Development of ErbB2-Targeting Liposomes for Enhancing Drug Delivery to ErbB2-Positive Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of ErbB2 Peptide
2.3. HPLC and Matrix-Assisted Laser Desorption Ionization-Time-of-Flight (MALDI-TOF) Mass Spectroscopy Analysis
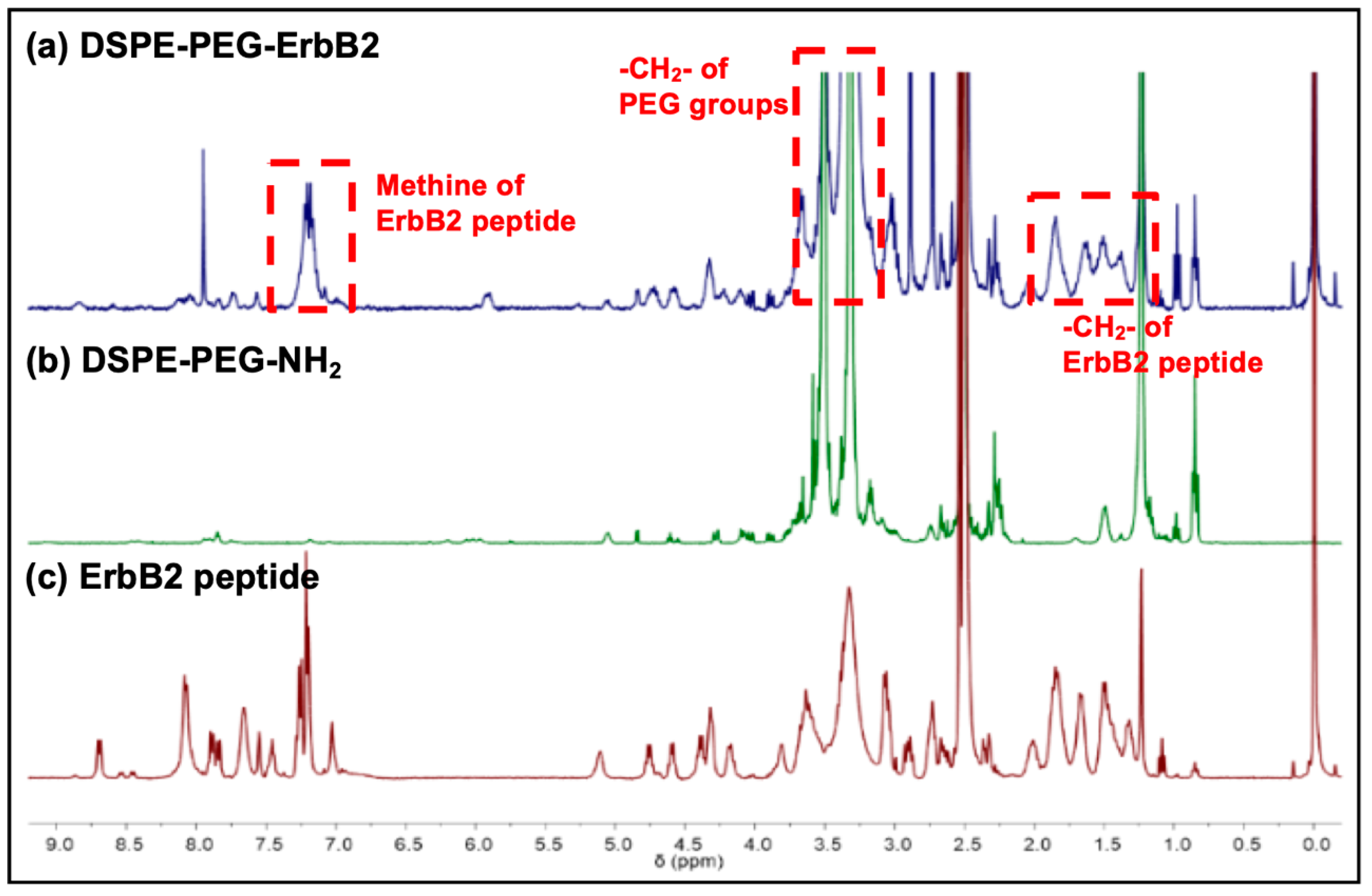
2.4. Peptide-Lipid Conjugation and Nuclear Magnetic Resonance (NMR) Analysis
2.5. Liposome Preparation and Characterization
2.6. Cell Culture
2.7. Western Blotting
2.8. Flow Cytometry
2.9. Confocal Microscopy
2.10. Cellular Toxicity
3. Results and Discussion
3.1. Characterizaation of the ErbB2 Peptide
3.2. Characterizaation of Peptide-Lipid Conjugates
3.3. Characterizaation of ErbB2-Targeting Liposomes
3.4. Cancer Cell-Specific Binding of ErbB2-Targeting Liposomes
3.5. Anti-Cancer Activity of ErbB2-Targeting Liposomes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ADC | antibody-drug conjugate |
| BareLipo | bare liposomes |
| CHCA | α-cyano-4-hydroxycinnamic acid |
| CHEMS | cholesteryl hemisuccinate |
| DCM | dichloromethane |
| DLS | dynamic light scattering |
| DMF | N,N-dimethylformamide |
| DSPE-PEG | 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000 |
| EDC | 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide |
| ErbB2Lipo | ErbB2-targeting liposomes |
| HBTU | 3-[bis(dimethylamino)methyliumyl]-3H-benzotriazol-1-oxide hexafluorophosphate |
| HOBt·H2O | 1- hydroxybenzotriazole hydrate |
| HPLC | high-performance liquid chromatography |
| MALDI-TOF | matrix-assisted laser desorption ionization-time-of-flight |
| mTOR | mammalian target of rapamycin |
| NHS | N-hydroxysuccinimide |
| PDC | peptide-drug conjugates |
| PDI | polydispersity index |
| PEG | polyethylene glycol |
| PFA | paraformaldehyde |
| PPD | piperidine |
| ScrErbB2 | scrambled peptide |
| ScrErbB2Lipo | non-targeting liposomes conjugated with ScrErbB2 |
| SPPS | solid-phase peptide synthesis |
| TEM | transmission electron microscopy |
| TIS | triisopropylsilane |
References
- DeSantis, C.E.; Ma, J.M.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Sauer, A.G.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. Ca-Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Harari, D.; Yarden, Y. Molecular mechanisms underlying ErbB2/HER2 action in breast cancer. Oncogene 2000, 19, 6102–6114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tai, W.; Mahato, R.; Cheng, K. The role of HER2 in cancer therapy and targeted drug delivery. J. Control. Release 2010, 146, 264–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tovey, S.M.; Brown, S.; Doughty, J.C.; Mallon, E.A.; Cooke, T.G.; Edwards, J. Poor survival outcomes in HER2-positive breast cancer patients with low-grade, node-negative tumours. Br. J. Cancer 2009, 100, 680–683. [Google Scholar] [CrossRef] [Green Version]
- Masoud, V.; Pages, G. Targeted therapies in breast cancer: New challenges to fight against resistance. World J. Clin. Oncol. 2017, 8, 120–134. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Zhou, L.L.; Wang, C.H.; Han, Y.; Lu, Y.L.; Liu, J.; Hu, X.C.; Yao, T.M.; Lin, Y.; Liang, S.J.; et al. Tumor-Targeted Drug and CpG Delivery System for Phototherapy and Docetaxel-Enhanced Immunotherapy with Polarization toward M1-Type Macrophages on Triple Negative Breast Cancers. Adv. Mater. 2019, 31. [Google Scholar] [CrossRef]
- Dadwal, A.; Baldi, A.; Narang, R.K. Nanoparticles as carriers for drug delivery in cancer. Artif. Cell Nanomed. Biotechnol. 2018, 46, 295–305. [Google Scholar] [CrossRef]
- Mi, P.; Cabral, H.; Kataoka, K. Ligand-Installed Nanocarriers toward Precision Therapy. Adv. Mater. 2020, 32, e1902604. [Google Scholar] [CrossRef]
- Sun, Q.Q.; Bi, H.T.; Wang, Z.; Li, C.X.; Wang, X.W.; Xu, J.T.; Zhu, H.; Zhao, R.X.; He, F.; Gai, S.L.; et al. Hyaluronic acid-targeted and pH-responsive drug delivery system based on metal-organic frameworks for efficient antitumor therapy. Biomaterials 2019, 223, 119473. [Google Scholar] [CrossRef]
- Yang, Y.; Zhao, W.H.; Tan, W.W.; Lai, Z.Q.; Fang, D.; Jiang, L.; Zuo, C.T.; Yang, N.; Lai, Y.R. An Efficient Cell-Targeting Drug Delivery System Based on Aptamer-Modified Mesoporous Silica Nanoparticles. Nanoscale Res. Lett. 2019, 14, 390. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Amari, T.; Semba, K.; Yamamoto, T.; Takeoka, S. Construction and evaluation of pH-sensitive immunoliposomes for enhanced delivery of anticancer drug to ErbB2 over-expressing breast cancer cells. Nanomedicine 2017, 13, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Suleman, K.; Mushtaq, A.; Haque, E.; Badran, A.; Ajarim, D.; Tweigeri, T.; Elashwah, A.; Shahin, A.; Khan, K.; Alsayed, A. Retrospective review of Her2 positive metastatic breast cancer patients who received Pertuzumab and Herceptin as a first line therapy at KFSH&RC (single institute experience) from 2013 to 2016. Breast 2019, 44, S64. [Google Scholar] [CrossRef]
- Chen, F.; Ma, K.; Madajewski, B.; Zhuang, L.; Zhang, L.; Rickert, K.; Marelli, M.; Yoo, B.; Turker, M.Z.; Overholtzer, M.; et al. Ultrasmall targeted nanoparticles with engineered antibody fragments for imaging detection of HER2-overexpressing breast cancer. Nat. Commun. 2018, 9, 4141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Li, M.; Liu, T.; Liu, J.; Xie, Y.; Zhang, J.; Xu, S.; Liu, H. A dual-functional HER2 aptamer-conjugated, pH-activated mesoporous silica nanocarrier-based drug delivery system provides in vitro synergistic cytotoxicity in HER2-positive breast cancer cells. Int. J. Nanomed. 2019, 14, 4029–4044. [Google Scholar] [CrossRef] [Green Version]
- Park, J.W.; Hong, K.; Kirpotin, D.B.; Colbern, G.; Shalaby, R.; Baselga, J.; Shao, Y.; Nielsen, U.B.; Marks, J.D.; Moore, D.; et al. Anti-HER2 immunoliposomes: Enhanced efficacy attributable to targeted delivery. Clin. Cancer Res. 2002, 8, 1172–1181. [Google Scholar] [PubMed]
- Gao, Z.; Li, G.; Li, X.; Zhou, J.; Duan, X.; Chen, J.; Joshi, B.P.; Kuick, R.; Khoury, B.; Thomas, D.G.; et al. In vivo near-infrared imaging of ErbB2 expressing breast tumors with dual-axes confocal endomicroscopy using a targeted peptide. Sci. Rep. 2017, 7, 14404. [Google Scholar] [CrossRef] [Green Version]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Yellen, P.; Saqcena, M.; Salloum, D.; Feng, J.; Preda, A.; Xu, L.; Rodrik-Outmezguine, V.; Foster, D.A. High-dose rapamycin induces apoptosis in human cancer cells by dissociating mTOR complex 1 and suppressing phosphorylation of 4E-BP1. Cell Cycle 2011, 10, 3948–3956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.H.; Chan, J.L.; Li, W. Rapamycin together with herceptin significantly increased anti-tumor efficacy compared to either alone in ErbB2 over expressing breast cancer cells. Int. J. Cancer 2007, 121, 157–164. [Google Scholar] [CrossRef]
- Decker, T.; Marschner, N.; Muendlein, A.; Welt, A.; Hagen, V.; Rauh, J.; Schroder, H.; Jaehnig, P.; Potthoff, K.; Lerchenmuller, C. VicTORia: A randomised phase II study to compare vinorelbine in combination with the mTOR inhibitor everolimus versus vinorelbine monotherapy for second-line chemotherapy in advanced HER2-negative breast cancer. Breast Cancer Res. Treat. 2019, 176, 637–647. [Google Scholar] [CrossRef]
- Pascual, J.; Turner, N.C. Targeting the PI3-kinase pathway in triple-negative breast cancer. Ann. Oncol. 2019, 30, 1051–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, P.; Zaiss, M.; Harper-Wynne, C.; Ferreira, M.; Dubey, S.; Chan, S.; Makris, A.; Nemsadze, G.; Brunt, A.M.; Kuemmel, S.; et al. Fulvestrant Plus Vistusertib vs Fulvestrant Plus Everolimus vs Fulvestrant Alone for Women With Hormone Receptor-Positive Metastatic Breast Cancer: The MANTA Phase 2 Randomized Clinical Trial. JAMA Oncol. 2019, 5, 1556–1563. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.W.; Jeong, H.Y.; Kang, S.J.; Jeong, I.H.; Choi, M.J.; You, Y.M.; Im, C.S.; Song, I.H.; Lee, T.S.; Lee, J.S.; et al. Anti-EGF Receptor Aptamer-Guided Co-Delivery of Anti-Cancer siRNAs and Quantum Dots for Theranostics of Triple-Negative Breast Cancer. Theranostics 2019, 9, 837–852. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, S.; Joo, H.; Tsai, J.; Jasti, B.; Li, X. A Rational Approach for Creating Peptides Mimicking Antibody Binding. Sci. Rep. 2019, 9, 997. [Google Scholar] [CrossRef]
- Amblard, M.; Fehrentz, J.A.; Martinez, J.; Subra, G. Methods and protocols of modern solid phase Peptide synthesis. Mol. Biotechnol. 2006, 33, 239–254. [Google Scholar] [CrossRef]
- Zhang, Y.; Mintzer, E.; Uhrich, K.E. Synthesis and characterization of PEGylated bolaamphiphiles with enhanced retention in liposomes. J. Colloid Interface Sci. 2016, 482, 19–26. [Google Scholar] [CrossRef]
- Fang, Y.; Xue, J.; Gao, S.; Lu, A.; Yang, D.; Jiang, H.; He, Y.; Shi, K. Cleavable PEGylation: A strategy for overcoming the “PEG dilemma” in efficient drug delivery. Drug Deliv. 2017, 24, 22–32. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, Z.; Shah, A.; Siddiq, M.; Kraatz, H.B. Polymeric micelles as drug delivery vehicles. Rsc Adv. 2014, 4, 17028–17038. [Google Scholar] [CrossRef]
- Eloy, J.O.; Petrilli, R.; Topan, J.F.; Antonio, H.M.R.; Barcellos, J.P.A.; Chesca, D.L.; Serafini, L.N.; Tiezzi, D.G.; Lee, R.J.; Marchetti, J.M. Co-loaded paclitaxel/rapamycin liposomes: Development, characterization and in vitro and in vivo evaluation for breast cancer therapy. Colloid Surface B 2016, 141, 74–82. [Google Scholar] [CrossRef] [Green Version]
- Lucas, A.T.; Price, L.S.L.; Schorzman, A.N.; Storrie, M.; Piscitelli, J.A.; Razo, J.; Zamboni, W.C. Factors Affecting the Pharmacology of Antibody-Drug Conjugates. Antibodies (Basel) 2018, 7, 10. [Google Scholar] [CrossRef] [Green Version]
- Vrettos, E.I.; Mezo, G.; Tzakos, A.G. On the design principles of peptide-drug conjugates for targeted drug delivery to the malignant tumor site. Beilstein J. Org. Chem. 2018, 14, 930–954. [Google Scholar] [CrossRef] [PubMed]
- Ferri, N.; Bellosta, S.; Baldessin, L.; Boccia, D.; Racagni, G.; Corsini, A. Pharmacokinetics interactions of monoclonal antibodies. Pharmacol. Res. 2016, 111, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Javier, D.J.; Nitin, N.; Levy, M.; Ellington, A.; Richards-Kortum, R. Aptamer-targeted gold nanoparticles as molecular-specific contrast agents for reflectance imaging. Bioconjug. Chem. 2008, 19, 1309–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danhier, F. To exploit the tumor microenvironment: Since the EPR effect fails in the clinic, what is the future of nanomedicine? J. Control. Release 2016, 244, 108–121. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Wang, X.; Proud, C.G. mTOR inhibitors in cancer therapy. F1000Research 2016, 5, 2078. [Google Scholar] [CrossRef] [Green Version]
- Kenny, P.A.; Lee, G.Y.; Myers, C.A.; Neve, R.M.; Semeiks, J.R.; Spellman, P.T.; Lorenz, K.; Lee, E.H.; Barcellos-Hoff, M.H.; Petersen, O.W.; et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 2007, 1, 84–96. [Google Scholar] [CrossRef]
- Kute, T.; Lack, C.M.; Willingham, M.; Bishwokama, B.; Williams, H.; Barrett, K.; Mitchell, T.; Vaughn, J.P. Development of Herceptin resistance in breast cancer cells. Cytom. A 2004, 57, 86–93. [Google Scholar] [CrossRef]
- Chen, Q.; Yu, S.; Zhang, D.; Zhang, W.; Zhang, H.; Zou, J.; Mao, Z.; Yuan, Y.; Gao, C.; Liu, R. Impact of Antifouling PEG Layer on the Performance of Functional Peptides in Regulating Cell Behaviors. J. Am. Chem. Soc. 2019, 141, 16772–16780. [Google Scholar] [CrossRef]
- Foldvari, M. Observations of membrane fusion in a liposome dispersion: The missing fusion intermediate? F1000Research 2015, 4, 4. [Google Scholar] [CrossRef]
- Telli, M.L.; Gradishar, W.J.; Ward, J.H. NCCN Guidelines Updates: Breast Cancer. J. Natl. Compr. Cancer Netw. 2019, 17, 552–555. [Google Scholar] [CrossRef]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ueno, S.; Kim, M.W.; Lee, G.; Park, Y.I.; Niidome, T.; Lee, R. Development of ErbB2-Targeting Liposomes for Enhancing Drug Delivery to ErbB2-Positive Breast Cancer. Pharmaceutics 2020, 12, 585. https://doi.org/10.3390/pharmaceutics12060585
Ueno S, Kim MW, Lee G, Park YI, Niidome T, Lee R. Development of ErbB2-Targeting Liposomes for Enhancing Drug Delivery to ErbB2-Positive Breast Cancer. Pharmaceutics. 2020; 12(6):585. https://doi.org/10.3390/pharmaceutics12060585
Chicago/Turabian StyleUeno, Sho, Min Woo Kim, Gibok Lee, Yong Il Park, Takuro Niidome, and Ruda Lee. 2020. "Development of ErbB2-Targeting Liposomes for Enhancing Drug Delivery to ErbB2-Positive Breast Cancer" Pharmaceutics 12, no. 6: 585. https://doi.org/10.3390/pharmaceutics12060585
APA StyleUeno, S., Kim, M. W., Lee, G., Park, Y. I., Niidome, T., & Lee, R. (2020). Development of ErbB2-Targeting Liposomes for Enhancing Drug Delivery to ErbB2-Positive Breast Cancer. Pharmaceutics, 12(6), 585. https://doi.org/10.3390/pharmaceutics12060585






