Transferrin-Conjugated Polymeric Nanoparticle for Receptor-Mediated Delivery of Doxorubicin in Doxorubicin-Resistant Breast Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Development of Dox-Resistant MDA-MB-231 Cells
2.3. Synthesis of Carboxylated Poloxamer 407
2.4. Preparation of Dox/F127&P123-Tf
2.5. Morphological Analysis and Characterization of Dox/F127&P123-Tf
2.6. Entrapment Efficiency and Loading Capacity of Dox in Dox/F127&P123-Tf
2.7. In Vitro Drug Release Study
2.8. In Vitro Cellular Uptake of Dox/F127&P123-Tf
2.9. Cell Migration Efficacy of Dox/F127&P123-Tf
2.10. In Vitro Cell Cytotoxicity
2.11. Live and Dead Cell Assay of Dox/F127&P123-Tf
2.12. Cell Cycle Arrest of Dox/F127&P123-Tf
2.13. Western Blot Analysis
2.14. In Vivo Imaging and Biodistribution Analysis
2.15. Statistical Analysis
3. Results and Discussion
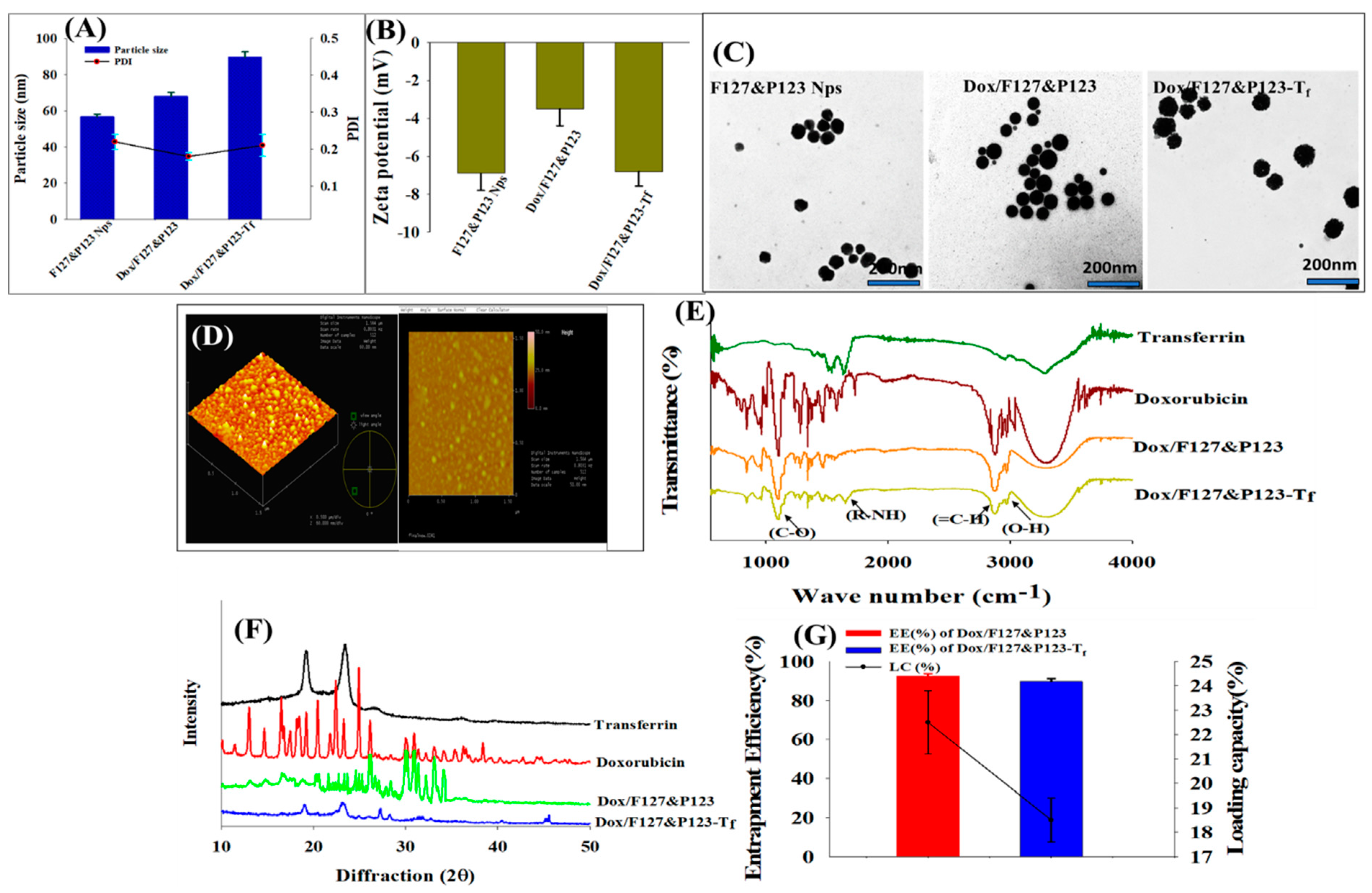
3.1. Physicochemical Characterization of Dox/F127&P123-Tf NPs
3.2. Entrapment Efficiency and Loading Capacity of Dox in Dox/F127&P123-Tf
3.3. In Vitro Drug Release Study
3.4. In Vitro Cellular Uptake of Dox/F127&P123-Tf
3.5. Cell Migration Efficacy of Dox/F127&P123-Tf
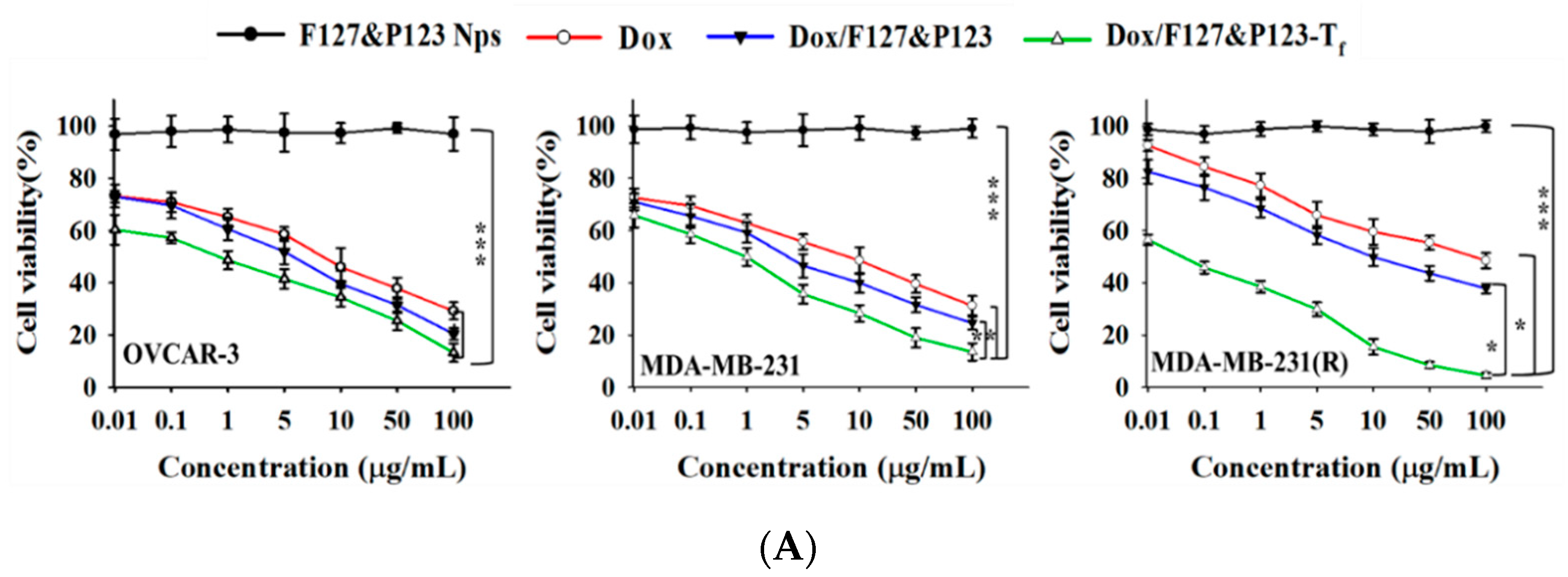
3.6. In Vitro Cellular Cytotoxicity of Dox/F127&P123-Tf
3.7. Live and Dead Cell Assay of Dox/F127&P123-Tf
3.8. Cell Cycle Arrest of Dox/F127&P123-Tf
3.9. Western Blot Analysis of Dox/F127&P123-Tf
3.10. In Vivo Imaging and Biodistribution Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Le, Q.-V.; Choi, J.; Oh, Y.-K. Nano delivery systems and cancer immunotherapy. J. Pharm. Investig. 2018, 48, 527–539. [Google Scholar] [CrossRef]
- Gupta, B.; Yong, C.S.; Kim, J.O. Solid matrix-based lipid nanoplatforms as carriers for combinational therapeutics in cancer. J. Pharm. Investig. 2017, 47, 461–473. [Google Scholar] [CrossRef]
- Bazak, R.; Houri, M.; Achy, S.E.; Hussein, W.; Refaat, T. Passive targeting of nanoparticles to cancer: A comprehensive review of the literature. Mol. Clin. Oncol. 2014, 2, 904–908. [Google Scholar] [CrossRef]
- Gullotti, E.; Yeo, Y. Extracellularly Activated Nanocarriers: A New Paradigm of Tumor Targeted Drug Delivery. Mol. Pharm. 2009, 6, 1041–1051. [Google Scholar] [CrossRef]
- Choi, Y.H.; Han, H.-K. Nanomedicines: Current status and future perspectives in aspect of drug delivery and pharmacokinetics. J. Pharm. Investig. 2018, 48, 43–60. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Williams, R.O. Polymeric nanomedicines for poorly soluble drugs in oral delivery systems: An update. J. Pharm. Investig. 2018, 48, 61–75. [Google Scholar] [CrossRef]
- Liu, X.; Li, Y.; He, J.; Zhao, T.; Chen, C.; Gu, H.; Wang, X. Paclitaxel-loaded pluronic F127/P123 silica nanocapsules with surface conjugated rhTRAIL for targeted cancer therapy. RSC Adv. 2017, 7, 30250–30261. [Google Scholar] [CrossRef]
- Peng, T.; Liu, K.; Gao, L.; Gao, L.; Chen, J.; Wang, J.; Liu, Y.; Wang, Y.; Yan, Z.; Yu, L. Poly (l-γ-glutamylglutamine) Polymer Enhances Doxorubicin Accumulation in Multidrug Resistant Breast Cancer Cells. Molecules 2016, 21, 720. [Google Scholar] [CrossRef]
- Piktel, E.; Niemirowicz, K.; Wątek, M.; Wollny, T.; Deptuła, P.; Bucki, R. Recent insights in nanotechnology-based drugs and formulations designed for effective anti-cancer therapy. J. Nanobiotechnol. 2016, 14, 39. [Google Scholar] [CrossRef]
- Xu, P.-Y.; Kankala, R.K.; Pan, Y.-J.; Yuan, H.; Wang, S.-B.; Chen, A.-Z. Overcoming multidrug resistance through inhalable siRNA nanoparticles-decorated porous microparticles based on supercritical fluid technology. Int. J. Nanomed. 2018, 13, 4685–4698. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, H.; Shi, J. Inorganic Nanoparticle-Based Drug Codelivery Nanosystems to Overcome the Multidrug Resistance of Cancer Cells. Mol. Pharm. 2014, 11, 2495–2510. [Google Scholar] [CrossRef] [PubMed]
- Kankala, R.K.; Tsai, P.-Y.; Kuthati, Y.; Wei, P.-R.; Liu, C.-L.; Lee, C.-H. Overcoming multidrug resistance through co-delivery of ROS-generating nano-machinery in cancer therapeutics. J. Mater. Chem. B 2017, 5, 1507–1517. [Google Scholar] [CrossRef]
- Bahrami, B.; Mohammadnia-Afrouzi, M.; Bakhshaei, P.; Yazdani, Y.; Ghalamfarsa, G.; Yousefi, M.; Sadreddini, S.; Jadidi-Niaragh, F.; Hojjat-Farsangi, M. Folate-conjugated nanoparticles as a potent therapeutic approach in targeted cancer therapy. Tumor Biol. 2015, 36, 5727–5742. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, T.; Ruttala, H.B.; Gupta, B.; Poudel, B.K.; Choi, H.G.; Yong, C.S.; Kim, J.O. Smart chemistry-based nanosized drug delivery systems for systemic applications: A comprehensive review. J. Control Release 2017, 258, 226–253. [Google Scholar] [CrossRef] [PubMed]
- Son, G.-H.; Lee, B.-J.; Cho, C.-W. Mechanisms of drug release from advanced drug formulations such as polymeric-based drug-delivery systems and lipid nanoparticles. J. Pharm. Investig. 2017, 47, 287–296. [Google Scholar] [CrossRef]
- Kim, C.H.; Lee, S.G.; Kang, M.J.; Lee, S.; Choi, Y.W. Surface modification of lipid-based nanocarriers for cancer cell-specific drug targeting. J. Pharm. Investig. 2017, 47, 203–227. [Google Scholar] [CrossRef]
- Bhushan, B.; Khanadeev, V.; Khlebtsov, B.; Khlebtsov, N.; Gopinath, P. Impact of albumin based approaches in nanomedicine: Imaging, targeting and drug delivery. Adv. Colloid Interface Sci. 2017, 246, 13–39. [Google Scholar] [CrossRef]
- Luria-Pérez, R.; Helguera, G.; Rodríguez, J.A. Antibody-mediated targeting of the transferrin receptor in cancer cells. Bol. Med. Hosp. Infant. Mex. 2016, 73, 372–379. [Google Scholar] [CrossRef]
- Sim, T.; Lim, C.; Hoang, N.H.; Oh, K.T. Recent advance of pH-sensitive nanocarriers targeting solid tumors. J. Pharm. Investig. 2017, 47, 383–394. [Google Scholar] [CrossRef]
- Phung, C.D.; Nguyen, H.T.; Tran, T.H.; Choi, H.-G.; Yong, C.S.; Kim, J.O. Rational combination immunotherapeutic approaches for effective cancer treatment. J. Control. Release 2019, 294, 114–130. [Google Scholar] [CrossRef]
- Li, Z.-L.; Chen, C.; Yang, Y.; Wang, C.; Yang, T.; Yang, X.; Liu, S.-C. Gamma secretase inhibitor enhances sensitivity to doxorubicin in MDA-MB-231 cells. Int. J. Clin. Exp. Pathol. 2015, 8, 4378–4387. [Google Scholar] [PubMed]
- Wang, S.; Konorev, E.A.; Kotamraju, S.; Joseph, J.; Kalivendi, S.; Kalyanaraman, B. Doxorubicin Induces Apoptosis in Normal and Tumor Cells via Distinctly Different Mechanisms: INTERMEDIACY OF H2O2- AND p53-DEPENDENT PATHWAYS. J. Biol. Chem. 2004, 279, 25535–25543. [Google Scholar] [CrossRef] [PubMed]
- AbuHammad, S.; Zihlif, M. Gene expression alterations in doxorubicin resistant MCF7 breast cancer cell line. Genomics 2013, 101, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Yue, W.; Xiao, W.; Liqian, X.; Xiaohong, Z.; Yunmei, Y. Chemoresistance is associated with overexpression of HAX-1, inhibition of which resensitizes drug-resistant breast cancer cells to chemotherapy. Tumor Biol. 2017, 39, 1010428317692228. [Google Scholar] [CrossRef]
- Sarisozen, C.; Pan, J.; Dutta, I.; Torchilin, V.P. Polymers in the co-delivery of siRNA and anticancer drugs to treat multidrug-resistant tumors. J. Pharm. Investig. 2017, 47, 37–49. [Google Scholar] [CrossRef]
- Kankala, R.K.; Liu, C.-G.; Chen, A.-Z.; Wang, S.-B.; Xu, P.-Y.; Mende, L.K.; Liu, C.-L.; Lee, C.-H.; Hu, Y.-F. Overcoming Multidrug Resistance through the Synergistic Effects of Hierarchical pH-Sensitive, ROS-Generating Nanoreactors. ACS Biomater. Sci. Eng. 2017, 3, 2431–2442. [Google Scholar] [CrossRef]
- Gupta, B.; Ramasamy, T.; Poudel, B.K.; Pathak, S.; Regmi, S.; Choi, J.Y.; Son, Y.; Thapa, R.K.; Jeong, J.-H.; Kim, J.R.; et al. Development of Bioactive PEGylated Nanostructured Platforms for Sequential Delivery of Doxorubicin and Imatinib to Overcome Drug Resistance in Metastatic Tumors. ACS Appl. Mater. Interfaces 2017, 9, 9280–9290. [Google Scholar] [CrossRef]
- Zhao, T.; Liu, X.; Li, Y.; Zhang, M.; He, J.; Zhang, X.; Liu, H.; Wang, X.; Gu, H. Fluorescence and drug loading properties of ZnSe:Mn/ZnS-Paclitaxel/SiO2 nanocapsules templated by F127 micelles. J. Colloid Interface Sci. 2017, 490, 436–443. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, M.; Zheng, Y.-g.; Tan, H.; Hsu, B.Y.-w.; Yang, Z.-c.; Wong, S.Y.; Chang, A.Y.-c.; Choolani, M.; Li, X.; et al. PEOlated Micelle/Silica as Dual-Layer Protection of Quantum Dots for Stable and Targeted Bioimaging. Chem. Mater. 2013, 25, 2976–2985. [Google Scholar] [CrossRef]
- Ruttala, H.B.; Ramasamy, T.; Poudal, B.K.; Choi, Y.; Choi, J.Y.; Kim, J.; Ku, S.K.; Choi, H.-G.; Yong, C.S.; Kim, J.O. Molecularly targeted co-delivery of a histone deacetylase inhibitor and paclitaxel by lipid-protein hybrid nanoparticles for synergistic combinational chemotherapy. Oncotarget 2017, 8, 14925–14940. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Phung, C.D.; Thapa, R.K.; Pham, T.T.; Tran, T.H.; Jeong, J.-H.; Ku, S.K.; Choi, H.-G.; Yong, C.S.; Kim, J.O. Multifunctional nanoparticles as somatostatin receptor-targeting delivery system of polyaniline and methotrexate for combined chemo–photothermal therapy. Acta Biomater. 2018, 68, 154–167. [Google Scholar] [CrossRef] [PubMed]
- Thapa, R.K.; Ku, S.K.; Choi, H.-G.; Yong, C.S.; Byeon, J.H.; Kim, J.O. Vibrating droplet generation to assemble zwitterion-coated gold-graphene oxide stealth nanovesicles for effective pancreatic cancer chemo-phototherapy. Nanoscale 2018, 10, 1742–1749. [Google Scholar] [CrossRef] [PubMed]
- Gupta, B.; Pathak, S.; Poudel, B.K.; Regmi, S.; Ruttala, H.B.; Gautam, M.; Lee, J.S.; Jeong, J.-H.; Choi, H.-G.; Yong, C.S.; et al. Folate receptor-targeted hybrid lipid-core nanocapsules for sequential delivery of doxorubicin and tanespimycin. Colloids Surf. B Biointerfaces 2017, 155, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, T.; Sundaramoorthy, P.; Ruttala, H.B.; Choi, Y.; Shin, W.H.; Jeong, J.-H.; Ku, S.K.; Choi, H.-G.; Kim, H.M.; Yong, C.S.; et al. Polyunsaturated fatty acid-based targeted nanotherapeutics to enhance the therapeutic efficacy of docetaxel. Drug Deliv. 2017, 24, 1262–1272. [Google Scholar] [CrossRef] [PubMed]
- Poudel, B.K.; Gupta, B.; Ramasamy, T.; Thapa, R.K.; Youn, Y.S.; Choi, H.G.; Yong, C.S.; Kim, J.O. Development of polymeric irinotecan nanoparticles using a novel lactone preservation strategy. Int. J. Pharm. 2016, 512, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Thapa, R.K.; Nguyen, H.T.; Jeong, J.-H.; Shin, B.S.; Ku, S.K.; Choi, H.-G.; Yong, C.S.; Kim, J.O. Synergistic anticancer activity of combined histone deacetylase and proteasomal inhibitor-loaded zein nanoparticles in metastatic prostate cancers. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Ruttala, H.B.; Ramasamy, T.; Gupta, B.; Choi, H.-G.; Yong, C.S.; Kim, J.O. Multiple polysaccharide–drug complex-loaded liposomes: A unique strategy in drug loading and cancer targeting. Carbohydr. Polym. 2017, 173, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Tran, T.H.; Thapa, R.K.; Phung, C.D.; Shin, B.S.; Jeong, J.-H.; Choi, H.-G.; Yong, C.S.; Kim, J.O. Targeted co-delivery of polypyrrole and rapamycin by trastuzumab-conjugated liposomes for combined chemo-photothermal therapy. Int. J. Pharm. 2017, 527, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Fu, L.; Li, X.; Zhang, Y.; Li, Z.; Wu, X.; Li, Y.; Bai, X.; Hu, D. Loss of CAR promotes migration and proliferation of HaCaT cells, and accelerates wound healing in rats via Src-p38 MAPK pathway. Sci. Rep. 2016, 6, 19735. [Google Scholar] [CrossRef] [PubMed]
- Thapa, R.K.; Soe, Z.C.; Ou, W.; Poudel, K.; Jeong, J.-H.; Jin, S.G.; Ku, S.K.; Choi, H.-G.; Lee, Y.M.; Yong, C.S.; et al. Palladium nanoparticle-decorated 2-D graphene oxide for effective photodynamic and photothermal therapy of prostate solid tumors. Colloids Surf. B Biointerfaces 2018, 169, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.T.; Nguyen, T.T.; Pathak, S.; Regmi, S.; Nguyen, H.T.; Tran, T.H.; Yong, C.S.; Kim, J.O.; Park, P.H.; Park, M.H.; et al. Tissue adhesive FK506–loaded polymeric nanoparticles for multi–layered nano–shielding of pancreatic islets to enhance xenograft survival in a diabetic mouse model. Biomaterials 2018, 154, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Thapa, R.K.; Nguyen, H.T.; Jeong, J.-H.; Kim, J.R.; Choi, H.-G.; Yong, C.S.; Kim, J.O. Progressive slowdown/prevention of cellular senescence by CD9-targeted delivery of rapamycin using lactose-wrapped calcium carbonate nanoparticles. Sci. Rep. 2017, 7, 43299. [Google Scholar] [CrossRef]
- Choi, J.Y.; Gupta, B.; Ramasamy, T.; Jeong, J.-H.; Jin, S.G.; Choi, H.-G.; Yong, C.S.; Kim, J.O. PEGylated polyaminoacid-capped mesoporous silica nanoparticles for mitochondria-targeted delivery of celastrol in solid tumors. Colloids Surf. B Biointerfaces 2018, 165, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Ou, W.; Thapa, R.K.; Jiang, L.; Soe, Z.C.; Gautam, M.; Chang, J.-H.; Jeong, J.-H.; Ku, S.K.; Choi, H.-G.; Yong, C.S.; et al. Regulatory T cell-targeted hybrid nanoparticles combined with immuno-checkpoint blockage for cancer immunotherapy. J. Control. Release 2018, 281, 84–96. [Google Scholar] [CrossRef]
- Mu hyun, J.; In joon, O. Targeted Drug Delivery of Transferrin-Conjugated Mesoporous Silica Nanoparticles. 약학회지 2017, 61, 241–247. [Google Scholar]
- Li, L.; Chen, W.; Zheng, J.; Wang, L.; Chen, Y. Characterization of Silver Nanoparticles Thin Films with Various Thicknesses by AFM. J. Mater. Sci. Chem. Eng. 2016, 4, 6. [Google Scholar] [CrossRef]
- Nguyen, M.N.-U.; Van Vo, T.; Tran, P.H.-L.; Tran, T.T.-D. Zein-based solid dispersion for potential application in targeted delivery. J. Pharm. Investig. 2017, 47, 357–364. [Google Scholar] [CrossRef]
- Dhamecha, D.; Jalalpure, S.; Jadhav, K. Doxorubicin functionalized gold nanoparticles: Characterization and activity against human cancer cell lines. Process Biochem. 2015, 50, 2298–2306. [Google Scholar] [CrossRef]
- Shilpi, S.; Vimal, V.D.; Soni, V. Assessment of lactoferrin-conjugated solid lipid nanoparticles for efficient targeting to the lung. Prog. Biomater. 2015, 4, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Danafar, H.; Rostamizadeh, K.; Hamidi, M. Polylactide/poly(ethylene glycol)/polylactide triblock copolymer micelles as carrier for delivery of hydrophilic and hydrophobic drugs: A comparison study. J. Pharm. Investig. 2018, 48, 381–391. [Google Scholar] [CrossRef]
- Ruttala, H.B.; Ramasamy, T.; Madeshwaran, T.; Hiep, T.T.; Kandasamy, U.; Oh, K.T.; Choi, H.-G.; Yong, C.S.; Kim, J.O. Emerging potential of stimulus-responsive nanosized anticancer drug delivery systems for systemic applications. Arch. Pharmacal. Res. 2018, 41, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-L.; Wang, L.; Liu, X.-Y.; Zhang, Z.-P.; Guo, H.-C.; Liu, W.-M.; Tang, S.-H. In vitro cancer cell imaging and therapy using transferrin-conjugated gold nanoparticles. Cancer Lett. 2009, 274, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, S.; Bhat, F.; Raja Singh, P.; Mukherjee, S.; Elumalai, P.; Das, S.; Patra, C.; Arunakaran, J. Gold nanoparticle–conjugated quercetin inhibits epithelial–mesenchymal transition, angiogenesis and invasiveness via EGFR/VEGFR-2-mediated pathway in breast cancer. Cell Prolif. 2016, 49, 678–697. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Weinman, S. Mechanisms of doxorubicin resistance in hepatocellular carcinoma. Hepatic Oncol. 2016, 3, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-Q.; Liu, K.; Huo, Z.-J.; Li, X.-C.; Wang, M.; Liu, P.; Pang, B.; Wang, S.-J. A cell-targeted chemotherapeutic nanomedicine strategy for oral squamous cell carcinoma therapy. J. Nanobiotechnol. 2015, 13, 63. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Sun, Z.-l. Transferrin-conjugated polymeric nanomedicine to enhance the anticancer efficacy of edelfosine in acute myeloid leukemia. Biomed. Pharm. 2016, 83, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhang, L.; Cao, H.; Yang, Y.; Zheng, Y.; Yang, X.-J. A Polyethylenimine-Containing and Transferrin-Conjugated Lipid Nanoparticle System for Antisense Oligonucleotide Delivery to AML. Biomed. Res. Int. 2016, 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Tsou, S.-H.; Chen, T.-M.; Hsiao, H.-T.; Chen, Y.-H. A Critical Dose of Doxorubicin Is Required to Alter the Gene Expression Profiles in MCF-7 Cells Acquiring Multidrug Resistance. PLoS ONE 2015, 10, e0116747. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soe, Z.C.; Kwon, J.B.; Thapa, R.K.; Ou, W.; Nguyen, H.T.; Gautam, M.; Oh, K.T.; Choi, H.-G.; Ku, S.K.; Yong, C.S.; et al. Transferrin-Conjugated Polymeric Nanoparticle for Receptor-Mediated Delivery of Doxorubicin in Doxorubicin-Resistant Breast Cancer Cells. Pharmaceutics 2019, 11, 63. https://doi.org/10.3390/pharmaceutics11020063
Soe ZC, Kwon JB, Thapa RK, Ou W, Nguyen HT, Gautam M, Oh KT, Choi H-G, Ku SK, Yong CS, et al. Transferrin-Conjugated Polymeric Nanoparticle for Receptor-Mediated Delivery of Doxorubicin in Doxorubicin-Resistant Breast Cancer Cells. Pharmaceutics. 2019; 11(2):63. https://doi.org/10.3390/pharmaceutics11020063
Chicago/Turabian StyleSoe, Zar Chi, Jun Bum Kwon, Raj Kumar Thapa, Wenquan Ou, Hanh Thuy Nguyen, Milan Gautam, Kyung Taek Oh, Han-Gon Choi, Sae Kwang Ku, Chul Soon Yong, and et al. 2019. "Transferrin-Conjugated Polymeric Nanoparticle for Receptor-Mediated Delivery of Doxorubicin in Doxorubicin-Resistant Breast Cancer Cells" Pharmaceutics 11, no. 2: 63. https://doi.org/10.3390/pharmaceutics11020063
APA StyleSoe, Z. C., Kwon, J. B., Thapa, R. K., Ou, W., Nguyen, H. T., Gautam, M., Oh, K. T., Choi, H.-G., Ku, S. K., Yong, C. S., & Kim, J. O. (2019). Transferrin-Conjugated Polymeric Nanoparticle for Receptor-Mediated Delivery of Doxorubicin in Doxorubicin-Resistant Breast Cancer Cells. Pharmaceutics, 11(2), 63. https://doi.org/10.3390/pharmaceutics11020063





