Postbiotic-Enabled Targeting of the Host-Microbiota-Pathogen Interface: Hints of Antibiotic Decline?
Abstract
:1. Introduction
2. Nonantibiotic Based Therapies
3. The Microbiota Relevance in Human Infections
3.1. The Host–Gut Microbiota Interface
3.2. Antibiotics and the Gut Microbiota
3.3. Microbiota-Based Therapeutics
3.3.1. Fecal Microbiota Transplantation
3.3.2. Bacteriophages
3.3.3. Probiotics and Prebiotics
4. Postbiotics: The Changing Paradigm
4.1. Major Postbiotic Groups
- Carbohydrate metabolites—anaerobic bacteria produce short-chain fatty acids (SCFAs) through carbohydrate fermentation in the intestine. They are formed starting from polysaccharide, oligosaccharide, protein, peptide, and glycoprotein precursors [193]. In particular, bacteria of the Bacteroidetes phylum are good producers of acetate and propionate SCFAs, whereas those in the Firmicutes phylum are efficient butyrate producers [194].
- Amino acid and related metabolites—proteins are metabolized by many bacterial species, such as Bacillus, Clostridium, Streptococcus, Lactobacillus, and Proteobacteria phyla.
- Lipid and bile acid metabolites—the GM alters bile acids through various modifications [195]. More than 20 different secondary bile acids are generated, including deoxycholic acid and lithocholic acid, as well as phosphatidylcholine is metabolized to produce trimethylamine-N-oxide.
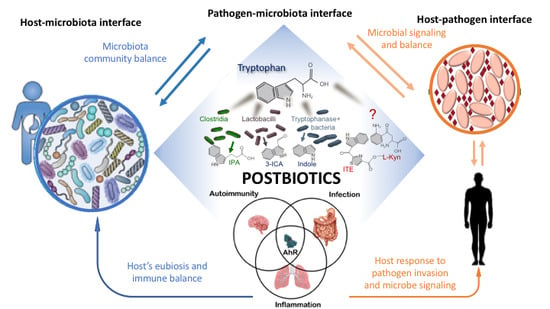
4.2. Targeting of the Host–Microbiota–Pathogen Interface
4.2.1. Signaling Molecules
4.2.2. Enhancement of Epithelial Barrier Function
4.2.3. Immunomodulatory Activity
4.2.4. Antimicrobial Activity
4.2.5. Antiproliferative Activity
5. Tryptophan-Derived Postbiotics: Indoles
5.1. Indoles at the Microbiota–Pathogen Interface-Indoles as Quorum Sensing Signals
5.2. Indole at the Host–Microbiota Interface-Indoles as an Intercellular Signal in Microbial Communities
Indoles as Ligands of the Aryl Hydrocarbon Receptor
5.3. Regulation of the Immune Response
5.4. Regulation of Intestinal Homeostasis
6. Nonantibiotic Indoles as Novel Therapeutic Tools
6.1. The Case of 3-Indole-Carboxaldehyde (3-ICA)
6.1.1. 3-ICA Enhances Epithelial Barrier Integrity
6.1.2. 3-ICA Reduces Intestinal Inflammation
6.1.3. 3-ICA Attenuates Inflammation in Patients with Atopic Dermatitis
6.1.4. 3-ICA Acts as a QS Signaling
6.1.5. 3-ICA Ameliorates Respiratory Allergic Bronchopulmonary Aspergillosis
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Amann, S.; Neef, K.; Kohl, S. Antimicrobial resistance (AMR). Eur. J. Hosp. Pharm. 2019, 26, 175–177. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Sifakis, F.; Harbarth, S.; Schrijver, R.; van Mourik, M.; Voss, A.; Sharland, M.; Rajendran, N.B.; Rodríguez-Baño, J.; Bielicki, J.; et al. Surveillance for control of antimicrobial resistance. Lancet Infect. Dis. 2018, 18, e99–e106. [Google Scholar] [CrossRef] [Green Version]
- Jacob, J.T.; Klein, E.; Laxminarayan, R.; Beldavs, Z.; Lynfield, R.; Kallen, A.J.; Ricks, P.; Edwards, J.; Srinivasan, A.; Fridkin, S.; et al. Vital signs: Carbapenem-resistant enterobacteriaceae. Morb. Mortal. Wkly. Rep. 2013, 62, 165–169. [Google Scholar]
- Spellberg, B.; Gilbert, D.N. The Future of Antibiotics and Resistance: A Tribute to a Career of Leadership by John Bartlett. Clin. Infect. Dis. 2014, 59, S71–S75. [Google Scholar] [CrossRef] [Green Version]
- WHO. Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation 2020; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Spellberg, B.; Bartlett, J.G.; Gilbert, D.N. The future of antibiotics and resistance. N. Engl. J. Med. 2013, 368, 299–302. [Google Scholar] [CrossRef] [Green Version]
- Gould, I.M.; Bal, A.M. New antibiotic agents in the pipeline and how hey can help overcome microbial resistance. Virulence 2013, 4, 185–191. [Google Scholar] [CrossRef] [Green Version]
- Murray, A.K. The Novel Coronavirus COVID-19 Outbreak: Global Implications for Antimicrobial Resistance. Front. Microbiol. 2020, 11, 1–4. [Google Scholar] [CrossRef]
- AMR Industry Alliance 2020 Progress Report. 2020. Available online: http://www.amrindustryalliance.org/wp-content/uploads/2020/01/AMR-2020-Progress-Report.pdf (accessed on 25 June 2020).
- Wang, W.; Arshad, M.I.; Khurshid, M.; Rasool, M.H.; Nisar, M.A.; Aslam, M.A.; Qamar, M.U. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 1645–1658. [Google Scholar]
- Martens, E.; Demain, A.L. The antibiotic resistance crisis, with a focus on the United States. J. Antibiot. (Tokyo) 2017, 70, 520–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Lancet The antimicrobial crisis: Enough advocacy, more action. Lancet 2020, 395, 247. [CrossRef] [Green Version]
- Ventola, C.L. The antibiotic resistance crisis: Part 2: Management strategies and new agents. Pharm. Ther. 2015, 40, 344–352. [Google Scholar]
- Ventola, C.L. The antibiotic resistance crisis: Causes and threats. Pharm. Ther. J. 2015. [Google Scholar]
- Modi, S.R.; Collins, J.J.; Relman, D.A. Antibiotics and the gut microbiota. J. Clin. Investig. 2014, 124, 4212–4218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, D.; Manna, S.; Mandal, S.M. Antibiotics Associated Disorders and Post-biotics Induced Rescue in Gut Health. Curr. Pharm. Des. 2017, 24, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Grigg, J.B.; Sonnenberg, G.F. Host-Microbiota Interactions Shape Local and Systemic Inflammatory Diseases. J. Immunol. 2017, 198, 564–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.V.; Leonardi, I.; Iliev, I.D. Gut Mycobiota in Immunity and Inflammatory Disease. Immunity 2019, 50, 1365–1379. [Google Scholar] [CrossRef] [PubMed]
- Francino, M.P. Antibiotics and the Human Gut Microbiome: Dysbioses and Accumulation of Resistances. Front. Microbiol. 2016, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Tagliabue, A.; Rappuoli, R. Changing Priorities in Vaccinology: Antibiotic Resistance Moving to the Top. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef]
- Abedon, S.T. Phage therapy of pulmonary infections. Bacteriophage 2015, 5, e1020260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasuyuki, M.; Kunihiro, K.; Kurissery, S.; Kanavillil, N.; Sato, Y.; Kikuchi, Y. Antibacterial properties of nine pure metals: A laboratory study using Staphylococcus aureus and Escherichia coli. Biofouling 2010, 26, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Beloin, C.; Renard, S.; Ghigo, J.-M.; Lebeaux, D. Novel approaches to combat bacterial biofilms. Curr. Opin. Pharmacol. 2014, 18, 61–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krachler, A.M.; Orth, K. Targeting the bacteria-host interface strategies in anti-adhesion therapy. Virulence 2013, 4, 284–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeFilipp, Z.; Bloom, P.P.; Soto, M.T.; Mansour, M.K.; Sater, M.R.A.; Huntley, M.H.; Turbett, S.; Chung, R.T.; Chen, Y.B.; Hohmann, E.L. Drug-resistant e. coli bacteremia transmitted by fecal microbiota transplant. N. Engl. J. Med. 2019, 381, 2043–2050. [Google Scholar] [CrossRef]
- Nielsen, T.B.; Brass, E.P.; Gilbert, D.N.; Bartlett, J.G.; Spellberg, B. Sustainable discovery and development of antibiotics - Is a nonprofit approach the future? N. Engl. J. Med. 2019, 381, 503–505. [Google Scholar] [CrossRef]
- Diel, R. Treatment of tuberculosis. Pneumologe 2019, 16, 117–130. [Google Scholar] [CrossRef]
- Kaufmann, S.H.E.; Dorhoi, A.; Hotchkiss, R.S.; Bartenschlager, R. Host-directed therapies for bacterial and viral infections. Nat. Rev. Drug Discov. 2018, 17, 35–56. [Google Scholar] [CrossRef]
- Andersson, J.A.; Sha, J.; Kirtley, M.L.; Reyes, E.; Fitts, E.C.; Dann, S.M.; Chopra, A.K. Combating Multidrug-Resistant Pathogens with Host-Directed Nonantibiotic Therapeutics. Antimicrob. Agents Chemother. 2017, 62. [Google Scholar] [CrossRef] [Green Version]
- Terme, M.; Ullrich, E.; Delahaye, N.F.; Chaput, N.; Zitvogel, L. Natural killer cell-directed therapies: Moving from unexpected results to successful strategies. Nat. Immunol. 2008, 9, 486–494. [Google Scholar] [CrossRef]
- Ulevitch, R.J. Therapeutics targeting the innate immune system. Nat. Rev. Immunol. 2004, 4, 512–520. [Google Scholar] [CrossRef]
- Montoya, D.; Inkeles, M.S.; Liu, P.T.; Realegeno, S.; Teles, R.M.B.; Vaidya, P.; Munoz, M.A.; Schenk, M.; Swindell, W.R.; Chun, R.; et al. IL-32 is a molecular marker of a host defense network in human tuberculosis. Sci. Transl. Med. 2014, 6, 250ra114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallis, R.S.; van Vuuren, C.; Potgieter, S. Adalimumab Treatment of Life-Threatening Tuberculosis. Clin. Infect. Dis. 2009, 48, 1429–1432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, B.L.; Mehra, S.; Ahsan, M.H.; Selman, M.; Khader, S.A.; Kaushal, D. LAG3 Expression in Active Mycobacterium tuberculosis Infections. Am. J. Pathol. 2015, 185, 820–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skrahin, A.; Ahmed, R.K.; Ferrara, G.; Rane, L.; Poiret, T.; Isaikina, Y.; Skrahina, A.; Zumla, A.; Maeurer, M.J. Autologous mesenchymal stromal cell infusion as adjunct treatment in patients with multidrug and extensively drug-resistant tuberculosis: An open-label phase 1 safety trial. Lancet Respir. Med. 2014, 2, 108–122. [Google Scholar] [CrossRef]
- Paik, S.; Kim, J.K.; Chung, C.; Jo, E.K. Autophagy: A new strategy for host-directed therapy of tuberculosis. Virulence 2019, 10, 448–459. [Google Scholar] [CrossRef] [Green Version]
- Atkin, T.A.; Maher, C.M.; Gerlach, A.C.; Gay, B.C.; Antonio, B.M.; Santos, S.C.; Padilla, K.M.; Rader, J.A.; Krafte, D.S.; Fox, M.A.; et al. A comprehensive approach to identifying repurposed drugs to treat SCN8A epilepsy. Epilepsia 2018, 59, 802–813. [Google Scholar] [CrossRef] [Green Version]
- Tharmalingam, N.; Port, J.; Castillo, D.; Mylonakis, E. Repurposing the anthelmintic drug niclosamide to combat Helicobacter pylori. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Abdelaleem, M.; Ezzat, H.; Osama, M.; Megahed, A.; Alaa, W.; Gaber, A.; Shafei, A.; Refaat, A. Prospects for repurposing CNS drugs for cancer treatment. Oncol. Rev. 2019, 13, 37–42. [Google Scholar] [CrossRef]
- Tobin, D.M.; Roca, F.J.; Ray, J.P.; Ko, D.C.; Ramakrishnan, L. An Enzyme That Inactivates the Inflammatory Mediator Leukotriene B4 Restricts Mycobacterial Infection. PLoS ONE 2013, 8, e67828. [Google Scholar] [CrossRef] [Green Version]
- Vilaplana, C.; Marzo, E.; Tapia, G.; Diaz, J.; Garcia, V.; Cardona, P.J. Ibuprofen therapy resulted in significantly decreased tissue bacillary loads and increased survival in a new murine experimental model of active tuberculosis. J. Infect. Dis. 2013, 208, 199–202. [Google Scholar] [CrossRef] [Green Version]
- Ivanyi, J.; Zumla, A. Nonsteroidal Antiinflammatory Drugs for Adjunctive Tuberculosis Treatment. J. Infect. Dis. 2013, 208, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Blum, C.A.; Nigro, N.; Briel, M.; Schuetz, P.; Ullmer, E.; Suter-Widmer, I.; Winzeler, B.; Bingisser, R.; Elsaesser, H.; Drozdov, D.; et al. Adjunct prednisone therapy for patients with community-acquired pneumonia: A multicentre, double-blind, randomised, placebo-controlled trial. Lancet 2015, 385, 1511–1518. [Google Scholar] [CrossRef]
- Ferrer, M.; Torres, A.; Baer, R.; Hernández, C.; Roca, J.; Rodriguez-Roisin, R. Effect of Acetylsalicylic Acid on Pulmonary Gas Exchange in Patients With Severe Pneumonia. Chest 1997, 111, 1094–1100. [Google Scholar] [CrossRef]
- Bernard, G.R.; Wheeler, A.P.; Russell, J.A.; Schein, R.; Summer, W.R.; Steinberg, K.P.; Fulkerson, W.J.; Wright, P.E.; Christman, B.W.; Dupont, W.D.; et al. The Effects of Ibuprofen on the Physiology and Survival of Patients with Sepsis. N. Engl. J. Med. 1997, 336, 912–918. [Google Scholar] [CrossRef]
- Halperin, S.A.; Vaudry, W.; Boucher, F.D.; Mackintosh, K.; Waggener, T.B.; Smith, B. Is pertussis immune globulin efficacious for the treatment of hospitalized infants with pertussis? No answer yet. Pediatr. Infect. Dis. J. 2007, 26, 79–81. [Google Scholar] [CrossRef]
- Bruss, J.B.; Siber, G.R. Protective effects of pertussis immunoglobulin (P-IGIV) in the aerosol challenge model. Clin. Diagn. Lab. Immunol. 1999, 6, 464–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scanlon, K.M.; Skerry, C.; Carbonetti, N.H. Novel therapies for the treatment of pertussis disease. Pathog. Dis. 2015, 73, ftv074. [Google Scholar] [CrossRef] [Green Version]
- Koh, G.C.K.W.; Weehuizen, T.A.; Breitbach, K.; Krause, K.; de Jong, H.K.; Kager, L.M.; Hoogendijk, A.J.; Bast, A.; Peacock, S.J.; van der Poll, T.; et al. Glyburide Reduces Bacterial Dissemination in a Mouse Model of Melioidosis. PLoS Negl. Trop. Dis. 2013, 7, e2500. [Google Scholar] [CrossRef] [Green Version]
- Skerry, C.; Scanlon, K.; Rosen, H.; Carbonetti, N.H. Sphingosine-1-phosphate Receptor Agonism Reduces Bordetella pertussis– mediated Lung Pathology. J. Infect. Dis. 2015, 211, 1883–1886. [Google Scholar] [CrossRef] [Green Version]
- Döcke, W.D.; Randow, F.; Syrbe, U.; Krausch, D.; Asadullah, K.; Reinke, P.; Volk, H.D.; Kox, W. Monocyte deactivation in septic patients: Restoration by IFN-γ treatment. Nat. Med. 1997, 3, 678–681. [Google Scholar] [CrossRef]
- Rosenbloom, A.J.; Linden, P.K.; Dorrance, A.; Penkosky, N.; Cohen-Melamed, M.H.; Pinsky, M.R. Effect of granulocyte-monocyte colony-stimulating factor therapy on leukocyte function and clearance of serious infection in nonneutropenic patients. Chest 2005, 127, 2139–2150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Presneill, J.J.; Harris, T.; Stewart, A.G.; Cade, J.F.; Wilson, J.W. A randomized phase II trial of granulocyte-macrophage colony-stimulating factor therapy in severe sepsis with respiratory dysfunction. Am. J. Respir. Crit. Care Med. 2002, 166, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Steinmann, J.; Halldórsson, S.; Agerberth, B.; Gudmundsson, G.H. Phenylbutyrate induces antimicrobial peptide expression. Antimicrob. Agents Chemother. 2009, 53, 5127–5133. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.-T.; Nestel, F.P.; Bourdeau, V.; Nagai, Y.; Wang, Q.; Liao, J.; Tavera-Mendoza, L.; Lin, R.; Hanrahan, J.W.; Mader, S.; et al. Cutting Edge: 1,25-Dihydroxyvitamin D 3 Is a Direct Inducer of Antimicrobial Peptide Gene Expression. J. Immunol. 2004, 173, 2909–2912. [Google Scholar] [CrossRef] [Green Version]
- Gombart, A.F.; Borregaard, N.; Koeffler, H.P. Human cathelicidin antimicrobial peptide (CAMP) gene is a direct target of the vitamin D receptor and is strongly up-regulated in myeloid cells by 1,25-dihydroxyvitamin D 3. FASEB J. 2005, 19, 1067–1077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sechet, E.; Telford, E.; Bonamy, C.; Sansonetti, P.J.; Sperandio, B. Natural molecules induce and synergize to boost expression of the human antimicrobial peptide β-defensin-3. Proc. Natl. Acad. Sci. USA 2018, 115, E9869–E9878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barber, D.L.; Sakai, S.; Kudchadkar, R.R.; Fling, S.P.; Day, T.A.; Vergara, J.A.; Ashkin, D.; Cheng, J.H.; Lundgren, L.M.; Raabe, V.N.; et al. Tuberculosis following PD-1 blockade for cancer immunotherapy. Sci. Transl. Med. 2019, 11. [Google Scholar]
- Munguia, J.; Nizet, V. Pharmacological Targeting of the Host–Pathogen Interaction: Alternatives to Classical Antibiotics to Combat Drug-Resistant Superbugs. Trends Pharmacol. Sci. 2017, 38, 473–488. [Google Scholar] [CrossRef] [Green Version]
- Roduit, C.; Scholtens, S.; De Jongste, J.C.; Wijga, A.H.; Gerritsen, J.; Postma, D.S.; Brunekreef, B.; Hoekstra, M.O.; Aalberse, R.; Smit, H.A. Asthma at 8 years of age in children born by caesarean section. Thorax 2009, 64, 107–113. [Google Scholar] [CrossRef] [Green Version]
- Mårild, K.; Ye, W.; Lebwohl, B.; Green, P.H.R.; Blaser, M.J.; Card, T.; Ludvigsson, J.F. Antibiotic exposure and the development of coeliac disease: A nationwide case–control study. BMC Gastroenterol. 2013, 13, 109. [Google Scholar] [CrossRef] [Green Version]
- Watson, J.; Jones, R.C.; Cortes, C.; Gerber, S.I.; Golash, R.G.; Price, J.; Bancroft, E.; Mascola, L.; Gorwitz, R.J.; Jernigan, D.B.; et al. Community-associated methicillin-resistant Staphylococcus aureus infection among healthy newborns - Chicago and Los Angeles County, 2004. Morb. Mortal. Wkly. Rep. 2006, 55, 329–332. [Google Scholar]
- Sekirov, I.; Tam, N.M.; Jogova, M.; Robertson, M.L.; Li, Y.; Lupp, C.; Finlay, B.B. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 2008, 76, 4726–4736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, J.Y.; Antonopoulos, D.A.; Kalra, A.; Tonelli, A.; Khalife, W.T.; Schmidt, T.M.; Young, V.B. Decreased Diversity of the Fecal Microbiome in Recurrent Clostridium difficile—Associated Diarrhea. J. Infect. Dis. 2008, 197, 435–438. [Google Scholar] [CrossRef] [Green Version]
- Buffie, C.G.; Jarchum, I.; Equinda, M.; Lipuma, L.; Gobourne, A.; Viale, A.; Ubeda, C.; Xavier, J.; Pamer, E.G. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 2012, 80, 62–73. [Google Scholar] [CrossRef] [Green Version]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fessler, J.; Matson, V.; Gajewski, T.F. Exploring the emerging role of the microbiome in cancer immunotherapy. J. Immunother. Cancer 2019, 7, 108. [Google Scholar] [CrossRef]
- Taylor, V.H. The microbiome and mental health: Hope or hype? J. Psychiatry Neurosci. 2019, 44, 219–222. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Elinav, E. The remedy within: Will the microbiome fulfill its therapeutic promise? J. Mol. Med. 2017, 95, 1021–1027. [Google Scholar] [CrossRef]
- Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [CrossRef] [Green Version]
- Simon, G.L.; Gorbach, S.L. Intestinal flora in health and disease. Gastroenterology 1984, 86, 174–193. [Google Scholar] [CrossRef]
- Delhaes, L.; Monchy, S.; Fréalle, E.; Hubans, C.; Salleron, J.; Leroy, S.; Prevotat, A.; Wallet, F.; Wallaert, B.; Dei-Cas, E.; et al. The Airway Microbiota in Cystic Fibrosis: A Complex Fungal and Bacterial Community—Implications for Therapeutic Management. PLoS ONE 2012, 7, e36313. [Google Scholar] [CrossRef] [PubMed]
- Ott, S.J.; Kühbacher, T.; Musfeldt, M.; Rosenstiel, P.; Hellmig, S.; Rehman, A.; Drews, O.; Weichert, W.; Timmis, K.N.; Schreiber, S. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand. J. Gastroenterol. 2008, 43, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, C.; Tang, C.; He, Q.; Li, N.; Li, J. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in crohn’s disease. J. Clin. Gastroenterol. 2014, 48, 513–523. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Archaea and Fungi of the Human Gut Microbiome: Correlations with Diet and Bacterial Residents. PLoS ONE 2013, 8, e66019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, E.; Tanaka, T.; Tajima, M.; Tsuboi, R.; Nishikawa, A.; Sugita, T. Characterization of the skin fungal microbiota in patients with atopic dermatitis and in healthy subjects. Microbiol. Immunol. 2011, 55, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Smeekens, S.P.; Malireddi, R.K.; Plantinga, T.S.; Buffen, K.; Oosting, M.; Joosten, L.A.B.; Kullberg, B.J.; Perfect, J.R.; Scott, W.K.; Van De Veerdonk, F.L.; et al. Autophagy is redundant for the host defense against systemic Candida albicans infections. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 711–722. [Google Scholar] [CrossRef]
- Cully, M. Antibiotics alter the gut microbiome and host health. Nat. Milestones 2019, 1423, S19. [Google Scholar]
- Lavelle, A.; Hoffmann, T.W.; Pham, H.P.; Langella, P.; Guédon, E.; Sokol, H. Baseline microbiota composition modulates antibiotic-mediated effects on the gut microbiota and host. Microbiome 2019, 7, 111. [Google Scholar] [CrossRef]
- Bhalodi, A.A.; Van Engelen, T.S.R.; Virk, H.S.; Wiersinga, W.J. Impact of antimicrobial therapy on the gut microbiome. J. Antimicrob. Chemother. 2019, 74, I6–I15. [Google Scholar] [CrossRef] [Green Version]
- Baümler, A.J.; Sperandio, V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature 2016, 535, 85–93. [Google Scholar] [CrossRef] [Green Version]
- Becattini, S.; Taur, Y.; Pamer, E.G. Antibiotic-Induced Changes in the Intestinal Microbiota and Disease. Trends Mol. Med. 2016, 22, 458–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Chen, D.C.; Chen, L.M. Facing a new challenge: The adverse effects of antibiotics on gut microbiota and host immunity. Chin. Med. J. (Engl.) 2019, 132, 1135–1138. [Google Scholar] [CrossRef] [PubMed]
- Guarner, F.; Malagelada, J.R. Gut flora in health and disease. Lancet 2003, 361, 512–519. [Google Scholar] [CrossRef]
- Stefka, A.T.; Feehley, T.; Tripathi, P.; Qiu, J.; McCoy, K.; Mazmanian, S.K.; Tjota, M.Y.; Seo, G.Y.; Cao, S.; Theriault, B.R.; et al. Commensal bacteria protect against food allergen sensitization. Proc. Natl. Acad. Sci. USA 2014, 111, 13145–13150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsiao, E.Y.; McBride, S.W.; Hsien, S.; Sharon, G.; Hyde, E.R.; McCue, T.; Codelli, J.A.; Chow, J.; Reisman, S.E.; Petrosino, J.F.; et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013, 155, 1451–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teo, S.M.; Mok, D.; Pham, K.; Kusel, M.; Serralha, M.; Troy, N.; Holt, B.J.; Hales, B.J.; Walker, M.L.; Hollams, E.; et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015, 17, 704–715. [Google Scholar] [CrossRef] [Green Version]
- Cuthbertson, L.; Rogers, G.B.; Walker, A.W.; Oliver, A.; Green, L.E.; Daniels, T.W.V.; Carroll, M.P.; Parkhill, J.; Bruce, K.D.; Van Der Gast, C.J. Respiratory microbiota resistance and resilience to pulmonary exacerbation and subsequent antimicrobial intervention. ISME J. 2016, 10, 1081–1091. [Google Scholar] [CrossRef] [Green Version]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Chan, B.K.; Turner, P.E.; Kim, S.; Mojibian, H.R.; Elefteriades, J.A.; Narayan, D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Heal. 2018, 2018, 60–66. [Google Scholar] [CrossRef] [Green Version]
- Jault, P.; Leclerc, T.; Jennes, S.; Pirnay, J.P.; Que, Y.A.; Resch, G.; Rousseau, A.F.; Ravat, F.; Carsin, H.; Le Floch, R.; et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): A randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 2019, 19, 35–45. [Google Scholar] [CrossRef]
- Vahedi, A.; Soltan Dallal, M.M.; Douraghi, M.; Nikkhahi, F.; Rajabi, Z.; Yousefi, M.; Mousavi, M. Isolation and identification of specific bacteriophage against enteropathogenic Escherichia coli (EPEC) and in vitro and in vivo characterization of bacteriophage. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef] [Green Version]
- Chadha, P.; Katare, O.P.; Chhibber, S. Liposome loaded phage cocktail: Enhanced therapeutic potential in resolving Klebsiella pneumoniae mediated burn wound infections. Burns 2017, 43, 1532–1543. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.L.; Zulquernain, S.A.; Shanahan, F. Faecal Microbiota Transplantation (FMT)—Classical bedside-to-bench clinical research. QJM 2019, 112. [Google Scholar] [CrossRef]
- Paramsothy, S.; Kamm, M.A.; Kaakoush, N.O.; Walsh, A.J.; van den Bogaerde, J.; Samuel, D.; Leong, R.W.L.; Connor, S.; Ng, W.; Paramsothy, R.; et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: A randomised placebo-controlled trial. Lancet 2017, 389, 1218–1228. [Google Scholar] [CrossRef]
- Paramsothy, S.; Paramsothy, R.; Rubin, D.T.; Kamm, M.A.; Kaakoush, N.O.; Mitchell, H.M.; Castaño-Rodríguez, N. Faecal Microbiota Transplantation for Inflammatory Bowel Disease: A Systematic Review and Meta-analysis. J. Crohn’s Colitis 2017, 11, 1180–1199. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.U. Clinical Uses of Probiotics. Medicine (Baltimore) 2016, 95, e2658. [Google Scholar] [CrossRef]
- Kim, H.G.; Lee, S.Y.; Kim, N.R.; Lee, H.Y.; Ko, M.Y.; Jung, B.J.; Kim, C.M.; Lee, J.M.; Park, J.H.; Han, S.H.; et al. Lactobacillus plantarum lipoteichoic acid down-regulated Shigella flexneri peptidoglycan-induced inflammation. Mol. Immunol. 2011, 48, 382–391. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermudez-Humaran, L.G.; Gratadoux, J.-J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [Green Version]
- Hart, A.L.; Stagg, A.J.; Kamm, M.A. Use of Probiotics in the Treatment of Inflammatory Bowel Disease. J. Clin. Gastroenterol. 2003, 36, 111–119. [Google Scholar] [CrossRef]
- Stripling, J.; Kumar, R.; Baddley, J.W.; Nellore, A.; Dixon, P.; Howard, D.; Ptacek, T.; Lefkowitz, E.J.; Tallaj, J.A.; Benjamin, W.H.; et al. Loss of Vancomycin-Resistant Enterococcus Fecal Dominance in an Organ Transplant Patient With Clostridium difficile Colitis After Fecal Microbiota Transplant. Open Forum Infect. Dis. 2015, 2. [Google Scholar] [CrossRef]
- Cammarota, G.; Masucci, L.; Ianiro, G.; Bibbò, S.; Dinoi, G.; Costamagna, G.; Sanguinetti, M.; Gasbarrini, A. Randomised clinical trial: Faecal microbiota transplantation by colonoscopy vs. vancomycin for the treatment of recurrent Clostridium difficile infection. Aliment. Pharmacol. Ther. 2015, 41, 835–843. [Google Scholar] [CrossRef]
- Battipaglia, G.; Malard, F.; Rubio, M.T.; Ruggeri, A.; Mamez, A.C.; Brissot, E.; Giannotti, F.; Dulery, R.; Joly, A.C.; Baylatry, M.T.; et al. Fecal microbiota transplantation before or after allogeneic hematopoietic transplantation in patients with hematologic malignancies carrying multidrug-resistance bacteria. Haematologica 2019, 104, 1682–1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganeshan, S.D.; Hosseinidoust, Z. Phage therapy with a focus on the human microbiota. Antibiotics 2019, 8, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutton, T.D.S.; Hill, C. Gut Bacteriophage: Current Understanding and Challenges. Front. Endocrinol. (Lausanne) 2019. [Google Scholar] [CrossRef] [PubMed]
- Hsu, B.B.; Gibson, T.E.; Yeliseyev, V.; Liu, Q.; Lyon, L.; Bry, L.; Silver, P.A.; Gerber, G.K. Dynamic Modulation of the Gut Microbiota and Metabolome by Bacteriophages in a Mouse Model. Cell Host Microbe 2019, 25, 803–814.e5. [Google Scholar] [CrossRef] [Green Version]
- Paule, A.; Frezza, D.; Edeas, M. Microbiota and Phage Therapy: Future Challenges in Medicine. Med. Sci. 2018, 6, 86. [Google Scholar] [CrossRef] [Green Version]
- Sausset, R.; Petit, M.A.; Gaboriau-Routhiau, V.; De Paepe, M. New insights into intestinal phages. Mucosal Immunol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Wong, W.F.; Santiago, M. Microbial approaches for targeting antibiotic-resistant bacteria. Microb. Biotechnol. 2017, 10, 1047–1053. [Google Scholar] [CrossRef] [Green Version]
- Tkhilaishvili, T.; Lombardi, L.; Klatt, A.B.; Trampuz, A.; Di Luca, M. Bacteriophage Sb-1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus. Int. J. Antimicrob. Agents 2018, 52, 842–853. [Google Scholar] [CrossRef]
- Fong, S.A.; Drilling, A.; Morales, S.; Cornet, M.E.; Woodworth, B.A.; Fokkens, W.J.; Psaltis, A.J.; Vreugde, S.; Wormald, P.-J. Activity of Bacteriophages in Removing Biofilms of Pseudomonas aeruginosa Isolates from Chronic Rhinosinusitis Patients. Front. Cell. Infect. Microbiol. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Bourdin, G.; Navarro, A.; Sarker, S.A.; Pittet, A.-C.; Qadri, F.; Sultana, S.; Cravioto, A.; Talukder, K.A.; Reuteler, G.; Brüssow, H. Coverage of diarrhoea-associated E scherichia coli isolates from different origins with two types of phage cocktails. Microb. Biotechnol. 2014, 7, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Maciejewska, B.; Olszak, T.; Drulis-Kawa, Z. Applications of bacteriophages versus phage enzymes to combat and cure bacterial infections: An ambitious and also a realistic application? Appl. Microbiol. Biotechnol. 2018, 102, 2563–2581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furfaro, L.L.; Payne, M.S.; Chang, B.J. Bacteriophage Therapy: Clinical Trials and Regulatory Hurdles. Front. Cell. Infect. Microbiol. 2018, 8, 376. [Google Scholar] [CrossRef] [Green Version]
- Pirnay, J.P.; Verbeken, G.; Ceyssens, P.J.; Huys, I.; de Vos, D.; Ameloot, C.; Fauconnier, A. The magistral phage. Viruses 2018, 10, 64. [Google Scholar] [CrossRef] [Green Version]
- Lerner, A.; Matthias, T.; Aminov, R. Potential effects of horizontal gene exchange in the human gut. Front. Immunol. 2017, 8, 1630. [Google Scholar] [CrossRef] [Green Version]
- Wong, A.; Ngu, D.Y.S.; Dan, L.A.; Ooi, A.; Lim, R.L.H. Detection of antibiotic resistance in probiotics of dietary supplements. Nutr. J. 2015, 14, 95. [Google Scholar] [CrossRef]
- Lerner, A.; Matthias, T. Changes in intestinal tight junction permeability associated with industrial food additives explain the rising incidence of autoimmune disease. Autoimmun. Rev. 2015, 14, 479–489. [Google Scholar] [CrossRef] [Green Version]
- Lerner, A.; Aminov, R.; Matthias, T. Transglutaminases in Dysbiosis As Potential Environmental Drivers of Autoimmunity. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Matthias, T.; Jeremias, P.; Neidhöfer, S.; Lerner, A. The industrial food additive, microbial transglutaminase, mimics tissue transglutaminase and is immunogenic in celiac disease patients. Autoimmun. Rev. 2016, 15, 1111–1119. [Google Scholar] [CrossRef]
- Torsten, M.; Aaron, L. Microbial Transglutaminase Is Immunogenic and Potentially Pathogenic in Pediatric Celiac Disease. Front. Pediatr. 2018, 6. [Google Scholar] [CrossRef]
- Singh, A.; Sarangi, A.N.; Goel, A.; Srivastava, R.; Bhargava, R.; Gaur, P.; Aggarwal, A.; Aggarwal, R. Effect of administration of a probiotic preparation on gut microbiota and immune response in healthy women in India: An open-label, single-arm pilot study. BMC Gastroenterol. 2018, 18, 85. [Google Scholar] [CrossRef]
- Laursen, M.F.; Laursen, R.P.; Larnkjær, A.; Michaelsen, K.F.; Bahl, M.I.; Licht, T.R. Administration of two probiotic strains during early childhood does not affect the endogenous gut microbiota composition despite probiotic proliferation. BMC Microbiol. 2017, 17, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kristensen, N.B.; Bryrup, T.; Allin, K.H.; Nielsen, T.; Hansen, T.H.; Pedersen, O. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: A systematic review of randomized controlled trials. Genome Med. 2016, 8, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz, J.A.M.; Chenoll, E.; Casinos, B.; Bataller, E.; Ramón, D.; Genovés, S.; Montava, R.; Ribes, J.M.; Buesa, J.; Fàbrega, J.; et al. Novel probiotic Bifidobacterium longum subsp. infantis CECT 7210 strain active against rotavirus infections. Appl. Environ. Microbiol. 2011, 77, 8775–8783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jalali, M.; Abedi, D.; Varshosaz, J.; Najjarzadeh, M.; Mirlohi, M.; Tavakoli, N. Stability evaluation of freeze-dried Lactobacillus paracasei subsp. tolerance and Lactobacillus delbrueckii subsp. bulgaricus in oral capsules. Res. Pharm. Sci. 2012, 7, 31–36. [Google Scholar]
- Markowiak, P.; Ślizewska, K. Effects of Probiotics, Prebiotics, and Synbiotics on Human Health. Nutrients 2017, 9, 1021. [Google Scholar] [CrossRef] [PubMed]
- Pandey, K.R.; Naik, S.R.; Vakil, B.V. Probiotics, prebiotics and synbiotics—A review. J. Food Sci. Technol. 2015, 52, 7577–7587. [Google Scholar] [CrossRef]
- McFarland, L.V.; Goh, S. Are probiotics and prebiotics effective in the prevention of travellers’ diarrhea: A systematic review and meta-analysis. Travel Med. Infect. Dis. 2019, 27, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Deng, L.; Wu, W.; Wang, Z.; Shao, W.; Liu, J. Systematic review and meta-analysis of the effect of probiotic supplementation on functional constipation in children. Medicine (USA) 2018, 97, e12174. [Google Scholar] [CrossRef]
- Rouhani, M.H.; Hadi, A.; Ghaedi, E.; Salehi, M.; Mahdavi, A.; Mohammadi, H. Do probiotics, prebiotics and synbiotics affect adiponectin and leptin in adults? A systematic review and meta-analysis of clinical trials. Clin. Nutr. 2019, 38, 2031–2037. [Google Scholar] [CrossRef]
- Tan-Lim, C.S.C.; Esteban-Ipac, N.A.R. Probiotics as treatment for food allergies among pediatric patients: A meta-analysis. World Allergy Organ. J. 2018, 11, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upadhyay, R.P.; Taneja, S.; Chowdhury, R.; Strand, T.A.; Bhandari, N. Effect of prebiotic and probiotic supplementation on neurodevelopment in preterm very low birth weight infants: Findings from a meta-analysis. Pediatr. Res. 2018. [Google Scholar] [CrossRef]
- Aqaeinezhad Rudbane, S.M.; Rahmdel, S.; Abdollahzadeh, S.M.; Zare, M.; Bazrafshan, A.; Mazloomi, S.M. The efficacy of probiotic supplementation in rheumatoid arthritis: A meta-analysis of randomized, controlled trials. Inflammopharmacology 2018, 26, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Hassan, H.; Rompola, M.; Glaser, A.W.; Kinsey, S.E.; Phillips, R.S. Systematic review and meta-analysis investigating the efficacy and safety of probiotics in people with cancer. Support. Care Cancer 2018, 26, 2503–2509. [Google Scholar] [CrossRef]
- Vieira, A.T.; Fukumori, C.; Ferreira, C.M. New insights into therapeutic strategies for gut microbiota modulation in inflammatory diseases. Clin. Transl. Immunol. 2016, 5, e87. [Google Scholar] [CrossRef] [PubMed]
- Descamps, H.C.; Herrmann, B.; Wiredu, D.; Thaiss, C.A. The path toward using microbial metabolites as therapies. EBioMedicine 2019, 44, 747–754. [Google Scholar] [CrossRef] [Green Version]
- Klemashevich, C.; Wu, C.; Howsmon, D.; Alaniz, R.C.; Lee, K.; Jayaraman, A. Rational identification of diet-derived postbiotics for improving intestinal microbiota function. Curr. Opin. Biotechnol. 2014, 26, 85–90. [Google Scholar] [CrossRef]
- Shoaie, S.; Ghaffari, P.; Kovatcheva-Datchary, P.; Mardinoglu, A.; Sen, P.; Pujos-Guillot, E.; De Wouters, T.; Juste, C.; Rizkalla, S.; Chilloux, J.; et al. Quantifying Diet-Induced Metabolic Changes of the Human Gut Microbiome. Cell Metab. 2015, 22, 320–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeevi, D.; Korem, T.; Zmora, N.; Israeli, D.; Rothschild, D.; Weinberger, A.; Ben-Yacov, O.; Lador, D.; Avnit-Sagi, T.; Lotan-Pompan, M.; et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015, 163, 1079–1094. [Google Scholar] [CrossRef] [Green Version]
- Umu, Ö.C.O.; Rudi, K.; Diep, D.B. Modulation of the gut microbiota by prebiotic fibres and bacteriocins. Microb. Ecol. Health Dis. 2017, 28, 1348886. [Google Scholar] [CrossRef] [Green Version]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Xie, G.; Zhao, A.; Zhao, L.; Yao, C.; Chiu, N.H.L.; Zhou, Z.; Bao, Y.; Jia, W.; Nicholson, J.K.; et al. The footprints of gut microbial-mammalian co-metabolism. J. Proteome Res. 2011, 10, 5512–5522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Z.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009, 58, 1509–1517. [Google Scholar] [CrossRef] [Green Version]
- Rouse, M.; Singh, N.P.; Nagarkatti, P.S.; Nagarkatti, M. Indoles mitigate the development of experimental autoimmune encephalomyelitis by induction of reciprocal differentiation of regulatory T cells and Th17 cells. Br. J. Pharmacol. 2013, 169, 1305–1321. [Google Scholar] [CrossRef] [Green Version]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [Green Version]
- Wegh, C.A.M.; Geerlings, S.Y.; Knol, J.; Roeselers, G.; Belzer, C. Postbiotics and Their Potential Applications in Early Life Nutrition and Beyond. Int. J. Mol. Sci. 2019, 20, 4673. [Google Scholar] [CrossRef] [Green Version]
- Aguilar-Toalá, J.E.; Garcia-Varela, R.; Garcia, H.S.; Mata-Haro, V.; González-Córdova, A.F.; Vallejo-Cordoba, B.; Hernández-Mendoza, A. Postbiotics: An evolving term within the functional foods field. Trends Food Sci. Technol. 2018, 75, 105–114. [Google Scholar] [CrossRef]
- Shenderov, B.A. Metabiotics: Novel idea or natural development of probiotic conception. Microb. Ecol. Heal. Dis. 2013, 24. [Google Scholar] [CrossRef]
- Zhang, L.S.; Davies, S.S. Microbial metabolism of dietary components to bioactive metabolites: Opportunities for new therapeutic interventions. Genome Med. 2016, 8, 46. [Google Scholar] [CrossRef] [Green Version]
- Kho, Z.Y.; Lal, S.K. The Human Gut Microbiome—A Potential Controller of Wellness and Disease. Front. Microbiol. 2018, 9, 1835. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Tang, H.; Chen, P.; Xie, H.; Tao, Y. Demystifying the manipulation of host immunity, metabolism, and extraintestinal tumors by the gut microbiome. Signal Transduct. Target. Ther. 2019, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Garge, N.; Zhang, X.; Sun, W.; O’Connell, T.M.; Bunger, M.K.; Bultman, S.J. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011, 13, 517–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaiko, G.E.; Ryu, S.H.; Koues, O.I.; Collins, P.L.; Solnica-Krezel, L.; Pearce, E.J.; Pearce, E.L.; Oltz, E.M.; Stappenbeck, T.S. The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell 2016, 165, 1708–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Round, J.L.; Mazmanian, S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA 2010, 107, 12204–12209. [Google Scholar] [CrossRef] [Green Version]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic T reg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [Green Version]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [Green Version]
- Arpaia, N.; Green, J.A.; Moltedo, B.; Arvey, A.; Hemmers, S.; Yuan, S.; Treuting, P.M.; Rudensky, A.Y. A Distinct Function of Regulatory T Cells in Tissue Protection. Cell 2015, 162, 1078–1089. [Google Scholar] [CrossRef] [Green Version]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef]
- Gaudier, E.; Jarry, A.; Blottière, H.M.; de Coppet, P.; Buisine, M.P.; Aubert, J.P.; Laboisse, C.; Cherbut, C.; Hoebler, C. Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. Am. J. Physiol. Liver Physiol. 2004, 287, G1168–G1174. [Google Scholar] [CrossRef] [Green Version]
- Macia, L.; Tan, J.; Vieira, A.T.; Leach, K.; Stanley, D.; Luong, S.; Maruya, M.; Ian McKenzie, C.; Hijikata, A.; Wong, C.; et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 2015, 6, 6734. [Google Scholar] [CrossRef] [Green Version]
- Vinolo, M.A.R.; Rodrigues, H.G.; Hatanaka, E.; Sato, F.T.; Sampaio, S.C.; Curi, R. Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils. J. Nutr. Biochem. 2011, 22, 849–855. [Google Scholar] [CrossRef] [PubMed]
- Usami, M.; Kishimoto, K.; Ohata, A.; Miyoshi, M.; Aoyama, M.; Fueda, Y.; Kotani, J. Butyrate and trichostatin A attenuate nuclear factor κB activation and tumor necrosis factor α secretion and increase prostaglandin E2 secretion in human peripheral blood mononuclear cells. Nutr. Res. 2008, 28, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef] [Green Version]
- Singh, N.; Thangaraju, M.; Prasad, P.D.; Martin, P.M.; Lambert, N.A.; Boettger, T.; Offermanns, S.; Ganapathy, V. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J. Biol. Chem. 2010, 285, 27601–27608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Bakdash, G.; Vogelpoel, L.T.; van Capel, T.M.; Kapsenberg, M.L.; de Jong, E.C. Retinoic acid primes human dendritic cells to induce gut-homing, IL-10-producing regulatory T cells. Mucosal Immunol. 2015, 8, 265–278. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Li, J.; Zheng, W.; Zhao, G.; Zhang, H.; Wang, X.; Guo, Y.; Qin, C.; Shi, Y. Peripheral Lymphoid Volume Expansion and Maintenance Are Controlled by Gut Microbiota via RALDH+ Dendritic Cells. Immunity 2016, 44, 330–342. [Google Scholar] [CrossRef] [Green Version]
- Mucida, D.; Park, Y.; Kim, G.; Turovskaya, O.; Scott, I.; Kronenberg, M.; Cheroutre, H. Reciprocal TH17 and Regulatory T Cell Differentiation Mediated by Retinoic Acid. Science 2007, 317, 256–260. [Google Scholar] [CrossRef] [Green Version]
- Hall, J.A.; Cannons, J.L.; Grainger, J.R.; Dos Santos, L.M.; Hand, T.W.; Naik, S.; Wohlfert, E.A.; Chou, D.B.; Oldenhove, G.; Robinson, M.; et al. Essential role for retinoic acid in the promotion of CD4+ T cell effector responses via retinoic acid receptor alpha. Immunity 2011, 34, 435–447. [Google Scholar] [CrossRef] [Green Version]
- Round, J.L.; Lee, S.M.; Li, J.; Tran, G.; Jabri, B.; Chatila, T.A.; Mazmanian, S.K. The Toll-Like Receptor 2 Pathway Establishes Colonization by a Commensal of the Human Microbiota. Science 2011, 332, 974–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazmanian, S.K.; Round, J.L.; Kasper, D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008, 453, 620–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut Microbiota from Twins Discordant for Obesity Modulate Metabolism in Mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pols, T.W.H.; Nomura, M.; Harach, T.; Lo Sasso, G.; Oosterveer, M.H.; Thomas, C.; Rizzo, G.; Gioiello, A.; Adorini, L.; Pellicciari, R.; et al. TGR5 Activation Inhibits Atherosclerosis by Reducing Macrophage Inflammation and Lipid Loading. Cell Metab. 2011, 14, 747–757. [Google Scholar] [CrossRef] [Green Version]
- Devkota, S.; Wang, Y.; Musch, M.W.; Leone, V.; Fehlner-Peach, H.; Nadimpalli, A.; Antonopoulos, D.A.; Jabri, B.; Chang, E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012, 487, 104–108. [Google Scholar] [CrossRef] [Green Version]
- Levy, M.; Thaiss, C.A.; Zeevi, D.; Dohnalová, L.; Zilberman-Schapira, G.; Mahdi, J.A.; David, E.; Savidor, A.; Korem, T.; Herzig, Y.; et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 2015, 163, 1428–1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; DeLuca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef] [Green Version]
- Qiu, J.; Heller, J.J.; Guo, X.; Chen, Z.E.; Fish, K.; Fu, Y.-X.; Zhou, L. The Aryl Hydrocarbon Receptor Regulates Gut Immunity through Modulation of Innate Lymphoid Cells. Immunity 2012, 36, 92–104. [Google Scholar] [CrossRef] [Green Version]
- Roager, H.M.; Dragsted, L.O. Diet-derived microbial metabolites in health and disease. Nutr. Bull. 2019, 44, 216–227. [Google Scholar] [CrossRef] [Green Version]
- Milligan, G.; Bolognini, D.; Sergeev, E. Ligands at the Free Fatty Acid Receptors 2/3 (GPR43/GPR41). Handb. Exp. Pharmacol. 2017, 236, 17–32. [Google Scholar]
- Thaiss, C.A.; Zmora, N.; Levy, M.; Elinav, E. The microbiome and innate immunity. Nature 2016, 535, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.D.; van den Brink, G.R. Selective inhibition of mucosal serotonin as treatment for IBD? Gut 2014, 63, 866–867. [Google Scholar] [CrossRef] [PubMed]
- Fiorucci, S.; Biagioli, M.; Zampella, A.; Distrutti, E. Bile Acids Activated Receptors Regulate Innate Immunity. Front. Immunol. 2018, 9, 1853. [Google Scholar] [CrossRef] [Green Version]
- Mertens, K.L.; Kalsbeek, A.; Soeters, M.R.; Eggink, H.M. Bile Acid Signaling Pathways from the Enterohepatic Circulation to the Central Nervous System. Front. Neurosci. 2017, 11. [Google Scholar] [CrossRef] [Green Version]
- Linsalata, M.; Giannini, R.; Notarnicola, M.; Cavallini, A. Peroxisome proliferator-activated receptor gamma and spermidine/spermine N1-acetyltransferase gene expressions are significantly correlated in human colorectal cancer. BMC Cancer 2006, 6, 191. [Google Scholar] [CrossRef] [Green Version]
- Ahern, G.P.; Wang, X.; Miyares, R.L. Polyamines are potent ligands for the capsaicin receptor TRPV1. J. Biol. Chem. 2006, 281, 8991–8995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Wang, H.; Tracey, K.J. Regulation of macrophage activation and inflammation by spermine: A new chapter in an old story. Crit. Care Med. 2000, 28, N60–N66. [Google Scholar] [CrossRef] [PubMed]
- Fennema, D.; Phillips, I.R.; Shephard, E.A. Trimethylamine and Trimethylamine N-Oxide, a Flavin-Containing Monooxygenase 3 (FMO3)-Mediated Host-Microbiome Metabolic Axis Implicated in Health and Disease. Drug Metab. Dispos. 2016, 44, 1839–1850. [Google Scholar] [CrossRef] [PubMed]
- Seldin, M.M.; Meng, Y.; Qi, H.; Zhu, W.; Wang, Z.; Hazen, S.L.; Lusis, A.J.; Shih, D.M. Trimethylamine N-Oxide Promotes Vascular Inflammation Through Signaling of Mitogen-Activated Protein Kinase and Nuclear Factor-κB. J. Am. Heart Assoc. 2016, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, E.-A.; Choo, M.-K.; Park, E.-K.; Park, S.-Y.; Shin, H.-Y.; Kim, D.-H. Metabolism of Ginsenoside Rc by Human Intestinal Bacteria and Its Related Antiallergic Activity. Biol. Pharm. Bull. 2002, 25, 743–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stark, A.H.; Madar, Z. In vitro production of short-chain fatty acids by bacterial fermentation of dietary fiber compared with effects of those fibers on hepatic sterol synthesis in rats. J. Nutr. 1993, 123, 2166–2173. [Google Scholar] [PubMed]
- Macfarlane, S.; Macfarlane, G.T. Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 2003, 62, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Gérard, P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens 2013, 3, 14–24. [Google Scholar] [CrossRef] [Green Version]
- Surprenant, A.; Rassendren, F.; Kawashima, E.; North, R.A.; Buell, G. The cytolytic P2Z receptor for extracellular ATP identified as a P2X receptor (P2X7). Science 1996, 272, 735–738. [Google Scholar] [CrossRef]
- Wang, H.; Chen, J.; Hollister, K.; Sowers, L.C.; Forman, B.M. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell 1999, 3, 543–553. [Google Scholar] [CrossRef]
- Maruyama, T.; Miyamoto, Y.; Nakamura, T.; Tamai, Y.; Okada, H.; Sugiyama, E.; Nakamura, T.; Itadani, H.; Tanaka, K. Identification of membrane-type receptor for bile acids (M-BAR). Biochem. Biophys. Res. Commun. 2002, 298, 714–719. [Google Scholar] [CrossRef]
- Roberts, E.; Golas, C.; Okey, A. Ah Receptor Mediating Induction of Aryl Hydrocarbon Hydroxylase: Detection in Human Lung by Binding of 2,3,7,8-[3H]Tetrachlorodibenzo–p-dioxin. Cancer Res. 1986, 46, 3739–3743. [Google Scholar]
- Kliewer, S.A.; Moore, J.T.; Wade, L.; Staudinger, J.L.; Watson, M.A.; Jones, S.A.; McKee, D.D.; Oliver, B.B.; Willson, T.M.; Zetterström, R.H.; et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 1998, 92, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Venkatesh, M.; Mukherjee, S.; Wang, H.; Li, H.; Sun, K.; Benechet, A.P.; Qiu, Z.; Maher, L.; Redinbo, M.R.; Phillips, R.S.; et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and toll-like receptor 4. Immunity 2014, 41, 296–310. [Google Scholar] [CrossRef] [Green Version]
- Holmes, E.; Loo, R.L.; Stamler, J.; Bictash, M.; Yap, I.K.S.; Chan, Q.; Ebbels, T.; De Iorio, M.; Brown, I.J.; Veselkov, K.A.; et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 2008, 453, 396–400. [Google Scholar] [CrossRef]
- Smith, M.I.; Yatsunenko, T.; Manary, M.J.; Trehan, I.; Mkakosya, R.; Cheng, J.; Kau, A.L.; Rich, S.S.; Concannon, P.; Mychaleckyj, J.C.; et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 2013, 339, 548–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, A.J.; Goldsworthy, S.M.; Barnes, A.A.; Eilert, M.M.; Tcheang, L.; Daniels, D.; Muir, A.I.; Wigglesworth, M.J.; Kinghorn, I.; Fraser, N.J.; et al. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 2003, 278, 11312–11319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Poul, E.; Loison, C.; Struyf, S.; Springael, J.Y.; Lannoy, V.; Decobecq, M.E.; Brezillon, S.; Dupriez, V.; Vassart, G.; Van Damme, J.; et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J. Biol. Chem. 2003, 278, 25481–25489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seljeset, S.; Siehler, S. Receptor-specific regulation of ERK1/2 activation by members of the “free fatty acid receptor” family. J. Recept. Signal Transduct. 2012, 32, 196–201. [Google Scholar] [CrossRef]
- Miyauchi, S.; Gopal, E.; Fei, Y.J.; Ganapathy, V. Functional Identification of SLC5A8, a Tumor Suppressor Down-regulated in Colon Cancer, as a Na+-coupled Transporter for Short-chain Fatty Acids. J. Biol. Chem. 2004, 279, 13293–13296. [Google Scholar] [CrossRef] [Green Version]
- Thangaraju, M.; Cresci, G.A.; Liu, K.; Ananth, S.; Gnanaprakasam, J.P.; Browning, D.D.; Mellinger, J.D.; Smith, S.B.; Digby, G.J.; Lambert, N.A.; et al. GPFM 09A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009, 69, 2826–2832. [Google Scholar] [CrossRef] [Green Version]
- Steliou, K.; Boosalis, M.S.; Perrine, S.P.; Sangerman, J.; Faller, D.V. Butyrate histone deacetylase inhibitors. Biores. Open Access 2012, 1, 192–198. [Google Scholar] [CrossRef] [Green Version]
- Donohoe, D.R.; Collins, L.B.; Wali, A.; Bigler, R.; Sun, W.; Bultman, S.J. The Warburg Effect Dictates the Mechanism of Butyrate-Mediated Histone Acetylation and Cell Proliferation. Mol. Cell 2012, 48, 612–626. [Google Scholar] [CrossRef] [Green Version]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [Green Version]
- Alex, S.; Lange, K.; Amolo, T.; Grinstead, J.S.; Haakonsson, A.K.; Szalowska, E.; Koppen, A.; Mudde, K.; Haenen, D.; Al-Lahham, S.; et al. Short-Chain Fatty Acids Stimulate Angiopoietin-Like 4 Synthesis in Human Colon Adenocarcinoma Cells by Activating Peroxisome Proliferator-Activated Receptor. Mol. Cell. Biol. 2013, 33, 1303–1316. [Google Scholar] [CrossRef] [Green Version]
- Elamin, E.E.; Masclee, A.A.; Dekker, J.; Pieters, H.-J.; Jonkers, D.M. Short-Chain Fatty Acids Activate AMP-Activated Protein Kinase and Ameliorate Ethanol-Induced Intestinal Barrier Dysfunction in Caco-2 Cell Monolayers. J. Nutr. 2013, 143, 1872–1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willemsen, L.E.M.; Koetsier, M.A.; Van Deventer, S.J.H.; Van Tol, E.A.F. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E1 and E2 production by intestinal myofibroblasts. Gut 2003, 52, 1442–1447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Assa, A.; Vong, L.; Pinnell, L.J.; Avitzur, N.; Johnson-Henry, K.C.; Sherman, P.M. Vitamin D deficiency promotes epithelial barrier dysfunction and intestinal inflammation. J. Infect. Dis. 2014, 210, 1296–1305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guttman, J.A.; Li, Y.; Wickham, M.E.; Deng, W.; Vogl, A.W.; Finlay, B.B. Attaching and effacing pathogen-induced tight junction disruption in vivo. Cell. Microbiol. 2006, 8, 634–645. [Google Scholar] [CrossRef]
- Scheppach, W.; Bartram, H.P.; Richter, F.; Müller, J.G.; Greinwald, K.; Tauschel, H.D.; Gierend, M.; Weber, A.; Hegemann, D.; Kubetzko, W.; et al. Treatment of distal ulcerative colitis with short-chain fatty acid enemas. A placebo-controlled trial. Dig. Dis. Sci. 1996, 41, 2254–2259. [Google Scholar] [CrossRef]
- Butzner, J.D.; Parmar, R.; Bell, C.J.; Dalal, V. Butyrate enema therapy stimulates mucosal repair in experimental colitis in the rat. Gut 1996, 38, 568–573. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Guryn, K.; Hubert, N.; Frazier, K.; Urlass, S.; Musch, M.W.; Ojeda, P.; Pierre, J.F.; Miyoshi, J.; Sontag, T.J.; Cham, C.M.; et al. Small Intestine Microbiota Regulate Host Digestive and Absorptive Adaptive Responses to Dietary Lipids. Cell Host Microbe 2018, 23, 458–469.e5. [Google Scholar] [CrossRef]
- Maciejewska, D.; Skonieczna-Zydecka, K.; Lukomska, A.; Gutowska, I.; Dec, K.; Kupnicka, P.; Palma, J.; Pilutin, A.; Marlicz, W.; Stachowska, E. The short chain fatty acids and lipopolysaccharides status in Sprague-Dawley rats fed with high-fat and high-cholesterol diet. J. Physiol. Pharmacol. 2018, 69. [Google Scholar]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, M.A.; Jackson, J.; Stanton, M.; Rojas-Triana, A.; Bober, L.; Laverty, M.; Yang, X.; Zhu, F.; Liu, J.; Wang, S.; et al. Short-chain fatty acids act as antiinflammatory mediators by regulating prostaglandin E2 and cytokines. World J. Gastroenterol. 2009, 15, 5549–5557. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Itav, S.; Rothschild, D.; Meijer, M.T.; Levy, M.; Moresi, C.; Dohnalová, L.; Braverman, S.; Rozin, S.; Malitsky, S.; et al. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature 2016, 540, 544–551. [Google Scholar] [CrossRef]
- Fraga, C.G.; Oteiza, P.I. Dietary flavonoids: Role of (−)-epicatechin and related procyanidins in cell signaling. Free Radic. Biol. Med. 2011, 51, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Rivera, L.; Morón, R.; Sánchez, M.; Zarzuelo, A.; Galisteo, M. Quercetin ameliorates metabolic syndrome and improves the inflammatory status in obese Zucker rats. Obesity 2008, 16, 2081–2087. [Google Scholar] [CrossRef] [PubMed]
- Son, M.; Ko, J.I.; Kim, W.B.; Kang, H.K.; Kim, B.K. Taurine Can Ameliorate Inflammatory Bowel Disease in Rats. In Advances in Experimental Medicine and Biology; Springer: Boston, MA, USA, 1998; Volume 442, pp. 291–298. [Google Scholar]
- Chery, J.; Dvoskin, D.; Morato, F.P.; Fahoum, B. Lactobacillus fermentum, a pathogen in documented cholecystitis. Int. J. Surg. Case Rep. 2013, 4, 662–664. [Google Scholar] [CrossRef] [Green Version]
- Zagato, E.; Mileti, E.; Massimiliano, L.; Fasano, F.; Budelli, A.; Penna, G.; Rescigno, M. Lactobacillus paracasei CBA l74 metabolic products and fermented milk for infant formula have anti-inflammatory activity on dendritic cells in Vitro and protective effects against colitis and an enteric pathogen in Vivo. PLoS ONE 2014, 9, e87615. [Google Scholar] [CrossRef]
- Jones, S.E.; Versalovic, J. Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC Microbiol. 2009, 9, 35. [Google Scholar] [CrossRef] [Green Version]
- Bermudez-Brito, M.; Muñoz-Quezada, S.; Gomez-Llorente, C.; Matencio, E.; Bernal, M.J.; Romero, F.; Gil, A. Human Intestinal Dendritic Cells Decrease Cytokine Release against Salmonella Infection in the Presence of Lactobacillus paracasei upon TLR Activation. PLoS ONE 2012, 7, e43197. [Google Scholar] [CrossRef] [Green Version]
- Ma, G.; Pan, B.; Chen, Y.; Guo, C.; Zhao, M.; Zheng, L.; Chen, B. Trimethylamine N-oxide in atherogenesis: Impairing endothelial self-repair capacity and enhancing monocyte adhesion. Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef] [Green Version]
- Preciado, G.M.; Michel, M.M.; Villarreal-Morales, S.L.; Flores-Gallegos, A.C.; Aguirre-Joya, J.; Morlett-Chávez, J.; Aguilar, C.N.; Rodríguez-Herrera, R. Bacteriocins and Its Use for Multidrug-Resistant Bacteria Control. In Antibiotic Resistance; Elsevier: Amsterdam, The Netherlands, 2016; pp. 329–349. ISBN 9780128036686. [Google Scholar]
- Lages, M.C.A.; Beilharz, K.; Morales Angeles, D.; Veening, J.W.; Scheffers, D.J. The localization of key Bacillus subtilis penicillin binding proteins during cell growth is determined by substrate availability. Environ. Microbiol. 2013, 15, 3272–3281. [Google Scholar] [CrossRef] [Green Version]
- Vilchèze, C.; Hartman, T.; Weinrick, B.; Jacobs, W.R. Mycobacterium tuberculosis is extraordinarily sensitive to killing by a vitamin C-induced Fenton reaction. Nat. Commun. 2013, 4, 1881. [Google Scholar] [CrossRef] [Green Version]
- Tukenmez, U.; Aktas, B.; Aslim, B.; Yavuz, S. The relationship between the structural characteristics of lactobacilli-EPS and its ability to induce apoptosis in colon cancer cells in vitro. Sci. Rep. 2019, 9, 8268. [Google Scholar] [CrossRef] [Green Version]
- Paulos, C.M.; Wrzesinski, C.; Kaiser, A.; Hinrichs, C.S.; Chieppa, M.; Cassard, L.; Palmer, D.C.; Boni, A.; Muranski, P.; Yu, Z.; et al. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J. Clin. Investig. 2007, 117, 2197–2204. [Google Scholar] [CrossRef] [PubMed]
- Abotaleb, M.; Samuel, S.; Varghese, E.; Varghese, S.; Kubatka, P.; Liskova, A.; Büsselberg, D. Flavonoids in Cancer and Apoptosis. Cancers (Basel) 2018, 11, 28. [Google Scholar] [CrossRef] [Green Version]
- Tin, A.S.; Park, A.H.; Sundar, S.N.; Firestone, G.L. Essential role of the cancer stem/progenitor cell marker nucleostemin for indole-3-carbinol anti-proliferative responsiveness in human breast cancer cells. BMC Biol. 2014, 12, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chinni, S.R.; Li, Y.; Upadhyay, S.; Koppolu, P.K.; Sarkar, F.H. Indole-3-carbinol (I3C) induced cell growth inhibition, G1 cell cycle arrest and apoptosis in prostate cancer cells. Oncogene 2001, 20, 2927–2936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, T.; Perlot, T.; Rehman, A.; Trichereau, J.; Ishiguro, H.; Paolino, M.; Sigl, V.; Hanada, T.; Hanada, R.; Lipinski, S.; et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012, 487, 477–481. [Google Scholar] [CrossRef]
- Taleb, S. Tryptophan Dietary Impacts Gut Barrier and Metabolic Diseases. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Clarke, G.; Grenham, S.; Scully, P.; Fitzgerald, P.; Moloney, R.D.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol. Psychiatry 2013, 18, 666–673. [Google Scholar] [CrossRef] [Green Version]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [Green Version]
- Smith, E.A.; Macfarlane, G.T. Enumeration of human colonie bacteria producing phenolic and indolic compounds: Effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J. Appl. Bacteriol. 1996, 81, 288–302. [Google Scholar] [CrossRef]
- Lee, J.-H.; Lee, J. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 2010, 34, 426–444. [Google Scholar] [CrossRef] [PubMed]
- Elsden, S.R.; Hilton, M.G.; Waller, J.M. The end products of the metabolism of aromatic amino acids by clostridia. Arch. Microbiol. 1976, 107, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Devlin, A.S.; Marcobal, A.; Dodd, D.; Nayfach, S.; Plummer, N.; Meyer, T.; Pollard, K.S.; Sonnenburg, J.L.; Fischbach, M.A. Modulation of a Circulating Uremic Solute via Rational Genetic Manipulation of the Gut Microbiota. Cell Host Microbe 2016, 20, 709–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, B.B.; Van Benschoten, A.H.; Cimermancic, P.; Donia, M.S.; Zimmermann, M.; Taketani, M.; Ishihara, A.; Kashyap, P.C.; Fraser, J.S.; Fischbach, M.A. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe 2014, 16, 495–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodd, D.; Spitzer, M.H.; Van Treuren, W.; Merrill, B.D.; Hryckowian, A.J.; Higginbottom, S.K.; Le, A.; Cowan, T.M.; Nolan, G.P.; Fischbach, M.A.; et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017, 551, 648–652. [Google Scholar] [CrossRef]
- Wilck, N.; Matus, M.G.; Kearney, S.M.; Olesen, S.W.; Forslund, K.; Bartolomaeus, H.; Haase, S.; Mahler, A.; Balogh, A.; Marko, L.; et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 2017, 551, 585–589. [Google Scholar] [CrossRef]
- Russell, W.R.; Duncan, S.H.; Scobbie, L.; Duncan, G.; Cantlay, L.; Calder, A.G.; Anderson, S.E.; Flint, H.J. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol. Nutr. Food Res. 2013, 57, 523–535. [Google Scholar] [CrossRef]
- Aragozzini, F.; Ferrari, A.; Pacini, N.; Gualandris, R. Indole-3-lactic acid as a tryptophan metabolite produced by Bifidobacterium spp. Appl. Environ. Microbiol. 1979, 38, 544–546. [Google Scholar] [CrossRef] [Green Version]
- Sonowal, R.; Swimm, A.; Sahoo, A.; Luo, L.; Matsunaga, Y.; Wu, Z.; Bhingarde, J.A.; Ejzak, E.A.; Ranawade, A.; Qadota, H.; et al. Indoles from commensal bacteria extend healthspan. Proc. Natl. Acad. Sci. USA 2017, 114, E7506–E7515. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-H.; Wood, T.K.; Lee, J. Roles of Indole as an Interspecies and Interkingdom Signaling Molecule. Trends Microbiol. 2015, 23, 707–718. [Google Scholar] [CrossRef]
- Lee, J.; Jayaraman, A.; Wood, T.K. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 2007, 7, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Attila, C.; Cirillo, S.L.G.; Cirillo, J.D.; Wood, T.K. Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb. Biotechnol. 2009, 2, 75–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, W.; Zere, T.R.; Weber, M.M.; Wood, T.K.; Whiteley, M.; Hidalgo-Romano, B.; Valenzuela, E.; McLean, R.J.C. Indole production promotes escherichia coli mixed-culture growth with Pseudomonas aeruginosa by inhibiting quorum signaling. Appl. Environ. Microbiol. 2012, 78, 411–419. [Google Scholar] [CrossRef] [Green Version]
- Nikaido, E.; Giraud, E.; Baucheron, S.; Yamasaki, S.; Wiedemann, A.; Okamoto, K.; Takagi, T.; Yamaguchi, A.; Cloeckaert, A.; Nishino, K. Effects of indole on drug resistance and virulence of Salmonella enterica serovar Typhimurium revealed by genome-wide analyses. Gut Pathog. 2012, 4, 5. [Google Scholar] [CrossRef] [Green Version]
- Vega, N.M.; Allison, K.R.; Khalil, A.S.; Collins, J.J. Signaling-mediated bacterial persister formation. Nat. Chem. Biol. 2012, 8, 431–433. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Hayashi-Nishino, M.; Yamaguchi, A.; Nishino, K. Indole enhances acid resistance in Escherichia coli. Microb. Pathog. 2010, 49, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Vega, N.M.; Allison, K.R.; Samuels, A.N.; Klempner, M.S.; Collins, J.J. Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc. Natl. Acad. Sci. USA 2013, 110, 14420–14425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina-Santiago, C.; Daddaoua, A.; Fillet, S.; Duque, E.; Ramos, J.L. Interspecies signalling: Pseudomonas putida efflux pump TtgGHI is activated by indole to increase antibiotic resistance. Environ. Microbiol. 2014, 16, 1267–1281. [Google Scholar] [CrossRef]
- Chimerel, C.; Field, C.M.; Piñero-Fernandez, S.; Keyser, U.F.; Summers, D.K. Indole prevents Escherichia coli cell division by modulating membrane potential. Biochim. Biophys. Acta-Biomembr. 2012, 1818, 1590–1594. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Hong, H.; Heo, A.; Park, W. Indole toxicity involves the inhibition of adenosine triphosphate production and protein folding in Pseudomonas putida. FEMS Microbiol. Lett. 2013, 343, 89–99. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Cho, H.S.; Kim, Y.; Kim, J.A.; Banskota, S.; Cho, M.H.; Lee, J. Indole and 7-benzyloxyindole attenuate the virulence of Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2013, 97, 4543–4552. [Google Scholar] [CrossRef] [PubMed]
- Plovier, H.; Cani, P.D. Enteroendocrine Cells: Metabolic Relays between Microbes and Their Host. In Endocrine Development; Karger Publishers: Basel, Switzerland, 2017; Volume 32, pp. 139–164. [Google Scholar]
- Levy, M.; Blacher, E.; Elinav, E. Microbiome, metabolites and host immunity. Curr. Opin. Microbiol. 2017, 35, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, T.D.; Murray, I.A.; Perdew, G.H. Special section on drug metabolism and the microbiome—Minireview indole and tryptophan metabolism: Endogenous and dietary routes to ah receptor activation. Drug Metab. Dispos. 2015, 43, 1522–1535. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Chen, L.; Yang, T.; Feng, Y.-L.; Vaziri, N.D.; Liu, B.-L.; Liu, Q.-Q.; Guo, Y.; Zhao, Y.-Y. Aryl hydrocarbon receptor activation mediates kidney disease and renal cell carcinoma. J. Transl. Med. 2019, 17, 302. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, B.; Di Meglio, P.; Gialitakis, M.; Duarte, J.H. The Aryl Hydrocarbon Receptor: Multitasking in the Immune System. Annu. Rev. Immunol. 2014, 32, 403–432. [Google Scholar] [CrossRef]
- Gutiérrez-Vázquez, C.; Quintana, F.J. Regulation of the Immune Response by the Aryl Hydrocarbon Receptor. Immunity 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stejskalova, L.; Dvorak, Z.; Pavek, P. Endogenous and Exogenous Ligands of Aryl Hydrocarbon Receptor: Current State of Art. Curr. Drug Metab. 2011, 12, 198–212. [Google Scholar] [CrossRef] [Green Version]
- Su, H.-H.; Lin, H.-T.; Suen, J.-L.; Sheu, C.C.; Yokoyama, K.K.; Huang, S.-K.; Cheng, C.M. Aryl hydrocarbon receptor-ligand axis mediates pulmonary fibroblast migration and differentiation through increased arachidonic acid metabolism. Toxicology 2016, 370, 116–126. [Google Scholar] [CrossRef]
- Chiaro, C.R.; Patel, R.D.; Perdew, G.H. 12(R)-hydroxy-5(Z),8(Z),10(E),14(Z)-eicosatetraenoic acid [12(R)-HETE], an arachidonic acid derivative, is an activator of the aryl hydrocarbon receptor. Mol. Pharmacol. 2008, 74, 1649–1656. [Google Scholar] [CrossRef] [Green Version]
- Quintana, F.J.; Sherr, D.H. Aryl hydrocarbon receptor control of adaptive immunity. Pharmacol. Rev. 2013, 65, 1148–1161. [Google Scholar] [CrossRef] [Green Version]
- Kerkvliet, N.I.; Shepherd, D.M.; Baecher-Steppan, L. T lymphocytes are direct, aryl hydrocarbon receptor (AhR)-dependent targets of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD): AhR expression in both CD4+ and CD8+ T cells is necessary for full suppression of a cytotoxic T lymphocyte response by TCDD. Toxicol. Appl. Pharmacol. 2002, 185, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.P.; Singh, U.P.; Rouse, M.; Zhang, J.; Chatterjee, S.; Nagarkatti, P.S.; Nagarkatti, M. Dietary Indoles Suppress Delayed-Type Hypersensitivity by Inducing a Switch from Proinflammatory Th17 Cells to Anti-Inflammatory Regulatory T Cells through Regulation of MicroRNA. J. Immunol. 2016, 196, 1108–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mezrich, J.D.; Fechner, J.H.; Zhang, X.; Johnson, B.P.; Burlingham, W.J.; Bradfield, C.A. An Interaction between Kynurenine and the Aryl Hydrocarbon Receptor Can Generate Regulatory T Cells. J. Immunol. 2010, 185, 3190–3198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veldhoen, M.; Hirota, K.; Westendorf, A.M.; Buer, J.; Dumoutier, L.; Renauld, J.C.; Stockinger, B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 2008, 453, 106–109. [Google Scholar] [CrossRef]
- Kimura, A.; Naka, T.; Nohara, K.; Fujii-Kuriyama, Y.; Kishimoto, T. Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc. Natl. Acad. Sci. USA 2008, 105, 9721–9726. [Google Scholar] [CrossRef] [Green Version]
- Cui, G.; Qin, X.; Wu, L.; Zhang, Y.; Sheng, X.; Yu, Q.; Sheng, H.; Xi, B.; Zhang, J.Z.; Zang, Y.Q. Liver X receptor (LXR) mediates negative regulation of mouse and human Th17 differentiation. J. Clin. Investig. 2011, 121, 658–670. [Google Scholar] [CrossRef] [Green Version]
- Quintana, F.J.; Basso, A.S.; Iglesias, A.H.; Korn, T.; Farez, M.F.; Bettelli, E.; Caccamo, M.; Oukka, M.; Weiner, H.L. Control of Treg and Th17 cell differentiation by the aryl hydrocarbon receptor. Nature 2008, 453, 65–71. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, J.; Takeuchi, M.; Usui, Y.; Hattori, T.; Okunuki, Y.; Yamakawa, N.; Kezuka, T.; Kuroda, M.; Goto, H. Suppression of experimental autoimmune uveoretinitis by inducing differentiation of regulatory T cells via activation of aryl hydrocarbon receptor. Investig. Ophthalmol. Vis. Sci. 2010, 51, 2109–2117. [Google Scholar] [CrossRef] [Green Version]
- Benson, J.M.; Shepherd, D.M. Aryl hydrocarbon receptor activation by TCDD reduces inflammation associated with Crohn’s disease. Toxicol. Sci. 2011, 120, 68–78. [Google Scholar] [CrossRef]
- Singh, N.P.; Singh, U.P.; Singh, B.; Price, R.L.; Nagarkatti, M.; Nagarkatti, P.S. Activation of Aryl hydrocarbon receptor (AhR) leads to reciprocal epigenetic regulation of Foxp3 and IL-17 expression and amelioration of experimental colitis. PLoS ONE 2011, 6, e23522. [Google Scholar] [CrossRef] [Green Version]
- Kerkvliet, N.I.; Steppan, L.B.; Vorachek, W.; Oda, S.; Farrer, D.; Wong, C.P.; Pham, D.; Mourich, D.V. Activation of aryl hydrocarbon receptor by TCDD prevents diabetes in NOD mice and increases Foxp3 + T cells in pancreatic lymph nodes. Immunotherapy 2009, 1, 539–547. [Google Scholar] [PubMed]
- Lanis, J.M.; Alexeev, E.E.; Curtis, V.F.; Kitzenberg, D.A.; Kao, D.J.; Battista, K.D.; Gerich, M.E.; Glover, L.E.; Kominsky, D.J.; Colgan, S.P. Tryptophan metabolite activation of the aryl hydrocarbon receptor regulates IL-10 receptor expression on intestinal epithelia. Mucosal Immunol. 2017, 10, 1133–1144. [Google Scholar] [CrossRef] [PubMed]
- Lamas, B.; Richard, M.L.; Leducq, V.; Pham, H.P.; Michel, M.L.; Da Costa, G.; Bridonneau, C.; Jegou, S.; Hoffmann, T.W.; Natividad, J.M.; et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016, 22, 598–605. [Google Scholar] [CrossRef]
- Monteleone, I.; Rizzo, A.; Sarra, M.; Sica, G.; Sileri, P.; Biancone, L.; MacDonald, T.T.; Pallone, F.; Monteleone, G. Aryl Hydrocarbon Receptor-Induced Signals Up-regulate IL-22 Production and Inhibit Inflammation in the Gastrointestinal Tract. Gastroenterology 2011, 141, 237–248.e1. [Google Scholar] [CrossRef]
- Lin, L.; Tan, B.; Pantapalangkoor, P.; Ho, T.; Baquir, B.; Tomaras, A.; Montgomery, J.I.; Reilly, U.; Barbacci, E.G.; Hujer, K.; et al. Inhibition of LpxC Protects Mice from Resistant Acinetobacter baumannii by Modulating Inflammation and Enhancing Phagocytosis. MBio 2012, 3. [Google Scholar] [CrossRef] [Green Version]
- Hauser, A.R.; Mecsas, J.; Moir, D.T. Beyond antibiotics: New therapeutic approaches for bacterial infections. Clin. Infect. Dis. 2016, 63, 89–95. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Q.; Chen, J.; Yang, C.; Yin, Y.; Yao, K. Quorum Sensing: A Prospective Therapeutic Target for Bacterial Diseases. Biomed Res. Int. 2019, 2019, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metidji, A.; Omenetti, S.; Crotta, S.; Li, Y.; Nye, E.; Ross, E.; Li, V.; Maradana, M.R.; Schiering, C.; Stockinger, B. The Environmental Sensor AHR Protects from Inflammatory Damage by Maintaining Intestinal Stem Cell Homeostasis and Barrier Integrity. Immunity 2018, 49, 353–362.e5. [Google Scholar] [CrossRef] [Green Version]
- Natividad, J.M.; Agus, A.; Planchais, J.; Lamas, B.; Jarry, A.C.; Martin, R.; Michel, M.L.; Chong-Nguyen, C.; Roussel, R.; Straube, M.; et al. Impaired Aryl Hydrocarbon Receptor Ligand Production by the Gut Microbiota Is a Key Factor in Metabolic Syndrome. Cell Metab. 2018, 28, 737–749.e4. [Google Scholar] [CrossRef] [Green Version]
- Lanktree, M.B.; Hegele, R.A. Metabolic Syndrome. In Genomic and Precision Medicine; Elsevier: Amsterdam, The Netherlands, 2017; pp. 283–299. ISBN 9780128006542. [Google Scholar]
- Romani, L.; Puccetti, P.; Zelante, T.; Ricci, M.; Giovagnoli, S. Indole-3-aldehyde for Treating Immune Dysreactive Diseases. European Patent EP3035967B1, 18 December 2019. [Google Scholar]
- Puccetti, M.; Giovagnoli, S.; Zelante, T.; Romani, L.; Ricci, M. Development of Novel Indole-3-Aldehyde–Loaded Gastro-Resistant Spray-Dried Microparticles for Postbiotic Small Intestine Local Delivery. J. Pharm. Sci. 2018, 107, 2341–2353. [Google Scholar] [CrossRef]
- Guo, X.; Qiu, J.; Tu, T.; Yang, X.; Deng, L.; Anders, R.A.; Zhou, L.; Fu, Y.X. Induction of innate lymphoid cell-derived interleukin-22 by the transcription factor STAT3 mediates protection against intestinal infection. Immunity 2014, 40, 25–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swimm, A.; Giver, C.R.; DeFilipp, Z.; Rangaraju, S.; Sharma, A.; Antonova, A.U.; Sonowal, R.; Capaldo, C.; Powell, D.; Qayed, M.; et al. Indoles derived from intestinal microbiota act via type I interferon signaling to limit graft-versus-host disease. Blood 2018, 132, 2506–2519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Luo, Y.; Zhu, Z.; Zhou, Y.; Sun, L.; Gao, J.; Sun, J.; Wang, G.; Yao, X.; Li, W. A tryptophan metabolite of the skin microbiota attenuates inflammation in patients with atopic dermatitis through the aryl hydrocarbon receptor. J. Allergy Clin. Immunol. 2019, 143, 2108–2119.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bommarius, B.; Anyanful, A.; Izrayelit, Y.; Bhatt, S.; Cartwright, E.; Wang, W.; Swimm, A.I.; Benian, G.M.; Schroeder, F.C.; Kalman, D. A Family of Indoles Regulate Virulence and Shiga Toxin Production in Pathogenic E. coli. PLoS ONE 2013, 8, e54456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romani, L.; Zelante, T.; De Luca, A.; Iannitti, R.G.; Moretti, S.; Bartoli, A.; Aversa, F.; Puccetti, P. Microbiota control of a tryptophan-AhR pathway in disease tolerance to fungi. Eur. J. Immunol. 2014, 44, 3192–3200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cicenia, A.; Scirocco, A.; Carabotti, M.; Pallotta, L.; Marignani, M.; Severi, C. Postbiotic Activities of Lactobacilli-derived Factors. J. Clin. Gastroenterol. 2014, 48, S18–S22. [Google Scholar] [CrossRef]
- Clayton, T.A.; Lindon, J.C.; Cloarec, O.; Antti, H.; Charuel, C.; Hanton, G.; Provost, J.P.; Le Net, J.L.; Baker, D.; Walley, R.J.; et al. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature 2006, 440, 1073–1077. [Google Scholar] [CrossRef]
- Holmes, E.; Kinross, J.; Gibson, G.R.; Burcelin, R.; Jia, W.; Pettersson, S.; Nicholson, J.K. Therapeutic Modulation of Microbiota-Host Metabolic Interactions. Sci. Transl. Med. 2012, 4, 137rv6. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [Green Version]


| Pathogen | HDTs | Mechanisms of Action | Ref | |
|---|---|---|---|---|
| Mycobacterium tuberculosis | Repurposed drug | Imatinib, verapamil, metformin, ibuprofen | Modulation of inflammation and activation of intracellular antimicrobial defenses | [29,37,38,39,40,41,42] |
| Cytokine therapy | IL-2, GM-CSF, INF-γ, IL-12 | Induction of pro-inflammatory cell signaling | [33] | |
| Monoclonal antibody | Anti-TNFα, anti-IL-6, anti-VEGF | Reduction of tissue-destructive inflammation by cytokine neutralization | [34] | |
| Monoclonal antibody | Anti-PD-1, anti-LAG3, anti-CTLA-4 | Activation and mobilization of antigen-specific T cells by immune checkpoint inhibition | [35,44] | |
| Vitamin | Vitamin D3 | Activation and augmentation of intracellular antimicrobial defenses (via IFN-γ and IL-15 signaling) | [41,42] | |
| Cellular therapy | Autologous mesenchymal stromal cells, T cells | Neutralization of tissue-destructive inflammation, enhancement of organ repair, and potentiation of antigen-specific immune responses | [36] | |
| Streptococcus pneumoniae | Repurposed drugs | Prednisone | Reduction of tissue-destructive inflammation by activating the glucocorticoid pathway | [44] |
| Ibuprofen, statins, indometacin, aspirin | Reduction of tissue-destructive inflammation by inhibiting prostaglandin release via cyclooxygenase inhibition, regulation of MHC molecules | [45,46] | ||
| Glibenclamide | An oral hypoglycemic agent that modulates voltage-gated calcium channels, leading to immunomodulatory effects | [50] | ||
| Bordetella pertussis | Repurposed drug | Fingolimod | Activates the sphingosine-1-phosphate pathway to improve antigen-specific lymphocyte responses, as well as reduced hyper-inflammation | [51] |
| Monoclonal antibody | Antipertussis toxin antibodies | Reduces toxin load via infusion of intravenous immunoglobulins | [47,48,49] | |
| Microbiota Based Therapies | Symptomatology | Trials | Reference | ||
|---|---|---|---|---|---|
| Bacteriophages | Muddy, BPs33ΔHTH-HRM10, and ZoeJΔ45 | Cystic fibrosis | Mycobacterial infection | Clinical case study | [90] |
| OMKO1 | Prosthetic vascular graft infections | Pseudomonas aeruginosa | Preclinical | [91] | |
| PP1131 cocktail | Burn wounds infected | Pseudomonas aeruginosa | Phase I-II | [92] | |
| Myoviridae | Enteric infection | Escherichia coli | Preclinical | [93] | |
| phage cocktail | Burn wound infections | Klebsiella pneumoniae | Preclinical | [94] | |
| FMT | Clostridium difficile | Preclinical | [95] | ||
| Ulcerative colitis | Phase II | [96] | |||
| Phase II | [97] | ||||
| Probiotics | Lactobacillus acidophilus Saccharomyces boulardii Lactococcus lactis, Lactobacillus rhamnosus, Lactobacillus plantarum, Lactobacillus casei, Lactobacillus reuteri, Lactobacillus plantarum, Bifidobacterium infantis | Traveler’s diarrhea, Antibiotic-associated diarrhea, Ulcerative colitis Crohn’s disease, Atopic dermatitis, Clostridium difficile Irritable bowel syndrome | Phase II | [98,99,100,101] | |
| Metabolite | Activities | Reference |
|---|---|---|
| Butyrate, acetate, propionate | Preserve mucosal immunity Enhance the regulatory function of Tregs in the large intestine Butyrate suppresses proliferation by acting as a HDAC inhibitor Enhance the protection against infections Activate GPR43 and GPR109a on intestinal epithelial cells, result in the activation of the NLRP3 inflammasome leading to production of IL-18. NF-kB inactivation and suppression of pro-inflammatory cytokines and nitric oxide in neutrophils and mononuclear cells through HDAC inhibition | [154,155,156,157,158,159,160,161,162,163,164,165,166,167,168] |
| Niacin | Induces anti-inflammatory properties in dendritic cells and macrophages in a GPR109a-dependent manner and suppresses colonic inflammation | [160] |
| Retinoic acid | Dendritic cell induction of gut-lymphocytes Supports the development of Tregs through TGF-β and suppresses the development of TH17 cells during inflammation, RA is required for the induction of a proinflammatory CD4+ helper T cell response | [169,170,171,172] |
| Polysaccharide A (PSA) | Suppresses the production of pro-inflammatory IL-17 and promotes expression of IL-10 by CD4+ T cells | [156,173,174] |
| Bile acids | Regulation of bacterial growth Inhibit the induction of pro-inflammatory genes through NF-kB | [175,176,177] |
| Taurine | Nlrp6 inflammasome activation and contribution to intestinal homeostasis | [178] |
| Indoles | Induce IL-22 secretion by ILCs, further driving the secretion of antimicrobial peptides and protection from infections by pathogens Epithelial barrier enhancement | [56,179,180] |
| Source | Bacteria | Microbial Metabolite | Family | Receptor | Reference |
|---|---|---|---|---|---|
| Fermentation of fibers/carbohydrate metabolism | Bacteroidetes, Firmicutes | Propionate, Acetate and Butyrate | SCFA | GPR43 GPR41 | [182] |
| Dietary Tryptophan (microbial origin) | Firmicutes, Lactobacillus, Clostridium, Bacteroides | 5-hydroxy-tryptophan Tryptamine Indoleacetic acid 3-methylindole (Skatole) 3-indole carboxaldehyde Indole-3-sulfate Indole propionic acid 3-indolelactic acid | Indole and Indole derivatives | AhR, PXR | [179,183,184] |
| Bile acids | Bacteroides, Bifidobacterium, Clostridium, Lactobacillus, Eubacterium | Deoxycholic and lithocholic acid | Secondary bile acids | GPBAR1, FXR | [185,186] |
| Nitrogenous compound | Escherichia Coli, Enterococcus faecalis, Bifidium bacterium and Bacteroides | Putrescine, Spermidine, spermine | Polyamines | PPARγ, TRPV1 | [187,188,189] |
| Dietary choline | Actinobacteria Bacteroidetes Firmicutes Proteobacteria | Trimethylamine-N-oxide | Amine oxides | FXR | [190,191] |
| Ginseng | Bacteroides | Compound K | Ginsenosides | GPBAR1 | [124,192] |
| Compound | Origin |
|---|---|
| Indole Metabolites [268,272] | |
| Indole | Dietary metabolite and microbiota metabolism |
| Indolo[3,2-b] carbazole | Dietary metabolite |
| 2-(Indol-3-ylmethyl)-3,3′-diindolylmethane | Dietary metabolite |
| 3,3′-Diindolylmethane | Dietary metabolite |
| Tryptophan Metabolites [268] | |
| Kynurenine | Host Metabolism |
| Kynyrenic acid | Host Metabolism |
| Zanthurenic acid | Host Metabolism |
| Cinnabarinic acid | Host Metabolism |
| 5-hydroxy-tryptophan | Host Metabolism |
| Tryptamine | Microbiota metabolism |
| Indol-3-acetic Acid | Microbiota metabolism |
| 3-methylindole (Skatole) | Microbiota metabolism |
| 3-indole-carboxaldehyde | Microbiota metabolism |
| Indoxyl-3-sulfate | Microbiota and Host Metabolite |
| Arachidonic Acid Metabolites [273,274] | |
| Lipoxin 4A | Host Metabolism |
| Prostaglandin -PGG2 | Host Metabolism |
| Hydroxyeicosatrienoic acid | Host Metabolism |
| Heme-derived [275] | |
| Indigorubin Bilirubin | Host Metabolism |
| Indigo Biliverdin | Host Metabolism |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puccetti, M.; Xiroudaki, S.; Ricci, M.; Giovagnoli, S. Postbiotic-Enabled Targeting of the Host-Microbiota-Pathogen Interface: Hints of Antibiotic Decline? Pharmaceutics 2020, 12, 624. https://doi.org/10.3390/pharmaceutics12070624
Puccetti M, Xiroudaki S, Ricci M, Giovagnoli S. Postbiotic-Enabled Targeting of the Host-Microbiota-Pathogen Interface: Hints of Antibiotic Decline? Pharmaceutics. 2020; 12(7):624. https://doi.org/10.3390/pharmaceutics12070624
Chicago/Turabian StylePuccetti, Matteo, Styliani Xiroudaki, Maurizio Ricci, and Stefano Giovagnoli. 2020. "Postbiotic-Enabled Targeting of the Host-Microbiota-Pathogen Interface: Hints of Antibiotic Decline?" Pharmaceutics 12, no. 7: 624. https://doi.org/10.3390/pharmaceutics12070624
APA StylePuccetti, M., Xiroudaki, S., Ricci, M., & Giovagnoli, S. (2020). Postbiotic-Enabled Targeting of the Host-Microbiota-Pathogen Interface: Hints of Antibiotic Decline? Pharmaceutics, 12(7), 624. https://doi.org/10.3390/pharmaceutics12070624