic virus. |
References
- FAO. FAO’s Plant Production and Protection Division; FAO: Rome, Italy, 2022. [Google Scholar] [CrossRef]
- Mansoor, S.; Zafar, Y.; Briddon, R.W. Geminivirus disease complexes: The threat is spreading. Trends Plant Sci. 2006, 11, 209–212. [Google Scholar] [CrossRef]
- Mansoor, S.; Briddon, R.W.; Zafar, Y.; Stanley, J. Geminivirus disease complexes: An emerging threat. Trends Plant Sci. 2003, 8, 128–134. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.J.; Yuan, W.; Wang, R.Q.; Ye, Q.J.; Ruan, M.Y.; Li, Z.M.; Zhou, G.Z.; Yao, Z.P.; Yang, Y.J. Assessment of the genetic diversity of tomato yellow leaf curl virus. Genet. Mol. Res. 2015, 14, 529–537. [Google Scholar] [CrossRef]
- Uke, A.; Tokunaga, H.; Utsumi, Y.; Vu, N.A.; Nhan, P.T.; Srean, P.; Hy, N.H.; Ham, L.H.; Lopez-Lavalle, L.A.B.; Ishitani, M.; et al. Cassava mosaic disease and its management in Southeast Asia. Plant Mol. Biol. 2022, 109, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, S.; Amin, I.; Iram, S.; Hussain, M.; Zafar, Y.; Malik, K.; Briddon, R. Breakdown of resistance in cotton to cotton leaf curl disease in Pakistan. Plant Pathol. 2003, 52, 784. [Google Scholar] [CrossRef]
- Mahmood, M.A.; Ahmed, N.; Hussain, A.; Naqvi, R.Z.; Amin, I.; Mansoor, S. Dominance of Cotton leaf curl Multan virus-Rajasthan strain associated with third epidemic of cotton leaf curl disease in Pakistan. Sci. Rep. 2024, 14, 13532. [Google Scholar] [CrossRef]
- Mahmood, M.A.; Ahmed, N.; Hussain, S.; Muntaha, S.T.; Amin, I.; Mansoor, S. Dominance of Asia II 1 species of Bemisia tabaci in Pakistan and beyond. Sci. Rep. 2022, 12, 1528. [Google Scholar] [CrossRef]
- Mahmood, M.A.; Naqvi, R.Z.; Rahman, S.U.; Amin, I.; Mansoor, S. Plant Virus-Derived Vectors for Plant Genome Engineering. Viruses 2023, 15, 531. [Google Scholar] [CrossRef]
- Abrahamian, P.; Hammond, R.W.; Hammond, J. Plant Virus-Derived Vectors: Applications in Agricultural and Medical Biotechnology. Annu. Rev. Virol. 2020, 7, 513–535. [Google Scholar] [CrossRef]
- Hefferon, K. Plant Virus Expression Vectors: A Powerhouse for Global Health. Biomedicines 2017, 5, 44. [Google Scholar] [CrossRef] [PubMed]
- Gil-Humanes, J.; Wang, Y.; Liang, Z.; Shan, Q.; Ozuna, C.V.; Sánchez-León, S.; Baltes, N.J.; Starker, C.; Barro, F.; Gao, C.; et al. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 2017, 89, 1251–1262. [Google Scholar] [CrossRef]
- Čermák, T.; Baltes, N.J.; Čegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef]
- Baltes, N.J.; Gil-Humanes, J.; Cermak, T.; Atkins, P.A.; Voytas, D.F. DNA replicons for plant genome engineering. Plant Cell. 2014, 26, 151–163. [Google Scholar] [CrossRef]
- Pasin, F.; Menzel, W.; Daròs, J.A. Harnessed viruses in the age of metagenomics and synthetic biology: An update on infectious clone assembly and biotechnologies of plant viruses. Plant Biotechnol. J. 2019, 17, 1010–1026. [Google Scholar] [CrossRef]
- Torti, S.; Schlesier, R.; Thümmler, A.; Bartels, D.; Römer, P.; Koch, B.; Werner, S.; Panwar, V.; Kanyuka, K.; Wirén, N.V.; et al. Transient reprogramming of crop plants for agronomic performance. Nat. Plants 2021, 7, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Hu, J.; Sun, Y.; Li, B.; Zhang, D.; Li, W.; Liu, J.; Li, D.; Gao, C.; Zhang, Y.; et al. Highly efficient heritable genome editing in wheat using an RNA virus and bypassing tissue culture. Mol. Plant 2021, 14, 1787–1798. [Google Scholar] [CrossRef] [PubMed]
- Uranga, M.; Aragonés, V.; García, A.; Mirabel, S.; Gianoglio, S.; Presa, S.; Granell, A.; Pasin, F.; Daròs, J.A. RNA virus-mediated gene editing for tomato trait breeding. Hortic. Res. 2024, 11, uhad279. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Lett, J.M.; Martin, D.P.; Roumagnac, P.; Varsani, A.; Zerbini, F.M.; Navas-Castillo, J. ICTV Virus Taxonomy Profile: Geminiviridae 2021. J. Gen. Virol. 2021, 102, 001696. [Google Scholar] [CrossRef]
- Shepherd, D.N.; Martin, D.P.; Van Der Walt, E.; Dent, K.; Varsani, A.; Rybicki, E.P. Maize streak virus: An old and complex ‘emerging’ pathogen. Mol. Plant Pathol. 2010, 11, 1–12. [Google Scholar] [CrossRef]
- Navas-Castillo, J.; Fiallo-Olivé, E.; Sánchez-Campos, S. Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 2011, 49, 219–248. [Google Scholar] [CrossRef]
- Lozano, G.; Trenado, H.P.; Fiallo-Olivé, E.; Chirinos, D.; Geraud-Pouey, F.; Briddon, R.W.; Navas-Castillo, J. Characterization of Non-coding DNA Satellites Associated with Sweepoviruses (Genus Begomovirus, Geminiviridae)—Definition of a Distinct Class of Begomovirus-Associated Satellites. Front. Microbiol. 2016, 7, 162. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Mahmood, M.A.; Amin, I.; Mansoor, S. Geminiviruses also encode small proteins with specific functions. Trends Microbiol. 2021, 29, 1052–1054. [Google Scholar] [CrossRef]
- Trenado, H.P.; Orílio, A.F.; Márquez-Martín, B.; Moriones, E.; Navas-Castillo, J. Sweepoviruses cause disease in sweet potato and related Ipomoea spp.: Fulfilling Koch’s postulates for a divergent group in the genus begomovirus. PLoS ONE 2011, 6, e27329. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, M.; Qazi, J.; Mansoor, S.; Briddon, R.W. Molecular characterisation and infectivity of a “Legumovirus” (genus Begomovirus: Family Geminiviridae) infecting the leguminous weed Rhynchosia minima in Pakistan. Virus Res. 2009, 145, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Tan, H.; Zhao, S.; Li, H.; Liu, H.; Ma, Y.; Zhang, X.; Rong, J.; Fu, X.; Lozano-Durán, R.; et al. Geminiviruses encode additional small proteins with specific subcellular localizations and virulence function. Nat. Commun. 2021, 12, 4278. [Google Scholar] [CrossRef]
- Lozano-Durán, R.; Rosas-Díaz, T.; Luna, A.P.; Bejarano, E.R. Identification of host genes involved in geminivirus infection using a reverse genetics approach. PLoS ONE 2011, 6, e22383. [Google Scholar] [CrossRef]
- Wang, L.; Lozano-Durán, R. Manipulation of plant RNA biology by geminiviruses. J. Exp. Bot. 2023, 74, 2311–2322. [Google Scholar] [CrossRef]
- Mahmood, M.A.; Naqvi, R.Z.; Amin, I.; Mansoor, S. Salicylic acid-driven innate antiviral immunity in plants. Trends Plant Sci. 2024, 29, 715–717. [Google Scholar] [CrossRef]
- Settlage, S.B.; See, R.G.; Hanley-Bowdoin, L. Geminivirus C3 protein: Replication enhancement and protein interactions. J. Virol. 2005, 79, 9885–9895. [Google Scholar] [CrossRef]
- Gafni, Y.; Epel, B.L. The role of host and viral proteins in intra-and inter-cellular trafficking of geminiviruses. Physiol. Mol. Plant Pathol. 2002, 60, 231–241. [Google Scholar] [CrossRef]
- Neubauer, M.; Vollen, K.; Ascencio-Ibanez, J.T.; Hanley-Bowdoin, L.; Stepanova, A.N.; Alonso, J.M. Development of modular geminivirus-based vectors for high cargo expression and gene targeting in plants. BioRxiv 2024. [Google Scholar] [CrossRef]
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef]
- Mahmood, M.A.; Naqvi, R.Z.; Mansoor, S. Engineering crop resistance by manipulating disease susceptibility genes. Mol. Plant 2022, 15, 1511–1513. [Google Scholar] [CrossRef]
- Mahmood, M.A.; Greenwood, J.R. A prime example of precisely delivered DNA. Trends Genet. 2023, 39, 717–718. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, M.A.; Mansoor, S. PASTE: The way forward for large DNA insertions. CRISPR J. 2023, 6, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Awan, M.J.A.; Mahmood, M.A.; Naqvi, R.Z.; Mansoor, S. PASTE: A high-throughput method for large DNA insertions. Trends Plant Sci. 2023, 28, 509–511. [Google Scholar] [CrossRef]
- Chen, X.; Du, J.; Yun, S.; Xue, C.; Yao, Y.; Rao, S. Recent advances in CRISPR-Cas9-based genome insertion technologies. Mol. Ther. Nucleic Acids 2024, 35, 102138. [Google Scholar] [CrossRef]
- Zaidi, S.S.; Mansoor, S. Viral Vectors for Plant Genome Engineering. Front. Plant Sci. 2017, 8, 539. [Google Scholar] [CrossRef]
- Wang, M.; Lu, Y.; Botella, J.R.; Mao, Y.; Hua, K.; Zhu, J.K. Gene Targeting by Homology-Directed Repair in Rice Using a Geminivirus-Based CRISPR/Cas9 System. Mol. Plant 2017, 10, 1007–1010. [Google Scholar] [CrossRef]
- Mor, T.S.; Moon, Y.S.; Palmer, K.E.; Mason, H.S. Geminivirus vectors for high-level expression of foreign proteins in plant cells. Biotechnol. Bioeng. 2003, 81, 430–437. [Google Scholar] [CrossRef]
- Gleba, Y.; Marillonnet, S.; Klimyuk, V. Engineering viral expression vectors for plants: The ‘full virus’ and the ‘deconstructed virus’ strategies. Curr. Opin. Plant Biol. 2004, 7, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, W.E.; Sunter, G.; Brand, L.; Elmer, J.S.; Rogers, S.G.; Bisaro, D.M. Genetic analysis of tomato golden mosaic virus: The coat protein is not required for systemic spread or symptom development. EMBO J. 1988, 7, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Hayes, R.J.; Petty, I.T.D.; Coutts, R.H.A.; Buck, K.W. Gene amplification and expression in plants by a replicating geminivirus vector. Nature 1988, 334, 179–182. [Google Scholar] [CrossRef]
- Pooma, W.; Gillette, W.K.; Jeffrey, J.L.; Petty, I.T. Host and viral factors determine the dispensability of coat protein for bipartite geminivirus systemic movement. Virology 1996, 218, 264–268. [Google Scholar] [CrossRef][Green Version]
- Ward, A.; Etessami, P.; Stanley, J. Expression of a bacterial gene in plants mediated by infectious geminivirus DNA. Embo. J. 1988, 7, 1583–1587. [Google Scholar] [CrossRef] [PubMed]
- Muangsan, N.; Robertson, D. Geminivirus vectors for transient gene silencing in plants. Methods Mol. Biol. 2004, 265, 101–115. [Google Scholar] [CrossRef]
- Gilbertson, R.L.; Sudarshana, M.; Jiang, H.; Rojas, M.R.; Lucas, W.J. Limitations on geminivirus genome size imposed by plasmodesmata and virus-encoded movement protein: Insights into DNA trafficking. Plant Cell. 2003, 15, 2578–2591. [Google Scholar] [CrossRef]
- Yin, K.; Han, T.; Liu, G.; Chen, T.; Wang, Y.; Yu, A.Y.; Liu, Y. A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci. Rep. 2015, 5, 14926. [Google Scholar] [CrossRef]
- Butler, N.M.; Baltes, N.J.; Voytas, D.F.; Douches, D.S. Geminivirus-Mediated Genome Editing in Potato (Solanum tuberosum L.) Using Sequence-Specific Nucleases. Front. Plant Sci. 2016, 7, 1045. [Google Scholar] [CrossRef]
- Hummel, A.W.; Chauhan, R.D.; Cermak, T.; Mutka, A.M.; Vijayaraghavan, A.; Boyher, A.; Starker, C.G.; Bart, R.; Voytas, D.F.; Taylor, N.J. Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol. J. 2018, 16, 1275–1282. [Google Scholar] [CrossRef] [PubMed]
- Peyret, H.; Lomonossoff, G.P. When plant virology met Agrobacterium: The rise of the deconstructed clones. Plant Biotechnol. J. 2015, 13, 1121–1135. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, M.; Wang, X.; Zhang, Y.J.; Da, X.W.; Jia, L.Y.; Pang, H.L.; Feng, H.Q. Effects of plant growth regulators on transient expression of foreign gene in Nicotiana benthamiana L. leaves. Bioresour. Bioprocess. 2021, 8, 124. [Google Scholar] [CrossRef] [PubMed]
- Eini, O.; Schumann, N.; Niessen, M.; Varrelmann, M. Targeted mutagenesis in plants using Beet curly top virus for efficient delivery of CRISPR/Cas12a components. New Biotechnol. 2022, 67, 1–11. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, X.; Sun, H.; Liang, Q.; Wang, W.; Zhang, C.; Bian, X.; Cao, Q.; Li, Q.; Xie, Y.; et al. Improving CRISPR-Cas-mediated RNA targeting and gene editing using SPLCV replicon-based expression vectors in Nicotiana benthamiana. Plant Biotechnol. J. 2020, 18, 1993–1995. [Google Scholar] [CrossRef]
- Sarrion-Perdigones, A.; Falconi, E.E.; Zandalinas, S.I.; Juárez, P.; Fernández-del-Carmen, A.; Granell, A.; Orzaez, D. GoldenBraid: An iterative cloning system for standardized assembly of reusable genetic modules. PLoS ONE 2011, 6, e21622. [Google Scholar] [CrossRef]
- Naqvi, R.Z.; Siddiqui, H.A.; Mahmood, M.A.; Najeebullah, S.; Ehsan, A.; Azhar, M.; Farooq, M.; Amin, I.; Asad, S.; Mukhtar, Z.; et al. Smart breeding approaches in post-genomics era for developing climate-resilient food crops. Front Plant Sci. 2022, 13, 972164. [Google Scholar] [CrossRef]
- Naqvi, R.Z.; Mahmood, M.A.; Mansoor, S.; Amin, I.; Asif, M. Omics-driven exploration and mining of key functional genes for the improvement of food and fiber crops. Front Plant Sci. 2023, 14, 1273859. [Google Scholar] [CrossRef]
- Vu, T.V.; Nguyen, N.T.; Kim, J.; Song, Y.J.; Nguyen, T.H.; Kim, J.Y. Optimized dicot prime editing enables heritable desired edits in tomato and Arabidopsis. Nat. Plants 2024, 10, 1502–1513. [Google Scholar] [CrossRef]
- Butler, N.M.; Atkins, P.A.; Voytas, D.F.; Douches, D.S. Generation and Inheritance of Targeted Mutations in Potato (Solanum tuberosum L.) Using the CRISPR/Cas System. PLoS ONE 2015, 10, e0144591. [Google Scholar] [CrossRef]
- Dahan-Meir, T.; Filler-Hayut, S.; Melamed-Bessudo, C.; Bocobza, S.; Czosnek, H.; Aharoni, A.; Levy, A.A. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 2018, 95, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.-Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef]
- Pausch, P.; Al-Shayeb, B.; Bisom-Rapp, E.; Tsuchida, C.A.; Li, Z.; Cress, B.F.; Knott, G.J.; Jacobsen, S.E.; Banfield, J.F.; Doudna, J.A. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 2020, 369, 333–337. [Google Scholar] [CrossRef]
- Rauch, B.J.; DeLoughery, A.; Sper, R.; Chen, S.; Yunanda, S.; Masnaghetti, M.; Chai, N.; Lin, J.C.; Neckelmann, A.; Bjornson, Y.; et al. Single-AAV CRISPR editing of skeletal muscle in non-human primates with NanoCas, an ultracompact nuclease. BioRxiv 2025. [Google Scholar] [CrossRef]
- Ruhel, R.; Chakraborty, S. Multifunctional roles of geminivirus encoded replication initiator protein. Virus Dis. 2019, 30, 66–73. [Google Scholar] [CrossRef]
- Wu, L.; Yang, J.; Gu, Y.; Wang, Q.; Zhang, Z.; Guo, H.; Zhao, L.; Zhang, H.; Gu, L. Bamboo mosaic virus-mediated transgene-free genome editing in bamboo. New Phytol. 2025, 245, 1810–1816. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Ellison, E.E.; Myers, E.A.; Donahue, L.I.; Xuan, S.; Swanson, R.; Qi, S.; Prichard, L.E.; Starker, C.G.; Voytas, D.F. Heritable gene editing in tomato through viral delivery of isopentenyl transferase and single-guide RNAs to latent axillary meristematic cells. Proc. Natl. Acad. Sci. USA 2024, 121, e2406486121. [Google Scholar] [CrossRef]
- Uranga, M.; Aragonés, V.; Selma, S.; Vázquez-Vilar, M.; Orzáez, D.; Daròs, J.A. Efficient Cas9 multiplex editing using unspaced sgRNA arrays engineering in a Potato virus X vector. Plant J. 2021, 106, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Eid, A.; Ali, S.; Mahfouz, M.M. Pea early-browning virus-mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis. Virus Res. 2018, 244, 333–337. [Google Scholar] [CrossRef]
- Hu, J.; Li, S.; Li, Z.; Li, H.; Song, W.; Zhao, H.; Lai, J.; Xia, L.; Li, D.; Zhang, Y. A barley stripe mosaic virus-based guide RNA delivery system for targeted mutagenesis in wheat and maize. Mol. Plant Pathol. 2019, 20, 1463–1474. [Google Scholar] [CrossRef]
- Wang, W.; Yu, Z.; He, F.; Bai, G.; Trick, H.N.; Akhunova, A.; Akhunov, E. Multiplexed promoter and gene editing in wheat using a virus-based guide RNA delivery system. Plant Biotechnol. J. 2022, 20, 2332–2341. [Google Scholar] [CrossRef]
- Tamilselvan-Nattar-Amutha, S.; Hiekel, S.; Hartmann, F.; Lorenz, J.; Dabhi, R.V.; Dreissig, S.; Hensel, G.; Kumlehn, J.; Heckmann, S. Barley stripe mosaic virus-mediated somatic and heritable gene editing in barley (Hordeum vulgare L.). Front. Plant Sci. 2023, 14, 1201446. [Google Scholar] [CrossRef]
- Baysal, C.; Kausch, A.P.; Cody, J.P.; Altpeter, F.; Voytas, D.F. Rapid and efficient in planta genome editing in sorghum using foxtail mosaic virus-mediated sgRNA delivery. Plant J. 2025, 121, e17196. [Google Scholar] [CrossRef]
- Bouton, C.; King, R.C.; Chen, H.; Azhakanandam, K.; Bieri, S.; Hammond-Kosack, K.E.; Kanyuka, K. Foxtail mosaic virus: A Viral Vector for Protein Expression in Cereals. Plant Physiol. 2018, 177, 1352–1367. [Google Scholar] [CrossRef]
- Bredow, M.; Natukunda, M.I.; Beernink, B.M.; Chicowski, A.S.; Salas-Fernandez, M.G.; Whitham, S.A. Characterization of a foxtail mosaic virus vector for gene silencing and analysis of innate immune responses in Sorghum bicolor. Mol. Plant Pathol. 2023, 24, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Huang, C.; Luo, C.; Zhang, Y.; Zhang, B.; Xie, Z.; Hao, M.; Ling, H.; Cao, G.; Tian, B.; et al. A New Method for Rapid Subcellular Localization and Gene Function Analysis in Cotton Based on Barley Stripe Mosaic Virus. Plants 2022, 11, 1765. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.S.; Schneider, W.L.; Chaluvadi, S.R.; Mian, M.A.; Nelson, R.S. Characterization of a Brome mosaic virus strain and its use as a vector for gene silencing in monocotyledonous hosts. Mol. Plant Microbe Interact. 2006, 19, 1229–1239. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Xu, W.-Y.; Yan, T.; Fang, X.-D.; Cao, Q.; Zhang, Z.-J.; Ding, Z.-H.; Wang, Y.; Wang, X.-B. Rescue of a plant cytorhabdovirus as versatile expression platforms for planthopper and cereal genomic studies. New Phytol. 2019, 223, 2120–2133. [Google Scholar] [CrossRef]
- Hsieh, M.H.; Lu, H.C.; Pan, Z.J.; Yeh, H.H.; Wang, S.S.; Chen, W.H.; Chen, H.H. Optimizing virus-induced gene silencing efficiency with Cymbidium mosaic virus in Phalaenopsis flower. Plant Sci. 2013, 201–202, 25–41. [Google Scholar] [CrossRef]
- Liou, M.R.; Huang, Y.W.; Hu, C.C.; Lin, N.S.; Hsu, Y.H. A dual gene-silencing vector system for monocot and dicot plants. Plant Biotechnol. J. 2014, 12, 330–343. [Google Scholar] [CrossRef]
- Globus, R.; Qimron, U. A technological and regulatory outlook on CRISPR crop editing. J. Cell Biochem. 2018, 119, 1291–1298. [Google Scholar] [CrossRef] [PubMed]
- McGarry, R.C.; Klocko, A.L.; Pang, M.; Strauss, S.H.; Ayre, B.G. Virus-Induced Flowering: An Application of Reproductive Biology to Benefit Plant Research and Breeding. Plant Physiol. 2017, 173, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Liou, M.-R.; Hu, C.-C.; Lin, N.-S.; Hsu, Y.-H. Development and application of satellite-based vectors. In Viroids and Satellites; Elsevier: Amsterdam, The Netherlands, 2017; pp. 597–604. [Google Scholar]
- Wunderlich, S.; Gatto, K.A. Consumer perception of genetically modified organisms and sources of information. Adv. Nutr. 2015, 6, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Ewa, W.G.; Agata, T.; Milica, P.; Anna, B.; Dennis, E.; Nick, V.; Godelieve, G.; Selim, C.; Naghmeh, A.; Tomasz, T. Public perception of plant gene technologies worldwide in the light of food security. GM Crops Food 2022, 13, 218–241. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahmood, M.A.; Sajjad, M.W.; Imran, I.; Naqvi, R.Z.; Amin, I.; Shafiq, M.; Aslam, M.Q.; Mansoor, S. Attaining the Promise of Geminivirus-Based Vectors in Plant Genome Editing. Viruses 2025, 17, 631. https://doi.org/10.3390/v17050631
Mahmood MA, Sajjad MW, Imran I, Naqvi RZ, Amin I, Shafiq M, Aslam MQ, Mansoor S. Attaining the Promise of Geminivirus-Based Vectors in Plant Genome Editing. Viruses. 2025; 17(5):631. https://doi.org/10.3390/v17050631
Chicago/Turabian StyleMahmood, Muhammad Arslan, Muhammad Waseem Sajjad, Ifrah Imran, Rubab Zahra Naqvi, Imran Amin, Muhammad Shafiq, Muhammad Qasim Aslam, and Shahid Mansoor. 2025. "Attaining the Promise of Geminivirus-Based Vectors in Plant Genome Editing" Viruses 17, no. 5: 631. https://doi.org/10.3390/v17050631
APA StyleMahmood, M. A., Sajjad, M. W., Imran, I., Naqvi, R. Z., Amin, I., Shafiq, M., Aslam, M. Q., & Mansoor, S. (2025). Attaining the Promise of Geminivirus-Based Vectors in Plant Genome Editing. Viruses, 17(5), 631. https://doi.org/10.3390/v17050631





