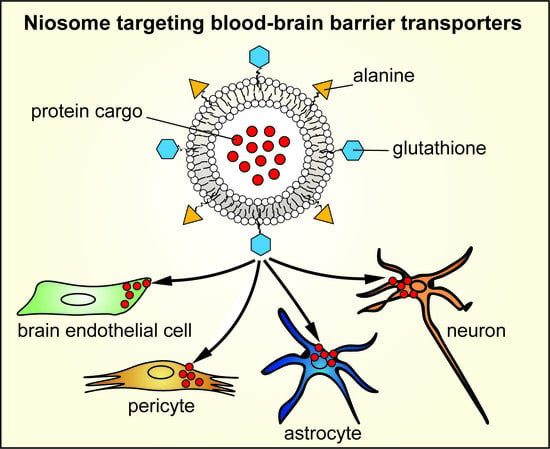
Combination of Alanine and Glutathione as Targeting Ligands of Nanoparticles Enhances Cargo Delivery into the Cells of the Neurovascular Unit
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Materials and Reagents
2.3. Cell Cultures
2.3.1. Primary Cell Cultures
2.3.2. Cell Lines
2.4. Immunohistochemistry
2.5. RNA Isolation and Quality Control
2.6. Quantitative Real-Time Polymerase Chain Reaction and Data Analysis
2.7. Synthesis of the Targeting Ligands of Niosomes
2.8. Preparation of Niosomes
2.9. Characterization of Niosomes
2.9.1. Atomic Force Microscopy
2.9.2. Size, Size Stability and Zeta Potential Measurements
2.10. Cell Viability Assay
2.11. Cellular Uptake Studies
2.12. Permeability Studies
2.13. Statistical Analysis
3. Results
3.1. Expression of Genes Encoding Alanine Transporters in the Cell Types of the Neurovascular Unit
3.2. Characterization of Niosomes
3.3. Effect of Niosomes on Cell Viability
3.4. Cellular Uptake of Cargo: Pericytes
3.5. Cellular Uptake of Cargo: Astrocytes
3.6. Cellular Uptake of Cargo: hCMEC/D3 Brain Endothelial Cells
3.7. Cellular Uptake of Cargo: Differentiated SH-SY5Y Neuronal Cells
3.8. Permeability of Cargo Across the Blood–Brain Barrier (BBB) Co-Culture Model
4. Discussion
4.1. Alanine-Glutathione Dual-Labeling of NPs as a Strategy for Entry in the Cells of the Neurovascular Unit
4.2. Alanine-Glutathione Targeted Niosomes Enchance the Delivery of a Large Protein Cargo into the Cells of the Neurovascular Unit
4.3. The Uptake of Targeted Niosomes is Energy-Dependent and is Partially Mediated by Endocytosis
4.4. Dual-Labeling of Niosomes with Alanine and Glutathione Enchances Cargo Delivery Across the BBB and Subsequently to Astrocytes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pardridge, W.M. Targeted delivery of protein and gene medicines through the blood-brain barrier. Clin. Pharmacol. Ther. 2015, 97, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Veszelka, S.; Bocsik, A.; Walter, F.R.; Hantosi, D.; Deli, M.A. Blood-brain barrier co-culture models to study nanoparticle penetration: Focus on co-culture systems. Acta Biol. Szeged. 2015, 59 (Suppl. S2), 157–168. [Google Scholar]
- Saraiva, C.; Praca, C.; Ferreira, R.; Santos, T.; Ferreira, L.; Bernardino, L. Nanoparticle-mediated brain drug delivery: Overcoming blood-brain barrier to treat neurodegenerative diseases. J. Control. Release 2016, 235, 34–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masserini, M. Nanoparticles for brain drug delivery. ISRN Biochem. 2013, 2013, 238428. [Google Scholar] [CrossRef] [Green Version]
- Bozzuto, G.; Molinari, A. Liposomes as nanomedical devices. Int. J. Nanomed. 2015, 10, 975–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelkader, H.; Alani, A.W.; Alany, R.G. Recent advances in non-ionic surfactant vesicles (niosomes): Self-assembly, fabrication, characterization, drug delivery applications and limitations. Drug Deliv. 2014, 21, 87–100. [Google Scholar] [CrossRef] [Green Version]
- Kreuter, J. Drug delivery to the central nervous system by polymeric nanoparticles: What do we know? Adv. Drug Deliv. Rev. 2014, 71, 2–14. [Google Scholar] [CrossRef]
- Neves, V.; Aires-da-Silva, F.; Corte-Real, S.; Castanho, M.A.R.B. Antibody Approaches to Treat Brain Diseases. Trends Biotechnol. 2016, 34, 36–48. [Google Scholar] [CrossRef]
- Campos-Bedolla, P.; Walter, F.R.; Veszelka, S.; Deli, M.A. Role of the blood-brain barrier in the nutrition of the central nervous system. Arch. Med. Res. 2014, 45, 610–638. [Google Scholar] [CrossRef]
- Pardridge, W.M. Blood-brain barrier endogenous transporters as therapeutic targets: A new model for small molecule CNS drug discovery. Expert Opin. Ther. Targets 2015, 19, 1059–1072. [Google Scholar] [CrossRef]
- Banks, W.A. From blood-brain barrier to blood-brain interface: New opportunities for CNS drug delivery. Nat. Rev. Drug Discov. 2016, 15, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Enerson, B.E.; Drewes, L.R. The rat blood-brain barrier transcriptome. J. Cereb. Blood Flow Metab. 2006, 26, 959–973. [Google Scholar] [CrossRef] [Green Version]
- Daneman, R.; Zhou, L.; Agalliu, D.; Cahoy, J.D.; Kaushal, A.; Barres, B.A. The mouse blood-brain barrier transcriptome: A new resource for understanding the development and function of brain endothelial cells. PLoS ONE 2010, 5, e13741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shawahna, R.; Uchida, Y.; Declèves, X.; Ohtsuki, S.; Yousif, S.; Dauchy, S.; Jacob, A.; Chassoux, F.; Daumas-Duport, C.; Couraud, P.O.; et al. Transcriptomic and quantitative proteomic analysis of transporters and drug metabolizing enzymes in freshly isolated human brain microvessels. Mol. Pharm. 2011, 8, 1332–1341. [Google Scholar] [CrossRef] [PubMed]
- Veszelka, S.; Tóth, A.; Walter, F.R.; Tóth, A.E.; Gróf, I.; Mészáros, M.; Bocsik, A.; Hellinger, E.; Vastag, M.; Rákhely, G.; et al. Comparison of a Rat Primary Cell-Based Blood-Brain Barrier Model With Epithelial and Brain Endothelial Cell Lines: Gene Expression and Drug Transport. Front. Mol. Neurosci. 2018, 11, 166. [Google Scholar] [CrossRef]
- Rask-Andersen, M.; Masuram, S.; Fredriksson, R.; Schioth, H.B. Solute carriers as drug targets: Current use, clinical trials and prospective. Mol. Asp. Med. 2013, 34, 702–710. [Google Scholar] [CrossRef]
- César-Razquin, A.; Snijder, B.; Frappier-Brinton, T.; Isserlin, R.; Gyimesi, G.; Bai, X.; Reithmeier, R.A.; Hepworth, D.; Hediger, M.A.; Edwards, A.M.; et al. A Call for Systematic Research on Solute Carriers. Cell 2015, 162, 478–487. [Google Scholar] [CrossRef] [Green Version]
- Gaillard, P.J.; Appeldoorn, C.C.; Rip, J.; Dorland, R.; van der Pol, S.M.; Kooij, G.; de Vries, H.E.; Reijerkerk, A. Enhanced brain delivery of liposomal methylprednisolone improved therapeutic efficacy in a model of neuroinflammation. J. Control. Release 2012, 164, 364–369. [Google Scholar] [CrossRef]
- Gaillard, P.J.; Appeldoorn, C.C.; Dorland, R.; van Kregten, J.; Manca, F.; Vugts, D.J.; Windhorst, B.; van Dongen, G.A.; de Vries, H.E.; Maussang, D.; et al. Pharmacokinetics, brain delivery, and efficacy in brain tumor-bearing mice of glutathione pegylated liposomal doxorubicin (2B3–101). PLoS ONE 2014, 9, e82331. [Google Scholar] [CrossRef] [Green Version]
- Lindqvist, A.; Rip, J.; van Kregten, J.; Gaillard, P.J.; Hammarlund-Udenaes, M. In vivo Functional Evaluation of Increased Brain Delivery of the Opioid Peptide DAMGO by Glutathione-PEGylated Liposomes. Pharm. Res. 2016, 33, 177–185. [Google Scholar] [CrossRef]
- Mészáros, M.; Porkoláb, G.; Kiss, L.; Pilbat, A.M.; Kóta, Z.; Kupihár, Z.; Kéri, A.; Galbács, G.; Siklós, L.; Tóth, A.; et al. Niosomes decorated with dual ligands targeting brain endothelial transporters increase cargo penetration across the blood-brain barrier. Eur. J. Pharm. Sci. 2018, 123, 228–240. [Google Scholar] [CrossRef]
- Neuwelt, E.A.; Bauer, B.; Fahlke, C.; Fricker, G.; Iadecola, C.; Janigro, D.; Leybaert, L.; Molnar, Z.; O’Donnell, M.E.; Povlishock, J.T.; et al. Engaging neuroscience to advance translational research in brain barrier biology. Nat. Rev. Neurosci. 2011, 12, 169–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sweeney, M.D.; Zhao, Z.; Montagne, A.; Nelson, A.R.; Zlokovic, B.V. Blood-Brain Barrier: From Physiology to Disease and Back. Physiol. Rev. 2019, 99, 21–78. [Google Scholar] [CrossRef] [PubMed]
- Gromnicova, R.; Davies, H.A.; Sreekanthreddy, P.; Romero, I.A.; Lund, T.; Roitt, I.M.; Phillips, J.B.; Male, D.K. Glucose-coated gold nanoparticles transfer across human brain endothelium and enter astrocytes in vitro. PLoS ONE 2013, 8, e81043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moscariello, P.; Ng, D.Y.W.; Jansen, M.; Weil, T.; Luhmann, H.J.; Hedrich, J. Brain Delivery of Multifunctional Dendrimer Protein Bioconjugates. Adv. Sci. 2018, 5, 1700897. [Google Scholar] [CrossRef]
- Surnar, B.; Basu, U.; Banik, B.; Ahmad, A.; Marples, B.; Kolishetti, N.; Dhar, S. Nanotechnology-mediated crossing of two impermeable membranes to modulate the stars of the neurovascular unit for neuroprotection. Proc. Natl. Acad. Sci. USA 2018, 115, E12333–E12342. [Google Scholar] [CrossRef]
- Nakagawa, S.; Deli, M.A.; Kawaguchi, H.; Shimizudani, T.; Shimono, T.; Kittel, Á.; Tanaka, K.; Niwa, M. A new blood-brain barrier model using primary rat brain endothelial cells, pericytes and astrocytes. Neurochem. Int. 2009, 54, 253–263. [Google Scholar] [CrossRef] [Green Version]
- Barna, L.; Walter, F.R.; Harazin, A.; Bocsik, A.; Kincses, A.; Tubak, V.; Jósvay, K.; Zvara, A.; Campos-Bedolla, P.; Deli, M.A. Simvastatin, edaravone and dexamethasone protect against kainate-induced brain endothelial cell damage. Fluids Barriers CNS 2020, 17, 5. [Google Scholar] [CrossRef] [Green Version]
- Perrière, N.; Demeuse, P.; Garcia, E.; Regina, A.; Debray, M.; Andreux, J.P.; Couvreur, P.; Scherrmann, J.M.; Temsamani, J.; Couraud, P.O.; et al. Puromycin-based purification of rat brain capillary endothelial cell cultures. Effect on the expression of blood-brain barrier-specific properties. J. Neurochem. 2005, 93, 279–289. [Google Scholar] [CrossRef]
- Walter, F.R.; Valkai, S.; Kincses, A.; Petneházi, A.; Czeller, T.; Veszelka, S.; Ormos, P.; Deli, M.A.; Dér, A. A versatile lab-on-a-chip tool for modeling biological barriers. Sens. Actuators B Chem. 2016, 222, 1209–1219. [Google Scholar] [CrossRef] [Green Version]
- Vandenhaute, E.; Dehouck, L.; Boucau, M.C.; Sevin, E.; Uzbekov, R.; Tardivel, M.; Gosselet, F.; Fenart, L.; Cecchelli, R.; Dehouck, M.P. Modelling the neurovascular unit and the blood-brain barrier with the unique function of pericytes. Curr. Neurovasc. Res. 2011, 8, 258–269. [Google Scholar] [CrossRef]
- Weksler, B.B.; Subileau, E.A.; Perrière, N.; Charneau, P.; Holloway, K.; Leveque, M.; Tricoire-Leignel, H.; Nicotra, A.; Bourdoulous, S.; Turowski, P.; et al. Blood-brain barrier-specific properties of a human adult brain endothelial cell line. FASEB J. 2005, 19, 1872–1874. [Google Scholar] [CrossRef]
- Liebner, S.; Corada, M.; Bangsow, T.; Babbage, J.; Taddei, A.; Czupalla, C.J.; Reis, M.; Felici, A.; Wolburg, H.; Fruttiger, M.; et al. Wnt/beta-catenin signaling controls development of the blood-brain barrier. J. Cell Biol. 2008, 183, 409–417. [Google Scholar] [CrossRef] [Green Version]
- Kovalevich, J.; Langford, D. Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol. Biol. 2013, 1078, 9–21. [Google Scholar] [CrossRef] [Green Version]
- Veszelka, S.; Mészáros, M.; Kiss, L.; Kóta, Z.; Páli, T.; Hoyk, Z.; Bozsó, Z.; Fülöp, L.; Tóth, A.; Rákhely, G.; et al. Biotin and Glutathione Targeting of Solid Nanoparticles to Cross Human Brain Endothelial Cells. Curr. Pharm. Des. 2017, 23, 4198–4205. [Google Scholar] [CrossRef] [Green Version]
- Toth, A.E.; Toth, A.; Walter, F.R.; Kiss, L.; Veszelka, S.; Ozsvari, B.; Puskas, L.G.; Heimesaat, M.M.; Dohgu, S.; Kataoka, Y.; et al. Compounds blocking methylglyoxal-induced protein modification and brain endothelial injury. Arch. Med. Res. 2014, 45, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Li, Y.; Deng, J.; Yang, W. Helix-sense-selective polymerization of achiral substituted acetylene in chiral micelles for preparing optically active polymer nanoparticles: Effects of chiral emulsifiers. Polymer 2014, 55, 840–847. [Google Scholar] [CrossRef]
- Hutter, J.L.; Bechhoefer, J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993, 64, 1868–1873. [Google Scholar] [CrossRef] [Green Version]
- Sader, J.E.; Chon, J.W.M.; Mulvaney, P. Calibration of rectangular atomic force microscope cantilevers. Rev. Sci. Instrum. 1999, 70, 3967–3969. [Google Scholar] [CrossRef] [Green Version]
- Aekbote, B.L.; Jacak, J.; Schütz, G.J.; Csányi, E.; Szegletes, Z.; Ormos, P.; Kelemen, L. Aminosilane-based functionalization of two-photon polymerized 3D SU-8 microstructures. Eur. Polym. J. 2012, 48, 1745–1754. [Google Scholar] [CrossRef]
- Harazin, A.; Bocsik, A.; Barna, L.; Kincses, A.; Váradi, J.; Fenyvesi, F.; Tubak, V.; Deli, M.A.; Vecsernyés, M. Protection of cultured brain endothelial cells from cytokine-induced damage by alpha-melanocyte stimulating hormone. PeerJ 2018, 6, e4774. [Google Scholar] [CrossRef] [Green Version]
- Kiss, L.; Walter, F.R.; Bocsik, A.; Veszelka, S.; Ozsvári, B.; Puskás, G.L.; Szabó-Révész, P.; Deli, M.A. Kinetic Analysis of the Toxicity of Pharmaceutical Excipients Cremophor EL and RH40 on Endothelial and Epithelial Cells. J. Pharm. Sci. 2013, 102, 1173–1181. [Google Scholar] [CrossRef]
- Bocsik, A.; Walter, F.R.; Gyebrovszki, A.; Fülöp, L.; Blasig, I.; Dabrowski, S.; Ötvös, F.; Tóth, A.; Rákhely, G.; Veszelka, S.; et al. Reversible Opening of Intercellular Junctions of Intestinal Epithelial and Brain Endothelial Cells With Tight Junction Modulator Peptides. J. Pharm. Sci. 2016, 105, 754–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiorentino, I.; Gualtieri, R.; Barbato, V.; Mollo, V.; Braun, S.; Angrisani, A.; Turano, M.; Furia, M.; Netti, P.A.; Guarnieri, D.; et al. Energy independent uptake and release of polystyrene nanoparticles in primary mammalian cell cultures. Exp. Cell Res. 2015, 330, 240–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanov, A.I. Pharmacological inhibition of endocytic pathways: Is it specific enough to be useful? Methods Mol. Biol. 2008, 440, 15–33. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, K.; Sloan, S.A.; Bennett, M.L.; Scholze, A.R.; O’Keeffe, S.; Phatnani, H.P.; Guarnieri, P.; Caneda, C.; Ruderisch, N.; et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014, 34, 11929–11947. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Vanlandewijck, M.; Raschperger, E.; Andaloussi Mae, M.; Jung, B.; Lebouvier, T.; Ando, K.; Hofmann, J.; Keller, A.; Betsholtz, C. Analysis of the brain mural cell transcriptome. Sci. Rep. 2016, 6, 35108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, L.; Vanlandewijck, M.; Mae, M.A.; Andrae, J.; Ando, K.; Del Gaudio, F.; Nahar, K.; Lebouvier, T.; Lavina, B.; Gouveia, L.; et al. Single-cell RNA sequencing of mouse brain and lung vascular and vessel-associated cell types. Sci. Data 2018, 5, 180160. [Google Scholar] [CrossRef] [Green Version]
- Chasseigneaux, S.; Moraca, Y.; Cochois-Guégan, V.; Boulay, A.C.; Gilbert, A.; Le Crom, S.; Blugeon, C.; Firmo, C.; Cisternino, S.; Laplanche, J.L.; et al. Isolation and differential transcriptome of vascular smooth muscle cells and mid-capillary pericytes from the rat brain. Sci. Rep. 2018, 8, 12272. [Google Scholar] [CrossRef]
- Yamamoto, T.; Nishizaki, I.; Nukada, T.; Kamegaya, E.; Furuya, S.; Hirabayashi, Y.; Ikeda, K.; Hata, H.; Kobayashi, H.; Sora, I.; et al. Functional identification of ASCT1 neutral amino acid transporter as the predominant system for the uptake of L-serine in rat neurons in primary culture. Neurosci. Res. 2004, 49, 101–111. [Google Scholar] [CrossRef]
- Lyck, R.; Ruderisch, N.; Moll, A.G.; Steiner, O.; Cohen, C.D.; Engelhardt, B.; Makrides, V.; Verrey, F. Culture-induced changes in blood-brain barrier transcriptome: Implications for amino-acid transporters in Vivo. J. Cereb. Blood Flow Metab. 2009, 29, 1491–1502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carl, S.M.; Lindley, D.J.; Das, D.; Couraud, P.O.; Weksler, B.B.; Romero, I.; Mowery, S.A.; Knipp, G.T. ABC and SLC transporter expression and Pot substrate characterization across the human CMEC/D3 blood-brain barrier cell line. Mol. Pharm. 2010, 7, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Kannan, R.; Kuhlenkamp, J.F.; Jeandidier, E.; Trinh, H.; Ookhtens, M.; Kaplowitz, N. Evidence for carrier-mediated transport of glutathione across the blood-brain barrier in the rat. J. Clin. Investig. 1990, 85, 2009–2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zlokovic, B.V.; Mackic, J.B.; McComb, J.G.; Weiss, M.H.; Kaplowitz, N.; Kannan, R. Evidence for transcapillary transport of reduced glutathione in vascular perfused guinea-pig brain. Biochem. Biophys. Res. Commun. 1994, 201, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kannan, R.; Chakrabarti, R.; Tang, D.; Kim, K.J.; Kaplowitz, N. GSH transport in human cerebrovascular endothelial cells and human astrocytes: Evidence for luminal localization of Na+-dependent GSH transport in HCEC. Brain Res. 2000, 852, 374–382. [Google Scholar] [CrossRef]
- Balabanov, R.; Washington, R.; Wagnerova, J.; Dore-Duffy, P. CNS microvascular pericytes express macrophage-like function, cell surface integrin alpha M, and macrophage marker ED-2. Microvasc. Res. 1996, 52, 127–142. [Google Scholar] [CrossRef]
- Winkler, E.A.; Bell, R.D.; Zlokovic, B.V. Central nervous system pericytes in health and disease. Nat. Neurosci. 2011, 14, 1398–1405. [Google Scholar] [CrossRef] [Green Version]
- Daneman, R.; Zhou, L.; Kebede, A.A.; Barres, B.A. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature 2010, 468, 562–566. [Google Scholar] [CrossRef] [Green Version]
- Armulik, A.; Genove, G.; Mae, M.; Nisancioglu, M.H.; Wallgard, E.; Niaudet, C.; He, L.; Norlin, J.; Lindblom, P.; Strittmatter, K.; et al. Pericytes regulate the blood-brain barrier. Nature 2010, 468, 557–561. [Google Scholar] [CrossRef] [Green Version]
- Bell, R.D.; Winkler, E.A.; Sagare, A.P.; Singh, I.; LaRue, B.; Deane, R.; Zlokovic, B.V. Pericytes control key neurovascular functions and neuronal phenotype in the adult brain and during brain aging. Neuron 2010, 68, 409–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, C.N.; Reynell, C.; Gesslein, B.; Hamilton, N.B.; Mishra, A.; Sutherland, B.A.; O’Farrell, F.M.; Buchan, A.M.; Lauritzen, M.; Attwell, D. Capillary pericytes regulate cerebral blood flow in health and disease. Nature 2014, 508, 55–60. [Google Scholar] [CrossRef] [Green Version]
- Guan, Y.Y.; Luan, X.; Xu, J.R.; Liu, Y.R.; Lu, Q.; Wang, C.; Liu, H.J.; Gao, Y.G.; Chen, H.Z.; Fang, C. Selective eradication of tumor vascular pericytes by peptide-conjugated nanoparticles for antiangiogenic therapy of melanoma lung metastasis. Biomaterials 2014, 35, 3060–3070. [Google Scholar] [CrossRef] [PubMed]
- Geldenhuys, W.; Mbimba, T.; Bui, T.; Harrison, K.; Sutariya, V. Brain-targeted delivery of paclitaxel using glutathione-coated nanoparticles for brain cancers. J. Drug Target 2011, 19, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Grover, A.; Hirani, A.; Pathak, Y.; Sutariya, V. Brain-targeted delivery of docetaxel by glutathione-coated nanoparticles for brain cancer. AAPS PharmSciTech 2014, 15, 1562–1568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, B.Y.; Liu, R.Y.; So, K.L.; Yu, A.C. Multi-lipofection efficiently transfected genes into astrocytes in primary culture. J. Neurosci. Methods 2000, 102, 133–141. [Google Scholar] [CrossRef]
- McKalip, D.; Grenier, M.; Choy, M. Liposomal mediated transfection of mature rat astrocytes in vitro: A comparison of ten reagents. Neurosci. Lett. 2000, 295, 64–66. [Google Scholar] [CrossRef]
- Tóth, A.E.; Nielsen, S.S.E.; Tomaka, W.; Abbott, N.J.; Nielsen, M.S. The endo-lysosomal system of bEnd.3 and hCMEC/D3 brain endothelial cells. Fluids Barriers CNS 2019, 16, 14. [Google Scholar] [CrossRef] [Green Version]
- Paka, G.D.; Ramassamy, C. Optimization of Curcumin-Loaded PEG-PLGA Nanoparticles by GSH Functionalization: Investigation of the Internalization Pathway in Neuronal Cells. Mol. Pharm. 2017, 14, 93–106. [Google Scholar] [CrossRef]
- Patel, P.J.; Acharya, N.S.; Acharya, S.R. Development and characterization of glutathione-conjugated albumin nanoparticles for improved brain delivery of hydrophilic fluorescent marker. Drug Deliv. 2013, 20, 143–155. [Google Scholar] [CrossRef]
- Maussang, D.; Rip, J.; van Kregten, J.; van den Heuvel, A.; van der Pol, S.; van der Boom, B.; Reijerkerk, A.; Chen, L.; de Boer, M.; Gaillard, P.; et al. Glutathione conjugation dose-dependently increases brain-specific liposomal drug delivery in vitro and in Vivo. Drug Discov. Today Technol. 2016, 20, 59–69. [Google Scholar] [CrossRef]
- Gromnicova, R.; Yilmaz, C.U.; Orhan, N.; Kaya, M.; Davies, H.; Williams, P.; Romero, I.A.; Sharrack, B.; Male, D. Localization and mobility of glucose-coated gold nanoparticles within the brain. Nanomedicine 2016, 11, 617–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dos Santos Rodrigues, B.; Oue, H.; Banerjee, A.; Kanekiyo, T.; Singh, J. Dual functionalized liposome-mediated gene delivery across triple co-culture blood brain barrier model and specific in vivo neuronal transfection. J. Control. Release 2018, 286, 264–278. [Google Scholar] [CrossRef] [PubMed]








© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Porkoláb, G.; Mészáros, M.; Tóth, A.; Szecskó, A.; Harazin, A.; Szegletes, Z.; Ferenc, G.; Blastyák, A.; Mátés, L.; Rákhely, G.; et al. Combination of Alanine and Glutathione as Targeting Ligands of Nanoparticles Enhances Cargo Delivery into the Cells of the Neurovascular Unit. Pharmaceutics 2020, 12, 635. https://doi.org/10.3390/pharmaceutics12070635
Porkoláb G, Mészáros M, Tóth A, Szecskó A, Harazin A, Szegletes Z, Ferenc G, Blastyák A, Mátés L, Rákhely G, et al. Combination of Alanine and Glutathione as Targeting Ligands of Nanoparticles Enhances Cargo Delivery into the Cells of the Neurovascular Unit. Pharmaceutics. 2020; 12(7):635. https://doi.org/10.3390/pharmaceutics12070635
Chicago/Turabian StylePorkoláb, Gergő, Mária Mészáros, András Tóth, Anikó Szecskó, András Harazin, Zsolt Szegletes, Györgyi Ferenc, András Blastyák, Lajos Mátés, Gábor Rákhely, and et al. 2020. "Combination of Alanine and Glutathione as Targeting Ligands of Nanoparticles Enhances Cargo Delivery into the Cells of the Neurovascular Unit" Pharmaceutics 12, no. 7: 635. https://doi.org/10.3390/pharmaceutics12070635
APA StylePorkoláb, G., Mészáros, M., Tóth, A., Szecskó, A., Harazin, A., Szegletes, Z., Ferenc, G., Blastyák, A., Mátés, L., Rákhely, G., Deli, M. A., & Veszelka, S. (2020). Combination of Alanine and Glutathione as Targeting Ligands of Nanoparticles Enhances Cargo Delivery into the Cells of the Neurovascular Unit. Pharmaceutics, 12(7), 635. https://doi.org/10.3390/pharmaceutics12070635