Protein Corona Fingerprints of Liposomes: New Opportunities for Targeted Drug Delivery and Early Detection in Pancreatic Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Liposomes Preparation
2.2. Size and Zeta-Potential Experiments
2.3. Proteomics Experiments
2.4. Cell Culture
2.5. Flow-Assisted Cell Sorting Experiments
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the united states. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, M. Pancreatic cancer. N. Engl. J. Med. 2010, 362, 1605–1617. [Google Scholar] [CrossRef]
- Kundranda, M.N.; Niu, J. Albumin-bound paclitaxel in solid tumors: Clinical development and future directions. Drug Des. Dev. Ther. 2015, 9, 3767–3777. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y. DNA topoisomerase i inhibitors: Chemistry, biology, and interfacial inhibition. Chem. Rev. 2009, 109, 2894–2902. [Google Scholar] [CrossRef] [PubMed]
- Ud Din, F.; Aman, W.; Ullah, I.; Qureshi, O.S.; Mustapha, O.; Shafique, S.; Zeb, A. Effective use of nanocarriers as drug delivery systems for the treatment of selected tumors. Int. J. Nanomed. 2017, 12, 7291–7309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DiGiulio, S. FDA approves onivyde combo regimen for advanced pancreatic cancer. Oncol. Times 2015, 37, 8. [Google Scholar] [CrossRef]
- Liu, D.; Liu, F.; Song, Y.K. Recognition and clearance of liposomes containing phosphatidylserine are mediated by serum opsonin. Biochim. Biophys. Acta (BBA)-Biomembr. 1995, 1235, 140–146. [Google Scholar] [CrossRef] [Green Version]
- Knop, K.; Hoogenboom, R.; Fischer, D.; Schubert, U.S. Poly(ethylene glycol) in drug delivery: Pros and cons as well as potential alternatives. Angew. Chem. Int. Edit. 2010, 49, 6288–6308. [Google Scholar] [CrossRef]
- Prencipe, G.; Tabakman, S.M.; Welsher, K.; Liu, Z.; Goodwin, A.P.; Zhang, L.; Henry, J.; Dai, H. Peg branched polymer for functionalization of nanomaterials with ultralong blood circulation. J. Am. Chem. Soc. 2009, 131, 4783–4787. [Google Scholar] [CrossRef]
- Pozzi, D.; Colapicchioni, V.; Caracciolo, G.; Piovesana, S.; Capriotti, A.L.; Palchetti, S.; De Grossi, S.; Riccioli, A.; Amenitsch, H.; Laganà, A. Effect of polyethyleneglycol (PEG) chain length on the bio-nano- interactions between pegylated lipid nanoparticles and biological fluids: From nanostructure to uptake in cancer cells. Nanoscale 2014, 6, 2782–2792. [Google Scholar] [CrossRef] [PubMed]
- Schöttler, S.; Becker, G.; Winzen, S.; Steinbach, T.; Mohr, K.; Landfester, K.; Mailänder, V.; Wurm, F.R. Protein adsorption is required for stealth effect of poly(ethylene glycol)-and poly(phosphoester)-coated nanocarriers. Nat. Nanotechnol. 2016, 11, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Harada, M.; Wang, X.Y.; Ichihara, M.; Irimura, K.; Kiwada, H. Accelerated blood clearance of pegylated liposomes following preceding liposome injection: Effects of lipid dose and peg surface-density and chain length of the first-dose liposomes. J. Control. Release 2005, 105, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Moghimi, S.M.; Hunter, A.C. Recognition by macrophages and liver cells of opsonized phospholipid vesicles and phospholipid headgroups. Pharm. Res. 2001, 18, 1–8. [Google Scholar] [CrossRef]
- Moghimi, S.M.; Hunter, A.C.; Murray, J.C. Long-circulating and target-specific nanoparticles: Theory to practice. Pharmacol. Rev. 2001, 53, 283–318. [Google Scholar] [PubMed]
- Moghimi, S.M.; Szebeni, J. Stealth liposomes and long circulating nanoparticles: Critical issues in pharmacokinetics, opsonization and protein-binding properties. Prog. Lipid Res. 2003, 42, 463–478. [Google Scholar] [CrossRef]
- Caracciolo, G. Liposome-protein corona in a physiological environment: Challenges and opportunities for targeted delivery of nanomedicines. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Caracciolo, G.; Farokhzad, O.C.; Mahmoudi, M. Biological identity of nanoparticles in vivo: Clinical implications of the protein corona. Trends Biotechnol. 2017, 35, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Caracciolo, G. Clinically approved liposomal nanomedicines: Lessons learned from the biomolecular corona. Nanoscale 2018, 10, 4167–4172. [Google Scholar] [CrossRef]
- Caracciolo, G.; Pozzi, D.; Capriotti, A.L.; Cavaliere, C.; Piovesana, S.; La Barbera, G.; Amici, A.; Laganà, A. The liposome-protein corona in mice and humans and its implications for in vivo delivery. J. Mater. Chem. B 2014, 2, 7419–7428. [Google Scholar] [CrossRef]
- Monopoli, M.P.; Aberg, C.; Salvati, A.; Dawson, K.A. Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotechnol. 2012, 7, 779–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walkey, C.D.; Chan, W.C. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 2012, 41, 2780–2799. [Google Scholar] [CrossRef] [PubMed]
- Tenzer, S.; Docter, D.; Kuharev, J.; Musyanovych, A.; Fetz, V.; Hecht, R.; Schlenk, F.; Fischer, D.; Kiouptsi, K.; Reinhardt, C. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 2013, 8, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Barrán-Berdón, A.L.; Pozzi, D.; Caracciolo, G.; Capriotti, A.L.; Caruso, G.; Cavaliere, C.; Riccioli, A.; Palchetti, S.; Laganaì, A. Time evolution of nanoparticle-protein corona in human plasma: Relevance for targeted drug delivery. Langmuir 2013, 29, 6485–6494. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, D.; Caracciolo, G.; Digiacomo, L.; Colapicchioni, V.; Palchetti, S.; Capriotti, A.L.; Cavaliere, C.; Zenezini Chiozzi, R.; Puglisi, A.; Laganà, A. The biomolecular corona of nanoparticles in circulating biological media. Nanoscale 2015, 7, 13958–13966. [Google Scholar] [CrossRef] [PubMed]
- Palchetti, S.; Digiacomo, L.; Pozzi, D.; Peruzzi, G.; Micarelli, E.; Mahmoudi, M.; Caracciolo, G. Nanoparticles-cell association predicted by protein corona fingerprints. Nanoscale 2016, 8, 12755–12763. [Google Scholar] [CrossRef] [PubMed]
- Mirshafiee, V.; Mahmoudi, M.; Lou, K.; Cheng, J.; Kraft, M.L. Protein corona significantly reduces active targeting yield. Chem. Commun. 2013, 49, 2557–2559. [Google Scholar] [CrossRef]
- Harashima, H.; Hiraiwa, T.; Ochi, Y.; Kiwada, H. Size dependent liposome degradation in blood: in vivo/in vitro correlation by kinetic modeling. J. Drug Target. 1995, 3, 253–261. [Google Scholar] [CrossRef]
- Jiang, W.; Kim, B.Y.; Rutka, J.T.; Chan, W.C. Nanoparticle-mediated cellular response is size-dependent. Nat. Nanotechnol. 2008, 3, 145–150. [Google Scholar] [CrossRef]
- Bigdeli, A.; Palchetti, S.; Pozzi, D.; Hormozi-Nezhad, M.R.; Baldelli Bombelli, F.; Caracciolo, G.; Mahmoudi, M. Exploring cellular interactions of liposomes using protein corona fingerprints and physicochemical properties. ACS Nano 2016, 10, 3723–3737. [Google Scholar] [CrossRef]
- Hajipour, M.J.; Laurent, S.; Aghaie, A.; Rezaee, F.; Mahmoudi, M. Personalized protein coronas: A “key” factor at the nanobiointerface. Biomater. Sci. 2014, 2, 1210–1221. [Google Scholar] [CrossRef]
- Monopoli, M.P.; Walczyk, D.; Campbell, A.; Elia, G.; Lynch, I.; Baldelli Bombelli, F.; Dawson, K.A. Physical—Chemical aspects of protein corona: Relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc. 2011, 133, 2525–2534. [Google Scholar] [CrossRef] [PubMed]
- Caracciolo, G.; Pozzi, D.; Caminiti, R.; Marchini, C.; Montani, M.; Amici, A.; Amenitsch, H. Transfection efficiency boost by designer multicomponent lipoplexes. Biochim. Biophys. Acta Biomembr. 2007, 1768, 2280–2292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pozzi, D.; Marchini, C.; Cardarelli, F.; Rossetta, A.; Colapicchioni, V.; Amici, A.; Montani, M.; Motta, S.; Brocca, P.; Cantù, L.; et al. Mechanistic understanding of gene delivery mediated by highly efficient multicomponent envelope-type nanoparticle systems. Mol. Pharm. 2013, 10, 4654–4665. [Google Scholar] [CrossRef]
- Caracciolo, G.; Pozzi, D.; Capriotti, A.L.; Cavaliere, C.; Piovesana, S.; Amenitsch, H.; Laganà, A. Lipid composition: A “key factor” for the rational manipulation of the liposome-protein corona by liposome design. RSC Adv. 2015, 5, 5967–5975. [Google Scholar] [CrossRef]
- Salvati, A.; Pitek, A.S.; Monopoli, M.P.; Prapainop, K.; Bombelli, F.B.; Hristov, D.R.; Kelly, P.M.; Åberg, C.; Mahon, E.; Dawson, K.A. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol. 2013, 8, 137–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahon, E.; Salvati, A.; Baldelli Bombelli, F.; Lynch, I.; Dawson, K.A. Designing the nanoparticle–biomolecule interface for “targeting and therapeutic delivery”. J. Control. Release 2012, 161, 164–174. [Google Scholar] [CrossRef]
- Caracciolo, G.; Cardarelli, F.; Pozzi, D.; Salomone, F.; Maccari, G.; Bardi, G.; Capriotti, A.L.; Cavaliere, C.; Papi, M.; Laganà, A. Selective targeting capability acquired with a protein corona adsorbed on the surface of 1,2-dioleoyl-3-trimethylammonium propane/dna nanoparticles. ACS Appl. Mater. Interfaces 2013, 5, 13171–13179. [Google Scholar] [CrossRef]
- Docter, D.; Westmeier, D.; Markiewicz, M.; Stolte, S.; Knauer, S.; Stauber, R. The nanoparticle biomolecule corona: Lessons learned–challenge accepted? Chem. Soc. Rev. 2015, 44, 6094–6121. [Google Scholar] [CrossRef]
- Treuel, L.; Docter, D.; Maskos, M.; Stauber, R.H. Protein corona–from molecular adsorption to physiological complexity. Beil. J. Nanotechnol. 2015, 6, 857–873. [Google Scholar] [CrossRef]
- Kelly, P.M.; Åberg, C.; Polo, E.; O’Connell, A.; Cookman, J.; Fallon, J.; Krpetić, Ž.; Dawson, K.A. Mapping protein binding sites on the biomolecular corona of nanoparticles. Nat. Nanotechnol. 2015, 10, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Gianneli, M.; Polo, E.; Lopez, H.; Castagnola, V.; Aastrup, T.; Dawson, K. Label-free in-flow detection of receptor recognition motifs on the biomolecular corona of nanoparticles. Nanoscale 2018, 10, 5474–5481. [Google Scholar] [CrossRef] [PubMed]
- Walkey, C.D.; Olsen, J.B.; Song, F.; Liu, R.; Guo, H.; Olsen, D.W.H.; Cohen, Y.; Emili, A.; Chan, W.C. Protein corona fingerprinting predicts the cellular interaction of gold and silver nanoparticles. ACS Nano 2014, 8, 2439–2455. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Jiang, W.; Walkey, C.D.; Chan, W.C.; Cohen, Y. Prediction of nanoparticles-cell association based on corona proteins and physicochemical properties. Nanoscale 2015, 7, 9664–9675. [Google Scholar] [CrossRef] [PubMed]
- Lorger, M.; Krueger, J.S.; O’Neal, M.; Staflin, K.; Felding-Habermann, B. Activation of tumor cell integrin αVβ3 controls angiogenesis and metastatic growth in the brain. Proc. Natl. Acad. Sci. USA 2009, 106, 10666–10671. [Google Scholar] [CrossRef]
- Weis, S.M.; Cheresh, D.A. αV integrins in angiogenesis and cancer. Cold Spring Harb. Perspect. Med. 2011, 1, a006478. [Google Scholar] [CrossRef]
- Hosotani, R.; Kawaguchi, M.; Masui, T.; Koshiba, T.; Ida, J.; Fujimoto, K.; Wada, M.; Doi, R.; Imamura, M. Expression of integrin αvβ3 in pancreatic carcinoma: Relation to MMP-2 activation and lymph node metastasis. Pancreas 2002, 25, e30–e35. [Google Scholar] [CrossRef]
- Trusolino, L.; Serini, G.; Cecchini, G.; Besati, C.; Ambesi-Impiombato, F.S.; Marchisio, P.C.; De Filippi, R. Growth factor–dependent activation of αvβ3 integrin in normal epithelial cells: Implications for tumor invasion. J. Cell Biol. 1998, 142, 1145–1156. [Google Scholar] [CrossRef]
- Neyen, C.; Plüddemann, A.; Mukhopadhyay, S.; Maniati, E.; Bossard, M.; Gordon, S.; Hagemann, T. Macrophage scavenger receptor a promotes tumor progression in murine models of ovarian and pancreatic cancer. J. Immunol. 2013, 190, 3798–3805. [Google Scholar] [CrossRef]
- Vasseur, S.; Guillaumond, F. LDL receptor: An open route to feed pancreatic tumor cells. Mol. Cell. Oncol. 2016, 3, e1033586. [Google Scholar] [CrossRef]
- Arcella, A.; Palchetti, S.; Digiacomo, L.; Pozzi, D.; Capriotti, A.L.; Frati, L.; Oliva, M.A.; Tsaouli, G.; Rota, R.; Screpanti, I. Brain targeting by liposome–biomolecular corona boosts anticancer efficacy of temozolomide in glioblastoma cells. ACS Chem. Neurosci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Papi, M.; Caputo, D.; Palmieri, V.; Coppola, R.; Palchetti, S.; Bugli, F.; Martini, C.; Digiacomo, L.; Pozzi, D.; Caracciolo, G. Clinically approved pegylated nanoparticles are covered by a protein corona that boosts the uptake by cancer cells. Nanoscale 2017, 9, 10327–10334. [Google Scholar] [CrossRef] [PubMed]
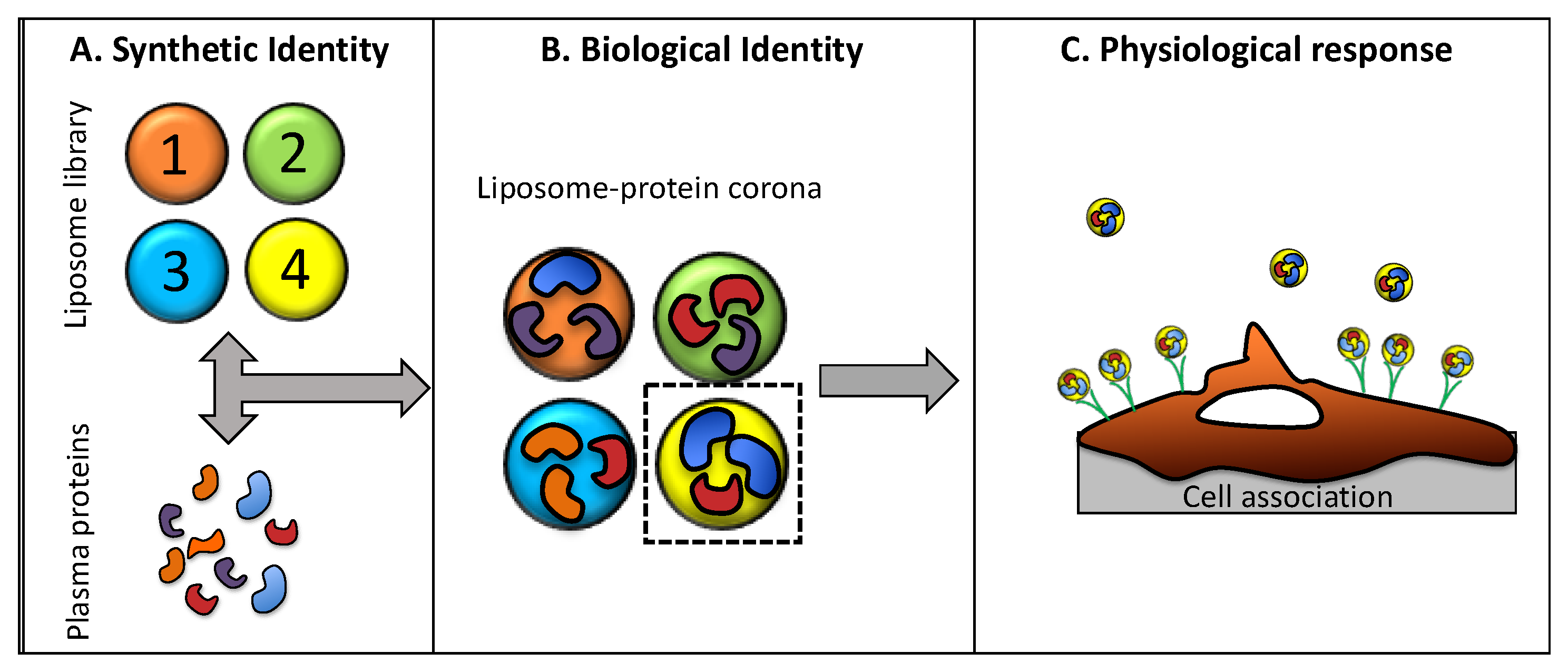

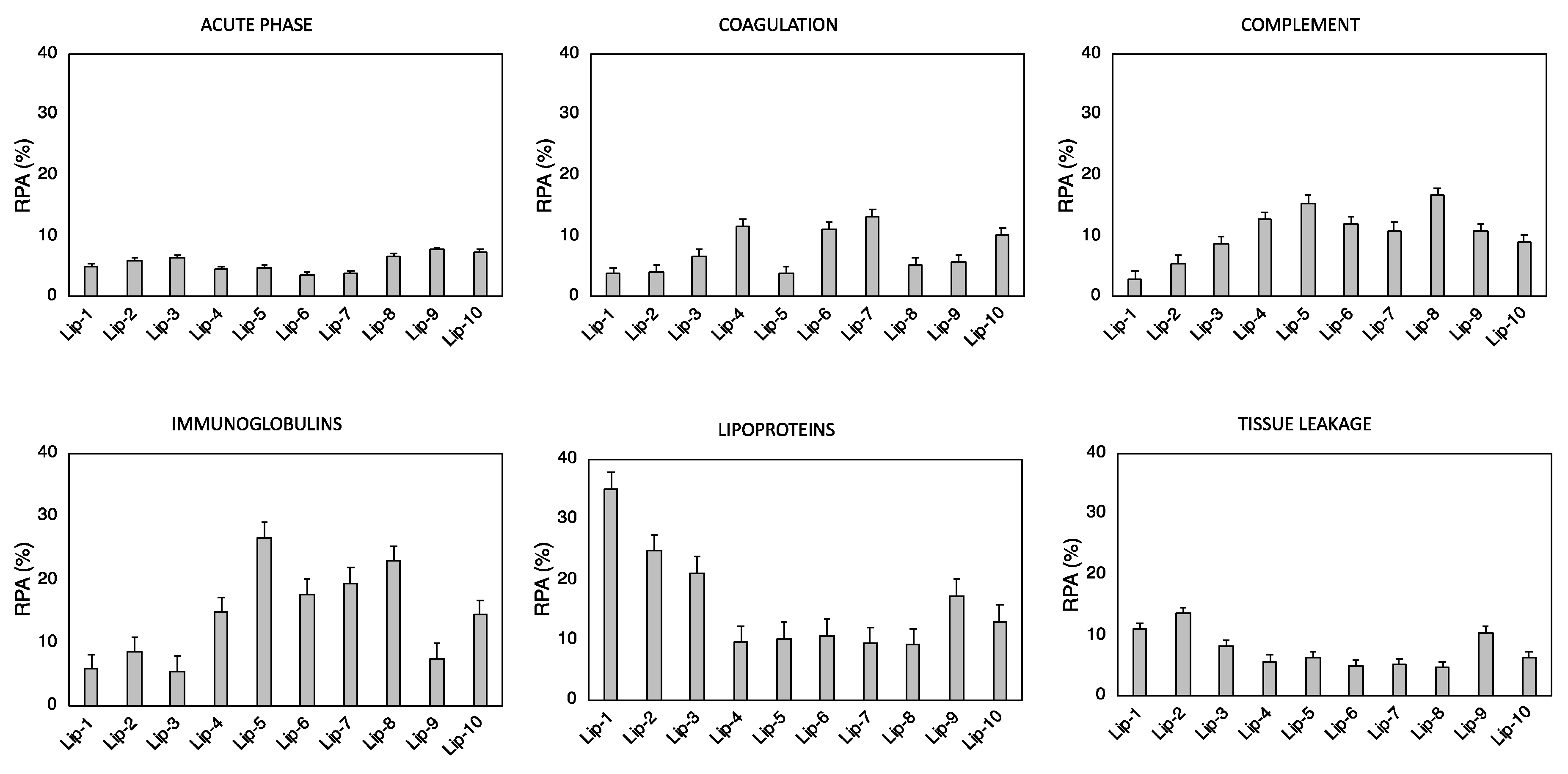

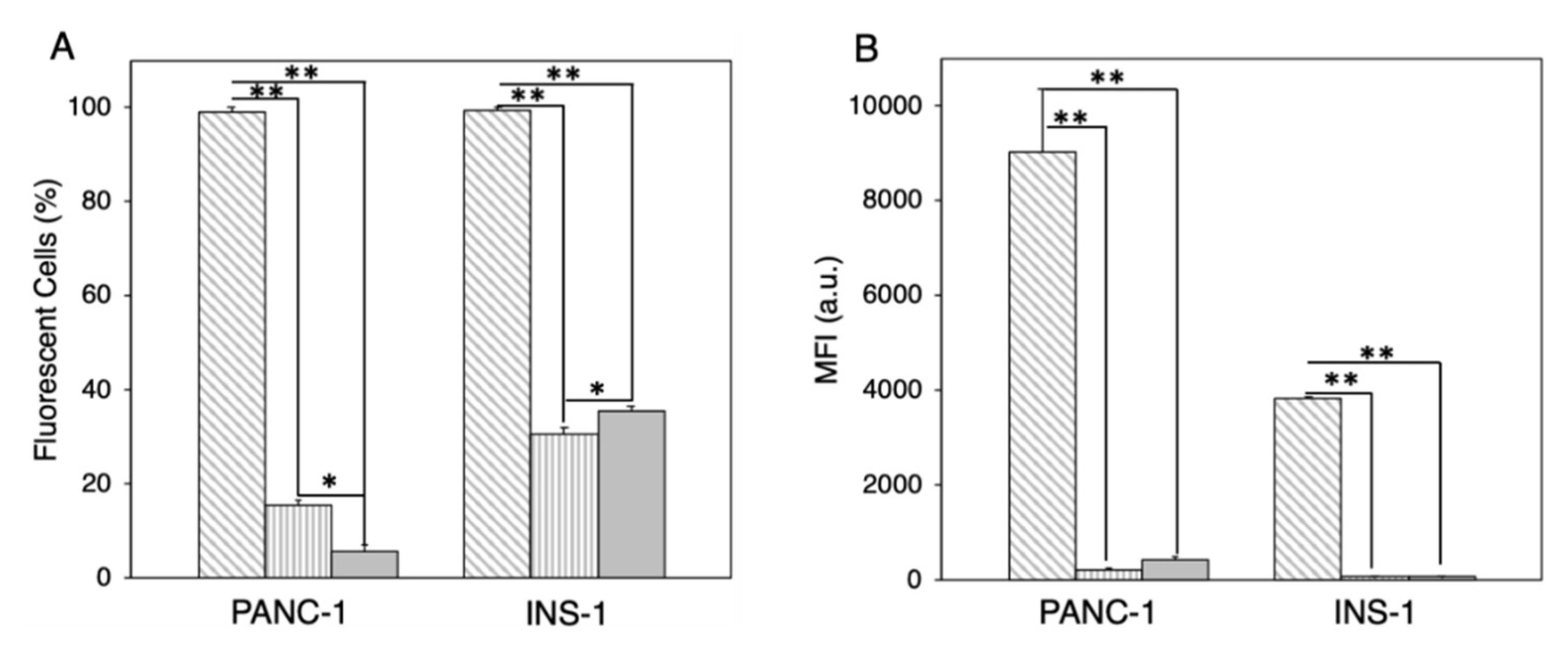
| Samples | CHOLESTEROL | DC-CHOL | DOPC | DOPE | DOTAP | DPPC | PC (20:0) | SPHINGOSINE |
|---|---|---|---|---|---|---|---|---|
| Lip-1 | 0 | 0 | 0 | 0.5 | 0.5 | 0 | 0 | 0 |
| Lip-2 | 0 | 0.5 | 0 | 0.5 | 0 | 0 | 0 | 0 |
| Lip-3 | 0 | 0.25 | 0.25 | 0.25 | 0.25 | 0 | 0 | 0 |
| Lip-4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Lip-5 | 0.2 | 0 | 0 | 0 | 0 | 0 | 0,8 | 0 |
| Lip-6 | 0.25 | 0 | 0 | 0 | 0.5 | 0 | 0.25 | 0 |
| Lip-7 | 0.25 | 0 | 0 | 0 | 0.5 | 0.25 | 0 | 0 |
| Lip-8 | 0.33 | 0 | 0 | 0 | 0 | 0 | 0.33 | 0.33 |
| Lip-9 | 0.5 | 0 | 0 | 0 | 0.5 | 0 | 0 | 0 |
| Lip-10 | 0.5 | 0.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| Samples | PDI | |
|---|---|---|
| −HP | +HP | |
| Lip-1 | 0.14 ± 0.04 | 0.12 ± 0.04 |
| Lip-2 | 0.11 ± 0.01 | 0.17 ± 0.05 |
| Lip-3 | 0.08 ± 0.01 | 0.13 ± 0.01 |
| Lip-4 | 0.12 ± 0.02 | 0.14 ± 0.01 |
| Lip-5 | 0.10 ± 0.02 | 0.16 ± 0.05 |
| Lip-6 | 0.10 ± 0.02 | 0.17 ± 0.01 |
| Lip-7 | 0.12 ± 0.02 | 0.20 ± 0.04 |
| Lip-8 | 0.11 ± 0.02 | 0.18 ± 0.03 |
| Lip-9 | 0.15 ± 0.01 | 0.22 ± 0.02 |
| Lip-10 | 0.14 ± 0.01 | 0.20 ± 0.01 |
| Samples | Protein (μg/μL) |
|---|---|
| +HP | |
| Lip-1 | 4.9 ± 0.4 |
| Lip-2 | 5.3 ± 0.6 |
| Lip-3 | 4.8 ± 0.5 |
| Lip-4 | 9.0 ± 0.9 |
| Lip-5 | 3.1 ± 0.4 |
| Lip-6 | 4.6 ± 0.3 |
| Lip-7 | 4.1 ± 0.4 |
| Lip-8 | 3.7 ± 0.4 |
| Lip-9 | 9.5 ± 0.9 |
| Lip-10 | 6.6 ± 0.5 |
| Samples | #Identified Proteins |
|---|---|
| Lip-1 | 220 |
| Lip-2 | 140 |
| Lip-3 | 205 |
| Lip-4 | 170 |
| Lip-5 | 174 |
| Lip-6 | 202 |
| Lip-7 | 206 |
| Lip-8 | 180 |
| Lip-9 | 222 |
| Lip-10 | 190 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palchetti, S.; Caputo, D.; Digiacomo, L.; Capriotti, A.L.; Coppola, R.; Pozzi, D.; Caracciolo, G. Protein Corona Fingerprints of Liposomes: New Opportunities for Targeted Drug Delivery and Early Detection in Pancreatic Cancer. Pharmaceutics 2019, 11, 31. https://doi.org/10.3390/pharmaceutics11010031
Palchetti S, Caputo D, Digiacomo L, Capriotti AL, Coppola R, Pozzi D, Caracciolo G. Protein Corona Fingerprints of Liposomes: New Opportunities for Targeted Drug Delivery and Early Detection in Pancreatic Cancer. Pharmaceutics. 2019; 11(1):31. https://doi.org/10.3390/pharmaceutics11010031
Chicago/Turabian StylePalchetti, Sara, Damiano Caputo, Luca Digiacomo, Anna Laura Capriotti, Roberto Coppola, Daniela Pozzi, and Giulio Caracciolo. 2019. "Protein Corona Fingerprints of Liposomes: New Opportunities for Targeted Drug Delivery and Early Detection in Pancreatic Cancer" Pharmaceutics 11, no. 1: 31. https://doi.org/10.3390/pharmaceutics11010031
APA StylePalchetti, S., Caputo, D., Digiacomo, L., Capriotti, A. L., Coppola, R., Pozzi, D., & Caracciolo, G. (2019). Protein Corona Fingerprints of Liposomes: New Opportunities for Targeted Drug Delivery and Early Detection in Pancreatic Cancer. Pharmaceutics, 11(1), 31. https://doi.org/10.3390/pharmaceutics11010031







