A Mixed-Surface Polyamidoamine Dendrimer for In Vitro and In Vivo Delivery of Large Plasmids
Abstract
:1. Introduction
2. Materials and Methods
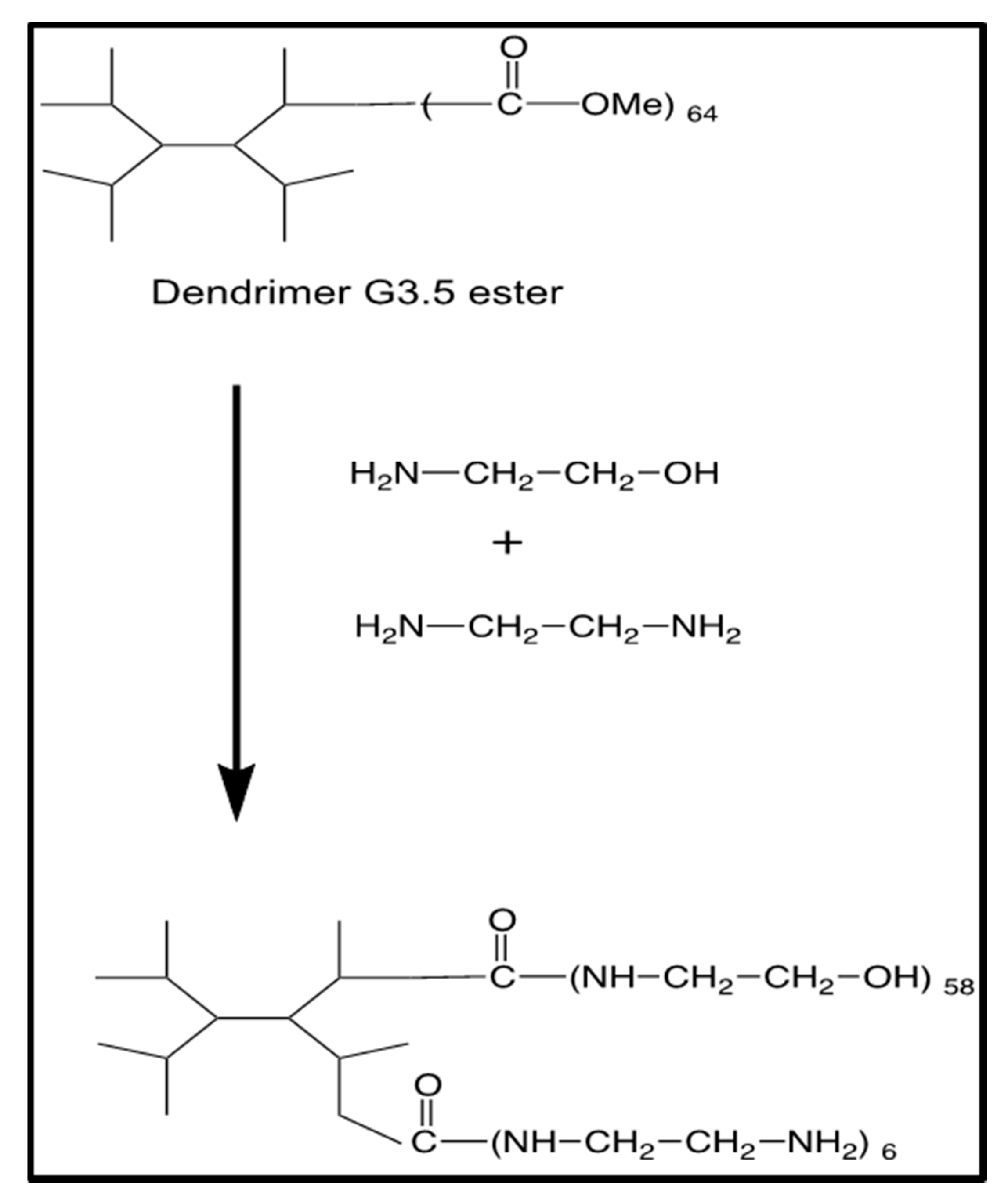
2.1. Synthesis of G4-90/10 and Fluorescent Conjugates
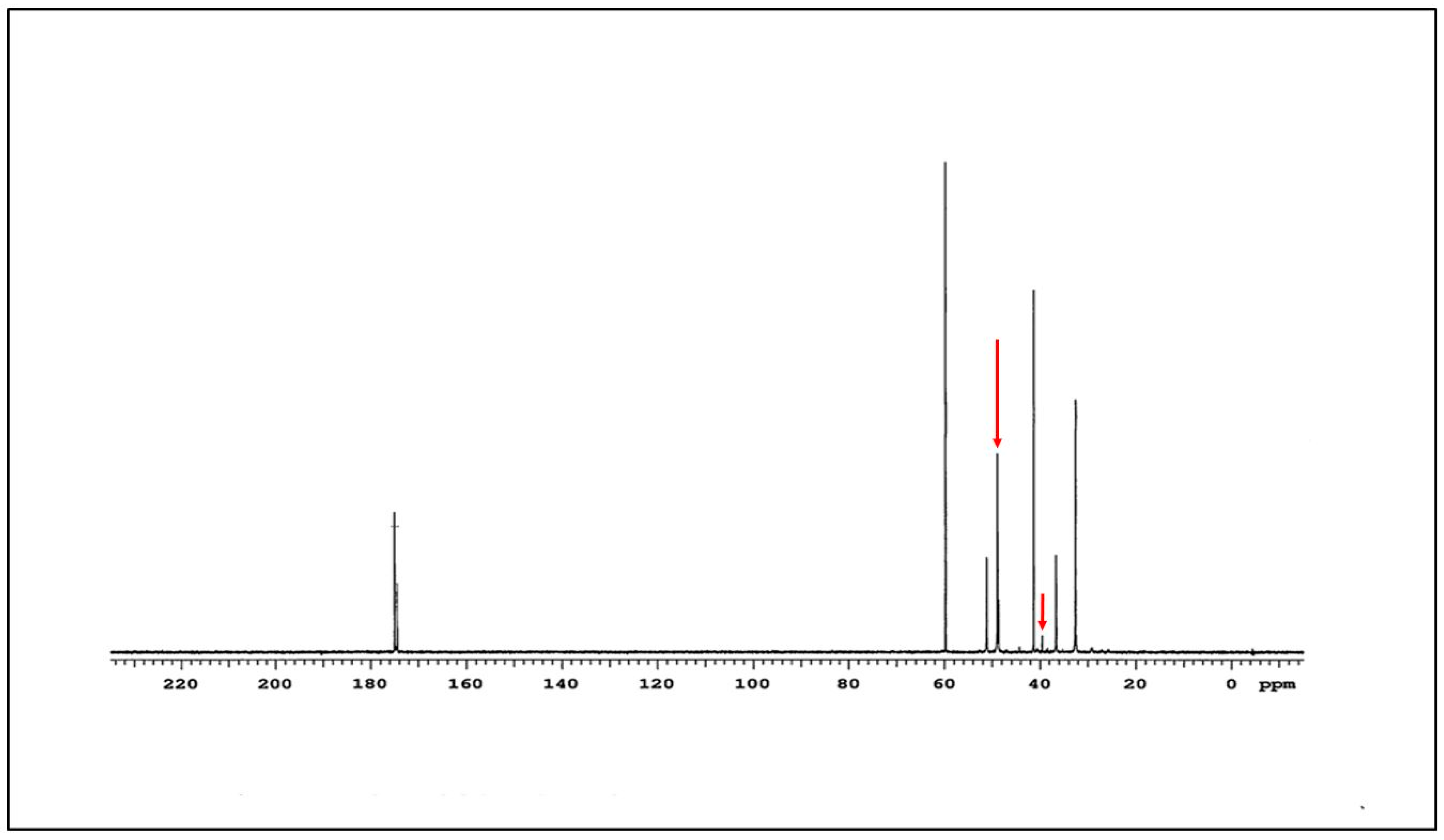
2.2. Characterization of G4-90/10 and Conjugates
2.3. Dendrimer Degradation
2.4. In Vitro Introduction of G4-90/10 into HEK293 Cells
2.5. Toxicity of Dendrimer in HEK293 Cells
2.6. In Vitro Introduction of Reporter Plasmid Dendriplex
2.7. In Vivo Introduction of hSOX2 Plasmid Dendriplex
3. Results
3.1. Synthesis and Characterization of G4-90/10
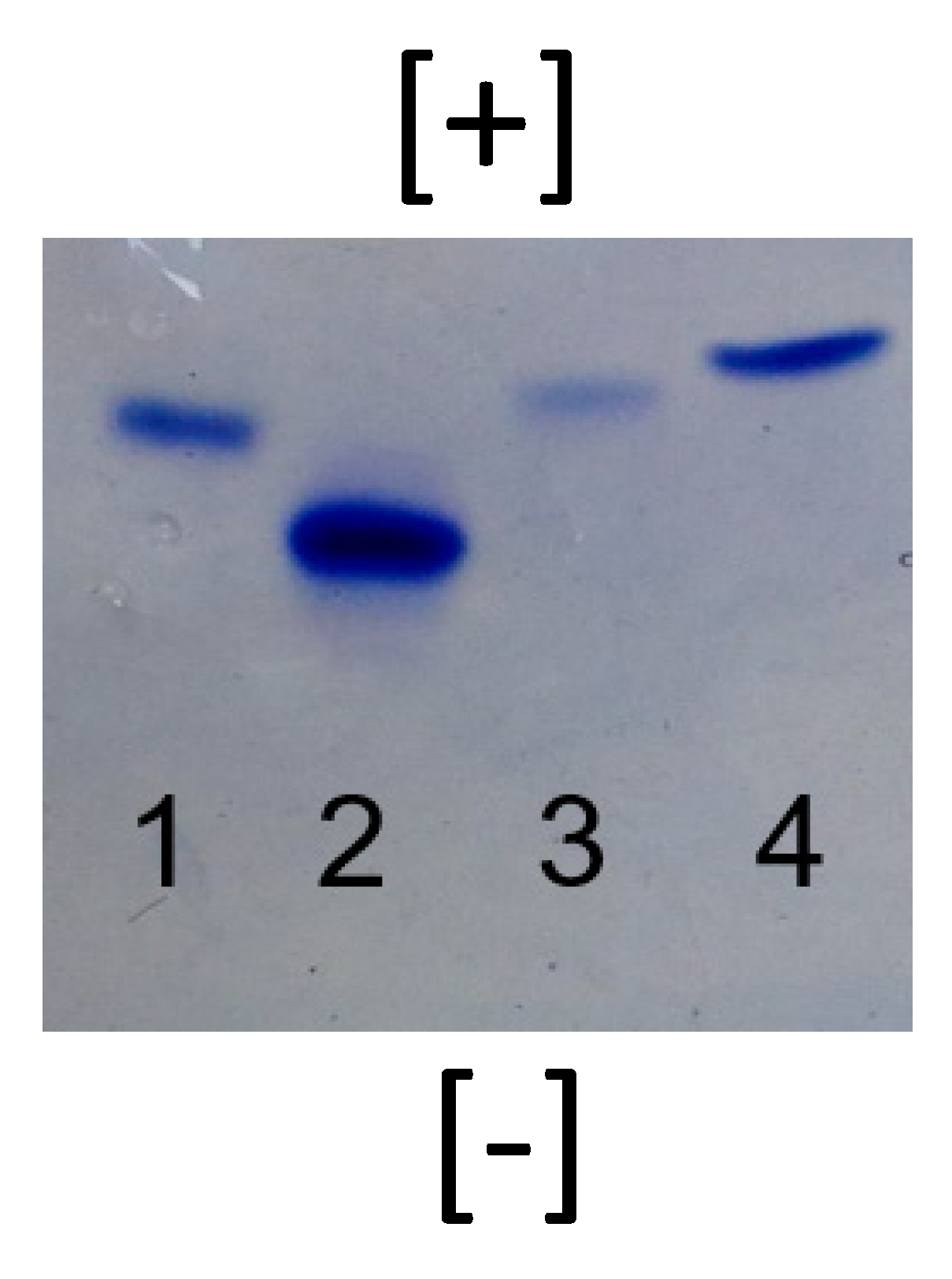
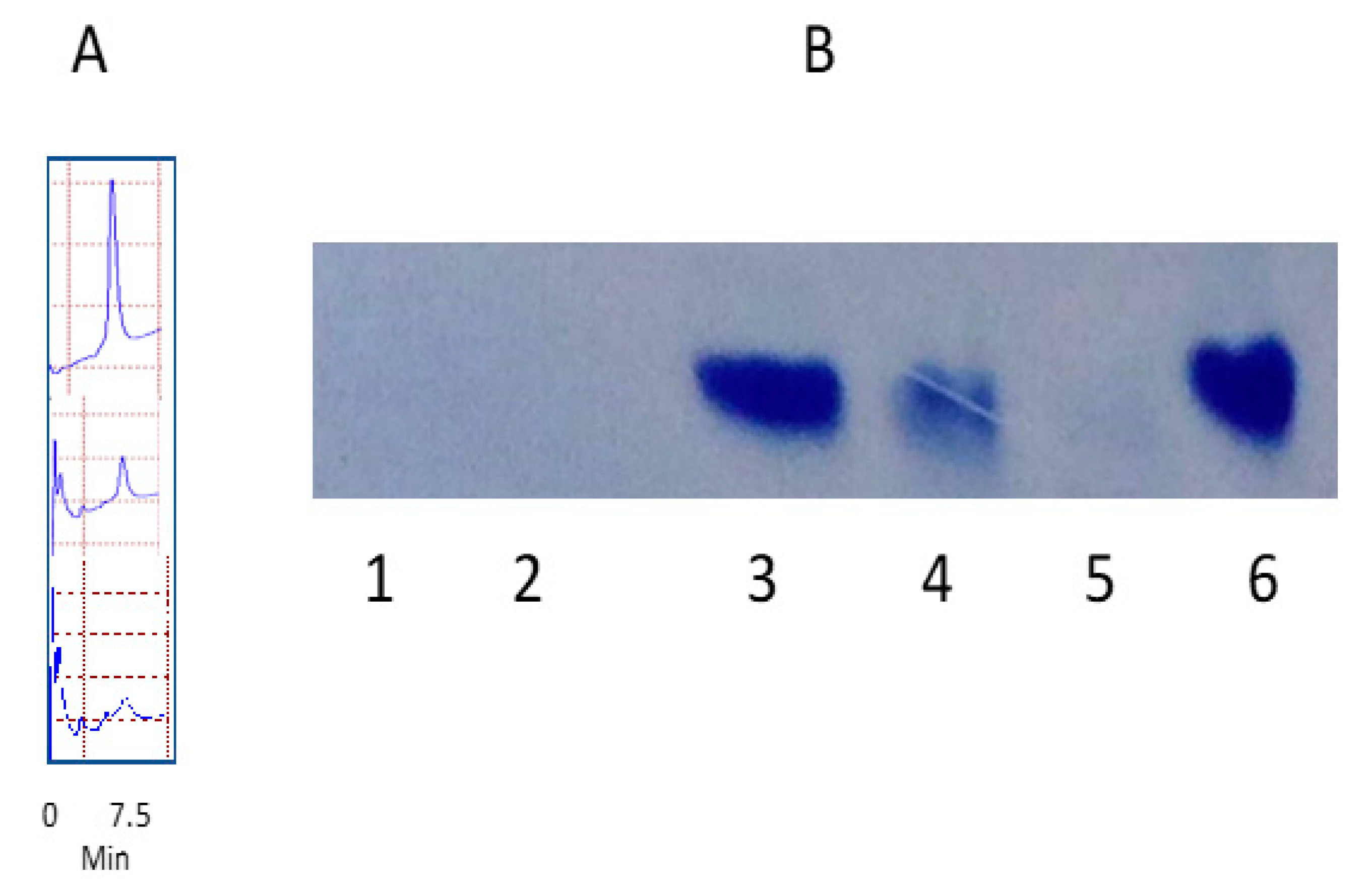
3.2. Degradation of Nanomolecules
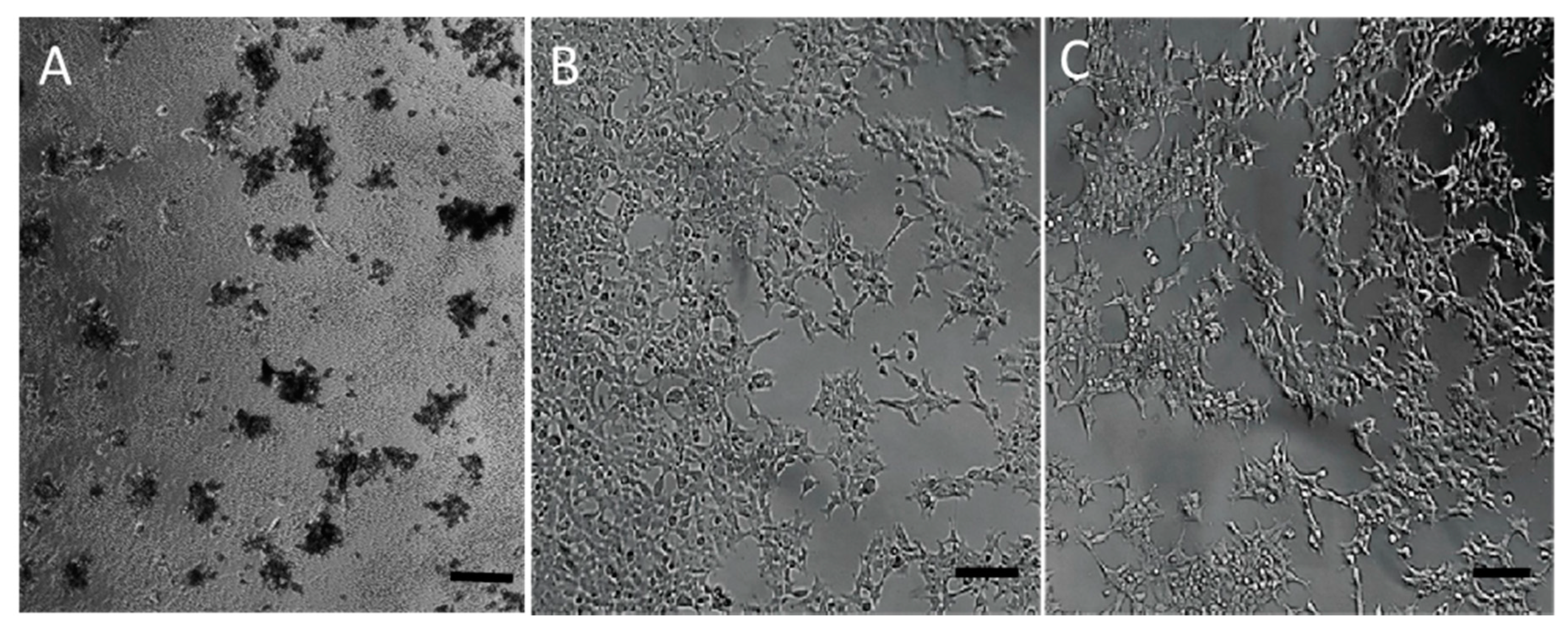
3.3. Toxicity of the Nanomolecule
3.4. Dendriplex Formation
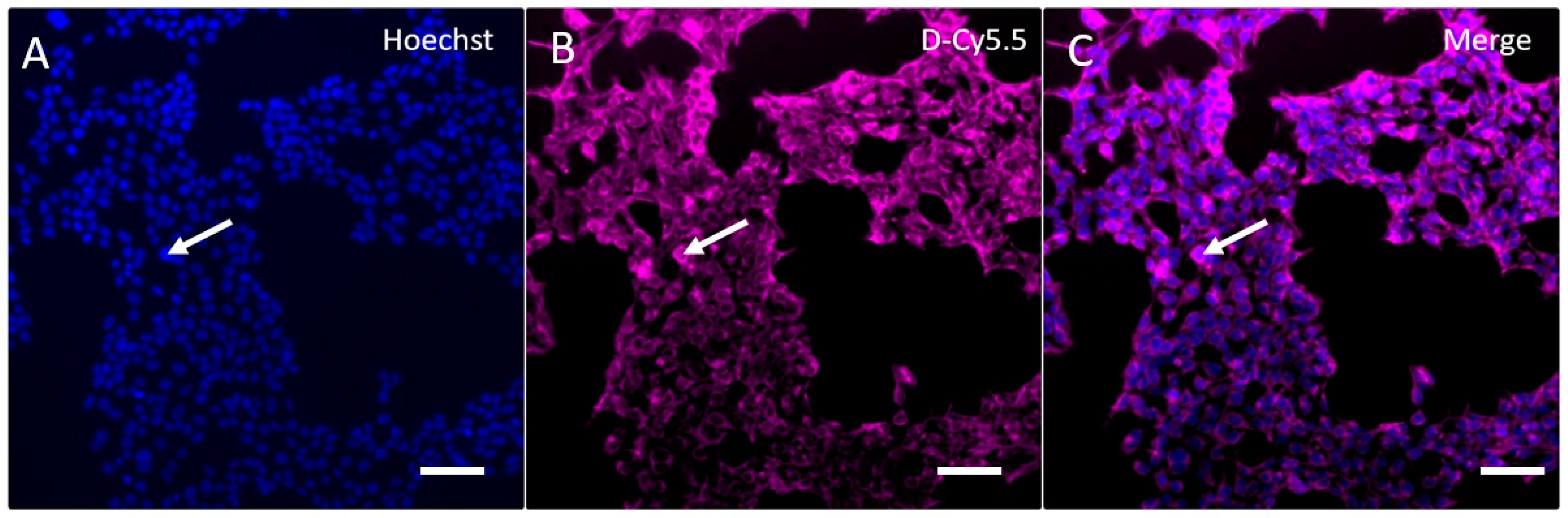
3.5. In Vitro Uptake of G4-90/10
3.6. In Vitro Introduction of RP1 and RP2 and Toxicity
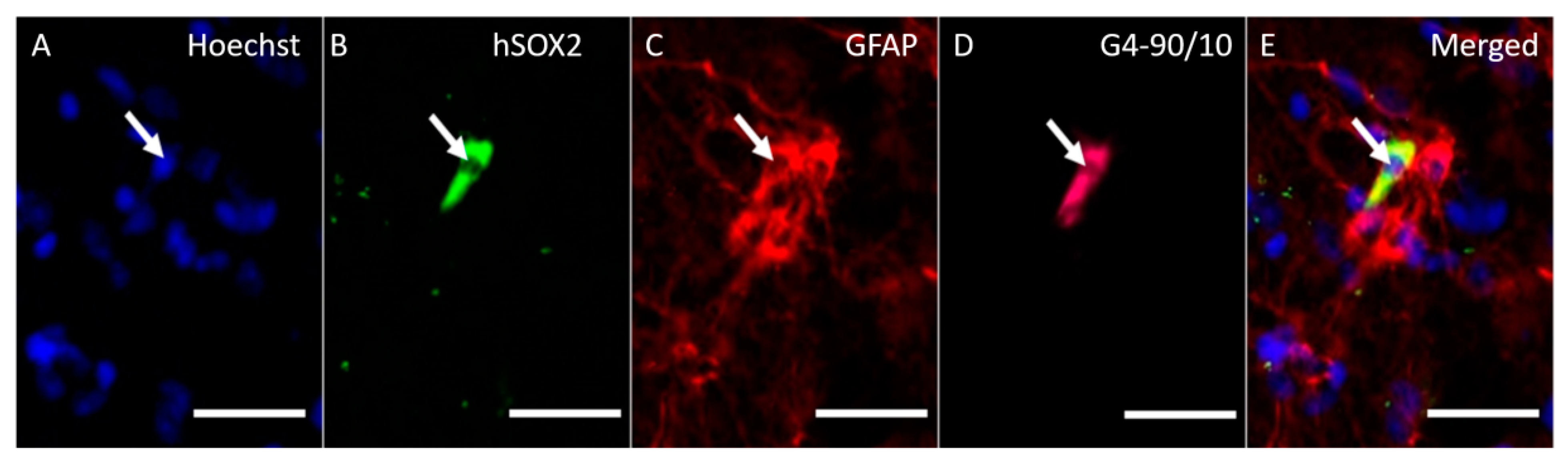
3.7. In Vivo Transfection
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Keeler, A.M.; ElMallah, M.K.; Flotte, T.R. Gene Therapy 2017: Progress and Future Directions. Clin. Transl. Sci. 2017, 10, 242–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kay, M.A. State-of-the-art gene-based therapies: The road ahead. Nat. Rev. Genet. 2011, 12, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Naldini, L. Gene therapy returns to centre stage. Nature 2015, 526, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.Y.; Jang, M.J.; Yoo, B.B.; Greenbaum, A.; Ravi, N.; Wu, W.-L.; Sánchez-Guardado, L.; Lois, C.; Mazmanian, S.K.; Deverman, B.E.; et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 2017, 20, 1172–1179. [Google Scholar] [CrossRef]
- Mali, S. Delivery systems for gene therapy. Indian J. Hum. Genet. 2013, 19, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Nayerossadat, N.; Maedeh, T.; Ali, P.A. Viral and nonviral delivery systems for gene delivery. Adv. Biomed. Res. 2012, 1. [Google Scholar] [CrossRef]
- Ramamoorth, M.; Narvekar, A. Non viral vectors in gene therapy—An overview. J. Clin. Diagn. Res. JCDR 2015, 9, GE01–GE06. [Google Scholar] [CrossRef]
- Glover, D.J.; Lipps, H.J.; Jans, D.A. Towards safe, non-viral therapeutic gene expression in humans. Nat. Rev. Genet. 2005, 6, 299–310. [Google Scholar] [CrossRef]
- Wang, W.; Li, W.; Ma, N.; Steinhoff, G. Non-viral gene delivery methods. Curr. Pharm. Biotechnol. 2013, 14, 46–60. [Google Scholar]
- Mingozzi, F.; High, K.A. Therapeutic in vivo gene transfer for genetic disease using AAV: Progress and challenges. Nat. Rev. Genet. 2011, 12, 341–355. [Google Scholar] [CrossRef]
- Al-Dosari, M.S.; Gao, X. Nonviral gene delivery: Principle, limitations, and recent progress. AAPS J. 2009, 11, 671–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kretzmann, J.A.; Ho, D.; Evans, C.W.; Plani-Lam, J.H.C.; Garcia-Bloj, B.; Mohamed, A.E.; O’Mara, M.L.; Ford, E.; Tan, D.E.K.; Lister, R.; et al. Synthetically controlling dendrimer flexibility improves delivery of large plasmid DNA. Chem. Sci. 2017, 8, 2923–2930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pack, D.W.; Hoffman, A.S.; Pun, S.; Stayton, P.S. Design and development of polymers for gene delivery. Nat. Rev. Drug Discov. 2005, 4, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef]
- Sułkowski, W.W.; Pentak, D.; Nowak, K.; Sułkowska, A. The influence of temperature, cholesterol content and pH on liposome stability. J. Mol. Struct. 2005, 744, 737–747. [Google Scholar] [CrossRef]
- Sharma, A.; Mohanty, D.K.; Desai, A.; Ali, R. A simple polyacrylamide gel electrophoresis procedure for separation of polyamidoamine dendrimers. Electrophoresis 2003, 24, 2733–2739. [Google Scholar] [CrossRef]
- Kannan, R.M.; Nance, E.; Kannan, S.; Tomalia, D.A. Emerging concepts in dendrimer-based nanomedicine: From design principles to clinical applications. J. Intern. Med. 2014, 276, 579–617. [Google Scholar] [CrossRef]
- Dufès, C.; Uchegbu, I.F.; Schätzlein, A.G. Dendrimers in gene delivery. Adv. Drug Deliv. Rev. 2005, 57, 2177–2202. [Google Scholar] [CrossRef] [Green Version]
- Mastorakos, P.; Kambhampati, S.P.; Mishra, M.K.; Wu, T.; Song, E.; Hanes, J.; Kannan, R.M. Hydroxyl PAMAM dendrimer-based gene vectors for transgene delivery to human retinal pigment epithelial cells. Nanoscale 2015, 7, 3845–3856. [Google Scholar] [CrossRef] [Green Version]
- Wood, K.C.; Little, S.R.; Langer, R.; Hammond, P.T. A family of hierarchically self-assembling linear-dendritic hybrid polymers for highly efficient targeted gene delivery. Angew. Chem. Int. Ed. 2005, 44, 6704–6708. [Google Scholar] [CrossRef]
- Kim, T.; Seo, H.J.; Choi, J.S.; Jang, H.-S.; Baek, J.; Kim, K.; Park, J.-S. PAMAM-PEG-PAMAM: Novel triblock copolymer as a biocompatible and efficient gene delivery carrier. Biomacromolecules 2004, 5, 2487–2492. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.S.; Ko, K.S.; Park, J.S.; Kim, Y.-H.; Kim, S.W.; Lee, M. Dexamethasone conjugated poly(amidoamine) dendrimer as a gene carrier for efficient nuclear translocation. Int. J. Pharm. 2006, 320, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lim, Y.; Choi, J.S.; Lee, Y.; Kim, T.; Kim, H.J.; Yoon, J.K.; Kim, K.; Park, J. Polyplexes assembled with internally quaternized PAMAM-OH dendrimer and plasmid DNA have a neutral surface and gene delivery potency. Bioconjug. Chem. 2003, 14, 1214–1221. [Google Scholar] [CrossRef] [PubMed]
- Kono, K.; Akiyama, H.; Takahashi, T.; Takagishi, T.; Harada, A. Transfection Activity of polyamidoamine dendrimers having hydrophobic amino acid residues in the periphery. Bioconjug. Chem. 2005, 16, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.S.; Nam, K.; Park, J.-Y.; Kim, J.-B.; Lee, J.-K.; Park, J.-S. Enhanced transfection efficiency of PAMAM dendrimer by surface modification with L-arginine. J. Control. Release 2004, 99, 445–456. [Google Scholar] [CrossRef]
- Mullen, D.G.; Borgmeier, E.L.; Fang, M.; McNerny, D.Q.; Desai, A.; Baker, J.R.; Orr, B.G.; Holl, M.M.B. Effect of mass transport in the synthesis of partially acetylated dendrimer: Implications for functional ligand–nanoparticle distributions. Macromolecules 2010, 43, 6577–6587. [Google Scholar] [CrossRef] [Green Version]
- Esfand, R.; Tomalia, D.A. Laboratory synthesis of poly(amidoamine)(PAMAM) dendrimers. Dendrimers Dendritic Polym. 2001, 1, 587–604. [Google Scholar]
- Srinageshwar, B.; Peruzzaro, S.; Andrews, M.; Johnson, K.; Hietpas, A.; Clark, B.; McGuire, C.; Petersen, E.; Kippe, J.; Stewart, A.; et al. PAMAM dendrimers cross the blood-brain barrier when administered through the carotid artery in C57BL/6J Mice. Int. J. Mol. Sci. 2017, 18, 628. [Google Scholar] [CrossRef] [Green Version]
- Karow, M.; Sánchez, R.; Schichor, C.; Masserdotti, G.; Ortega, F.; Heinrich, C.; Gascón, S.; Khan, M.A.; Lie, D.C.; Dellavalle, A.; et al. Reprogramming of pericyte-derived cells of the adult human brain into induced neuronal cells. Cell Stem Cell 2012, 11, 471–476. [Google Scholar] [CrossRef] [Green Version]
- Vierbuchen, T.; Ostermeier, A.; Pang, Z.P.; Kokubu, Y.; Südhof, T.C.; Wernig, M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010, 463, 1035–1041. [Google Scholar] [CrossRef] [Green Version]
- Heinrich, C.; Gascón, S.; Masserdotti, G.; Lepier, A.; Sanchez, R.; Simon-Ebert, T.; Schroeder, T.; Götz, M.; Berninger, B. Generation of subtype-specific neurons from postnatal astroglia of the mouse cerebral cortex. Nat. Protoc. 2011, 6, 214–228. [Google Scholar] [CrossRef]
- Liu, M.-L.; Zang, T.; Zou, Y.; Chang, J.C.; Gibson, J.R.; Huber, K.M.; Zhang, C.-L. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 2013, 4, 2183. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Brown, J.; Kanarek, A.; Rajagopal, J.; Melton, D.A. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 2008, 455, 627–632. [Google Scholar] [CrossRef]
- Song, K.; Nam, Y.-J.; Luo, X.; Qi, X.; Tan, W.; Huang, G.N.; Acharya, A.; Smith, C.L.; Tallquist, M.D.; Neilson, E.G.; et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 2012, 485, 599–604. [Google Scholar] [CrossRef] [Green Version]
- Qian, L.; Huang, Y.; Spencer, C.I.; Foley, A.; Vedantham, V.; Liu, L.; Conway, S.J.; Fu, J.; Srivastava, D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012, 485, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Niu, W.; Liu, M.-L.; Zou, Y.; Zhang, C.-L. In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nat. Commun. 2014, 5, 3338. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Desai, A.; Ali, R.; Tomalia, D. Polyacrylamide gel electrophoresis separation and detection of polyamidoamine dendrimers possessing various cores and terminal groups. J. Chromatogr. A 2005, 1081, 238–244. [Google Scholar] [CrossRef]
- Upadhaya, S.K.; Swanson, D.R.; Tomalia, D.A.; Sharma, A. Analysis of polyamidoamine dendrimers by isoelectric focusing. Anal. Bioanal. Chem. 2014, 406, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Switala, J.; Loewen, P.C. Diversity of properties among catalases. Arch. Biochem. Biophys. 2002, 401, 145–154. [Google Scholar] [CrossRef]
- Elsaesser, A.; Howard, C.V. Toxicology of nanoparticles. Adv. Drug Deliv. Rev. 2012, 64, 129–137. [Google Scholar] [CrossRef]
- Sadekar, S.; Ray, A.; Janàt-Amsbury, M.; Peterson, C.M.; Ghandehari, H. Comparative biodistribution of PAMAM dendrimers and HPMA copolymers in ovarian-tumor-bearing mice. Biomacromolecules 2011, 12, 88–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naha, P.C.; Mukherjee, S.P.; Byrne, H.J. Toxicology of engineered nanoparticles: Focus on poly(amidoamine) dendrimers. Int. J. Environ. Res. Public Health 2018, 15, 338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pryor, J.B.; Harper, B.J.; Harper, S.L. Comparative toxicological assessment of PAMAM and thiophosphoryl dendrimers using embryonic zebrafish. Int. J. Nanomed. 2014, 9, 1947–1956. [Google Scholar]
- Chauhan, A.S.; Jain, N.K.; Diwan, P.V. Pre-clinical and behavioural toxicity profile of PAMAM dendrimers in mice. Proc. R. Soc. A Math. Phys. Eng. Sci. 2010, 466, 1535–1550. [Google Scholar] [CrossRef]
- Naha, P.C.; Davoren, M.; Lyng, F.M.; Byrne, H.J. Reactive oxygen species (ROS) induced cytokine production and cytotoxicity of PAMAM dendrimers in J774A.1 cells. Toxicol. Appl. Pharmacol. 2010, 246, 91–99. [Google Scholar] [CrossRef]
- Heiden, T.C.K.; Dengler, E.; Kao, W.J.; Heideman, W.; Peterson, R.E. Developmental toxicity of low generation PAMAM dendrimers in zebrafish. Toxicol. Appl. Pharmacol. 2007, 225, 70–79. [Google Scholar] [CrossRef]
- Bodewein, L.; Schmelter, F.; Di Fiore, S.; Hollert, H.; Fischer, R.; Fenske, M. Differences in toxicity of anionic and cationic PAMAM and PPI dendrimers in zebrafish embryos and cancer cell lines. Toxicol. Appl. Pharmacol. 2016, 305, 83–92. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srinageshwar, B.; Florendo, M.; Clark, B.; Johnson, K.; Munro, N.; Peruzzaro, S.; Antcliff, A.; Andrews, M.; Figacz, A.; Swanson, D.; et al. A Mixed-Surface Polyamidoamine Dendrimer for In Vitro and In Vivo Delivery of Large Plasmids. Pharmaceutics 2020, 12, 619. https://doi.org/10.3390/pharmaceutics12070619
Srinageshwar B, Florendo M, Clark B, Johnson K, Munro N, Peruzzaro S, Antcliff A, Andrews M, Figacz A, Swanson D, et al. A Mixed-Surface Polyamidoamine Dendrimer for In Vitro and In Vivo Delivery of Large Plasmids. Pharmaceutics. 2020; 12(7):619. https://doi.org/10.3390/pharmaceutics12070619
Chicago/Turabian StyleSrinageshwar, Bhairavi, Maria Florendo, Brittany Clark, Kayla Johnson, Nikolas Munro, Sarah Peruzzaro, Aaron Antcliff, Melissa Andrews, Alexander Figacz, Douglas Swanson, and et al. 2020. "A Mixed-Surface Polyamidoamine Dendrimer for In Vitro and In Vivo Delivery of Large Plasmids" Pharmaceutics 12, no. 7: 619. https://doi.org/10.3390/pharmaceutics12070619
APA StyleSrinageshwar, B., Florendo, M., Clark, B., Johnson, K., Munro, N., Peruzzaro, S., Antcliff, A., Andrews, M., Figacz, A., Swanson, D., Dunbar, G. L., Sharma, A., & Rossignol, J. (2020). A Mixed-Surface Polyamidoamine Dendrimer for In Vitro and In Vivo Delivery of Large Plasmids. Pharmaceutics, 12(7), 619. https://doi.org/10.3390/pharmaceutics12070619







