Linking Pharmacogenomic Information on Drug Safety and Efficacy with Ethnic Minority Populations
Abstract
:1. Introduction
2. Methods
2.1. Data Collection and Processing
- NSF: Nonsynonymous frameshift. A coding region variation where one allele in the set changes all downstream amino acids.
- NSM: Nonsynonymous missense. A coding region variation where one allele in the set changes a protein peptide.
- NSN: Nonsynonymous nonsense. A coding region variation where one allele in the set changes to a stop codon, i.e., a termination codon.
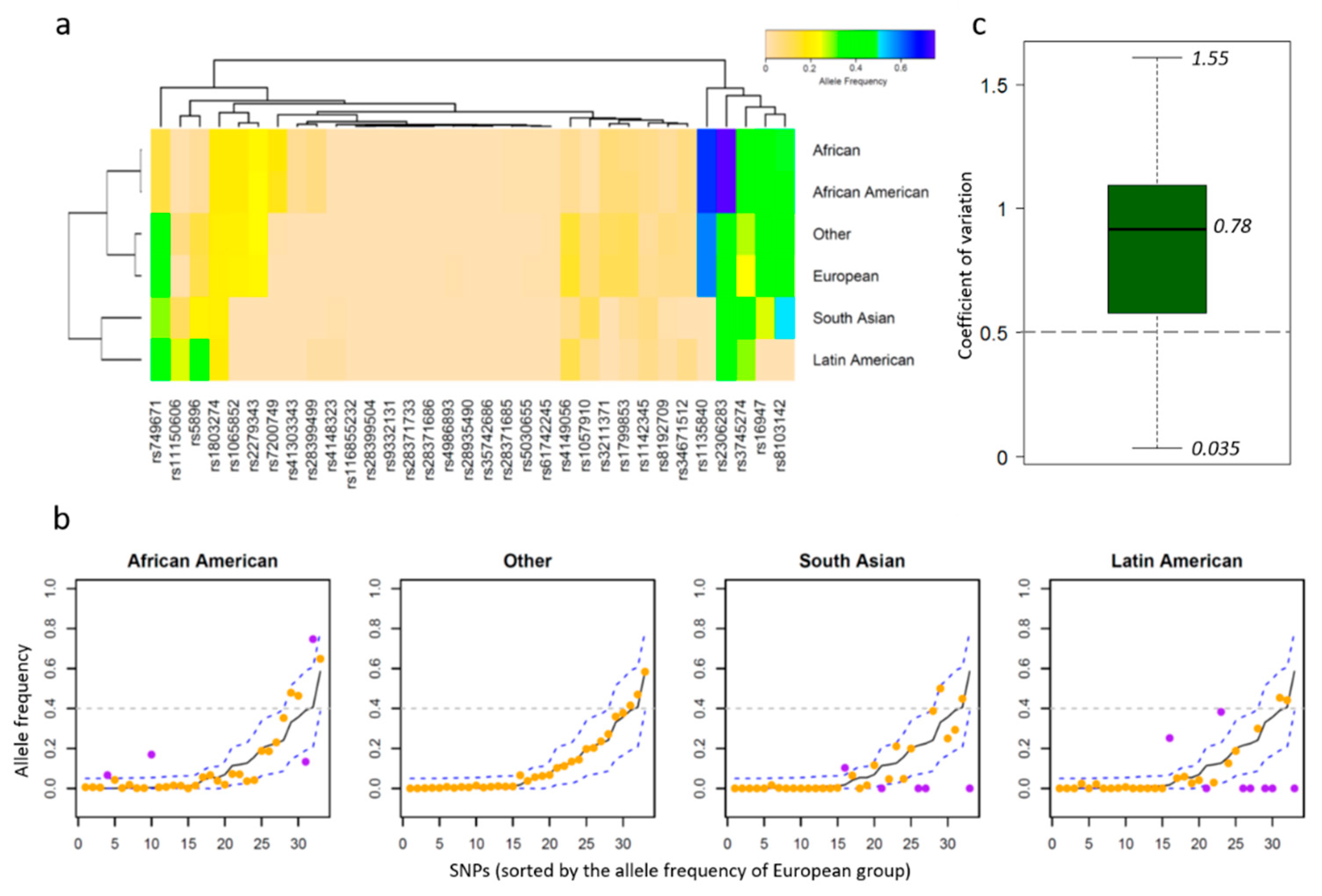
2.2. Allele Frequency Thresholds Based on the European Population
2.3. Literature Screening Process
3. Results
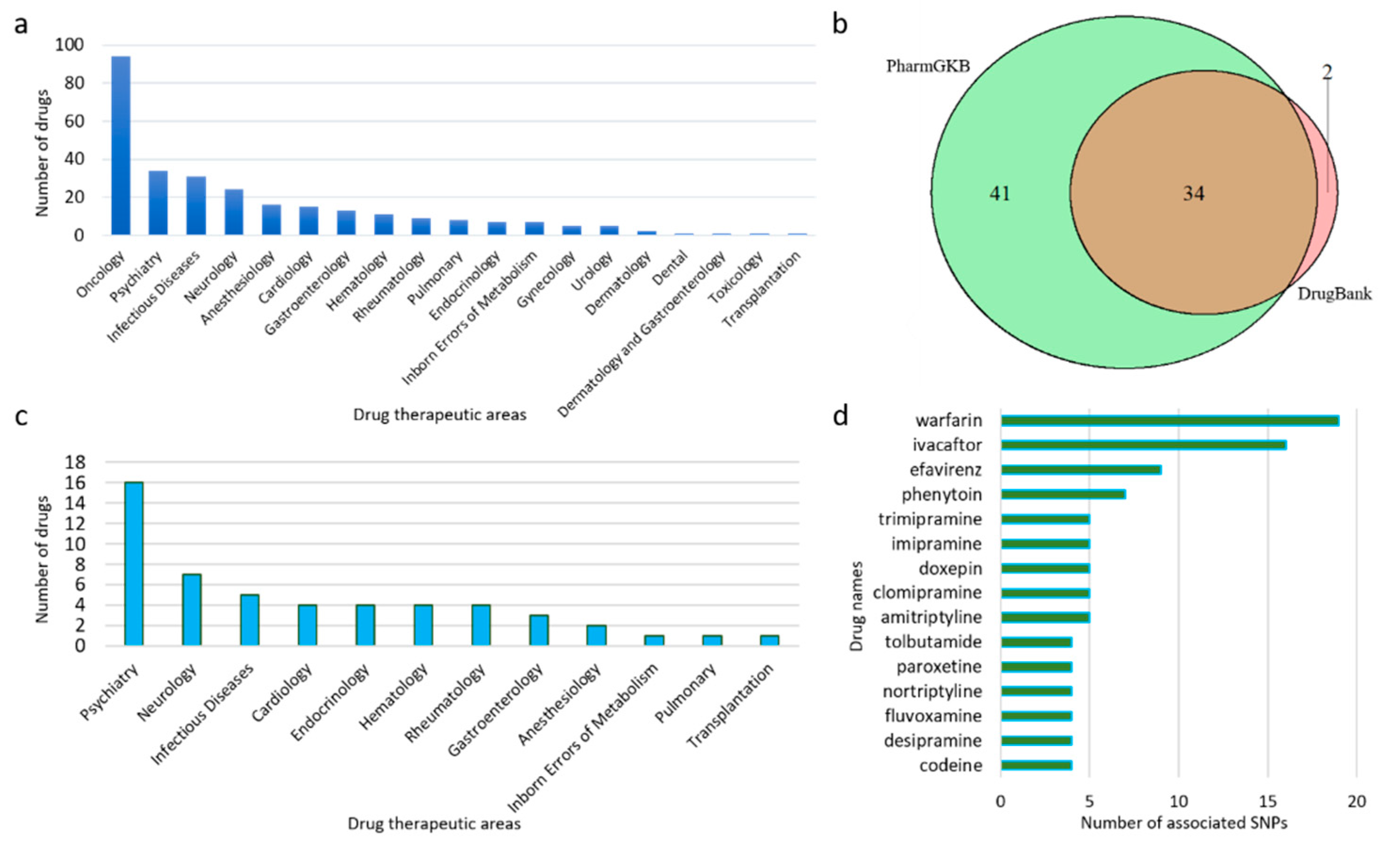
3.1. Data Collection by Drugs
3.2. PGx Information of Biomarkers
3.3. Literature Screening to Provide PGx PubMed IDs
3.4. Database Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Karczewski, J.K.; Daneshjou, R.; Altman, R.B. Pharmacogenomics. PLoS Comput. Biol. 2012, 8, e1002817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carr, D.F.; Alfirevic, A.; Pirmohamed, M. Pharmacogenomics: Current state-of-the-art. Genes 2014, 5, 430–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.W.; Aminkeng, F.; Bhavsar, A.P.; Shaw, K.; Carleton, B.C.; Hayden, M.R.; Ross, C.J.D. The emerging era of pharmacogenomics: Current successes, future potential, and challenges. Clin. Genet. 2014, 86, 21–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daly, A.K. Pharmacogenomics of adverse drug reactions. Genom. Med. 2013, 5, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, L.; Zhang, L.; Peng, Y.; Huang, R.S. Pharmacogenetics and pharmacogenomics: A bridge to individualized cancer therapy. Pharmacogenomics 2013, 14, 315–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hicks, J.K.; Stowe, D.; Willner, M.A.; Wai, M.; Daly, T.; Gordon, S.M.; Moss, T. Implementation of clinical pharmacogenomics within a large health system: From electronic health record decision support to consultation services. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2016, 36, 940–948. [Google Scholar] [CrossRef]
- Bell, G.C.; Crews, K.R.; Wilkinson, M.R.; Haidar, C.E.; Hicks, J.K.; Baker, D.K.; Freimuth, R.R. Development and use of active clinical decision support for preemptive pharmacogenomics. J. Am. Med. Inform. Assn. 2014, 21, e93–e99. [Google Scholar] [CrossRef]
- Van der Wouden, C.H.; Cambon-Thomsen, A.; Cecchin, E.; Cheung, K.C.; Dávila-Fajardo, C.L.; Deneer, V.H.; Kriek, M. Implementing pharmacogenomics in Europe: Design and implementation strategy of the ubiquitous pharmacogenomics consortium. Clin. Pharmacol. Ther. 2017, 101, 341–358. [Google Scholar] [CrossRef]
- Kaye, J.B.; Veenstra, D.L. Warfarin pharmacogenomics in diverse populations. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2017, 37, 1150–1163. [Google Scholar] [CrossRef]
- Verhoef TIRagia, G.; de Boer, A.; Barallon, R.; Kolovou, G.; Kolovou, V.; Redekop, W.K. A randomized trial of genotype-guided dosing of acenocoumarol and phenprocoumon. N. Engl. J. Med. 2013, 369, 2304–2312. [Google Scholar] [CrossRef] [Green Version]
- Frueh, F.W.; Amur, S.; Mummaneni, P.; Epstein, R.S.; Aubert, R.E.; DeLuca, T.M.; Lesko, L.J. Pharmacogenomic biomarker information in drug labels approved by the United States food and drug administration: Prevalence of related drug use. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2008, 28, 992–998. [Google Scholar] [CrossRef] [PubMed]
- Tutton, R. Pharmacogenomic biomarkers in drug labels: What do they tell us? Pharmacogenomics 2014, 15, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Assempour, N. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef] [PubMed]
- Whirl-Carrillo MMcDonagh, E.M.; Hebert, J.M.; Gong, L.; Sangkuhl, K.; Thorn, C.F.; Klein, T.E. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2012, 92, 414–417. [Google Scholar] [CrossRef]
- Griffith, M.; Spies, N.C.; Krysiak, K.; McMichael, J.F.; Coffman, A.C.; Danos, A.M.; Barnell, E.K. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat. Genet. 2017, 49, 170–174. [Google Scholar] [CrossRef] [Green Version]
- Dienstmann, R.; Jang, I.S.; Bot, B.; Friend, S.; Guinney, J. Database of genomic biomarkers for cancer drugs and clinical targetability in solid tumors. Cancer Discov. 2015, 5, 118–123. [Google Scholar] [CrossRef] [Green Version]
- Phan, L.; Jin, Y.; Zhang, H.; Qiang, W.; Shekhtman, E.; Shao, D.; Kimura, M. ALFA: Allele Frequency Aggregator. National Center for Biotechnology Information, US National Library of Medicine. Available online: www.ncbi.nlm.nih.gov/snp/docs/gsr/alfa/ (accessed on 10 March 2020).
- Cavallari, L.H.; Limdi, N.A. Warfarin pharmacogenomics. Curr. Opin. Mol. Ther. 2009, 11, 243. [Google Scholar]
- Johnson, J.A.; Cavallari, L.H. Warfarin pharmacogenetics. Trends Cardiovas. Med. 2015, 25, 33–41. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.-W.; Kim, J.Y.; Park, I.W.; Choi, B.W.; Kang, H.R. Genetic markers of severe cutaneous adverse reactions. Korean J. Intern. Med. 2018, 33, 867. [Google Scholar] [CrossRef] [Green Version]
- Fantini, D. easyPubMed: Search and Retrieve Scientific Publication Records from PubMed; PubMed Help: Bethesda, MD, USA, 2019; Available online: https://cran.r-project.org/web/packages/easyPubMed/vignettes/getting_started_with_easyPubMed.html (accessed on 15 October 2020).
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Sham, P.C. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Gauthier, L.D.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Bradford, L.D. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics 2002, 3, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Bijl, M.J.; Visser, L.E.; Hofman, A.; Vulto, A.G.; Van Gelder, T.; Stricker, B.H.C.; Van Schaik, R.H. Influence of the CYP2D6*4 polymorphism on dose, switching and discontinuation of antidepressants. Br. J. Clin. Pharmacol. 2008, 65, 558–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.; Chao, C.; Chitipiralla, S.; Jang, W. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2015, 44, D862–D868. [Google Scholar] [CrossRef] [Green Version]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Karapetyan, K. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, D.; Xie, A.H.; Liu, Z.; Li, D.; Ning, B.; Thakkar, S.; Tong, W.; Xu, J. Linking Pharmacogenomic Information on Drug Safety and Efficacy with Ethnic Minority Populations. Pharmaceutics 2020, 12, 1021. https://doi.org/10.3390/pharmaceutics12111021
Li D, Xie AH, Liu Z, Li D, Ning B, Thakkar S, Tong W, Xu J. Linking Pharmacogenomic Information on Drug Safety and Efficacy with Ethnic Minority Populations. Pharmaceutics. 2020; 12(11):1021. https://doi.org/10.3390/pharmaceutics12111021
Chicago/Turabian StyleLi, Dan, April Hui Xie, Zhichao Liu, Dongying Li, Baitang Ning, Shraddha Thakkar, Weida Tong, and Joshua Xu. 2020. "Linking Pharmacogenomic Information on Drug Safety and Efficacy with Ethnic Minority Populations" Pharmaceutics 12, no. 11: 1021. https://doi.org/10.3390/pharmaceutics12111021
APA StyleLi, D., Xie, A. H., Liu, Z., Li, D., Ning, B., Thakkar, S., Tong, W., & Xu, J. (2020). Linking Pharmacogenomic Information on Drug Safety and Efficacy with Ethnic Minority Populations. Pharmaceutics, 12(11), 1021. https://doi.org/10.3390/pharmaceutics12111021





