Targeting Germ Cell Tumors with the Newly Synthesized Flavanone-Derived Compound MLo1302 Efficiently Reduces Tumor Cell Viability and Induces Apoptosis and Cell Cycle Arrest
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Drugs, Cell Viability, and IC50
2.3. Apoptosis Assay
2.4. Proliferation Assay
2.5. Lactate Dehydrogenase (LDH) Release Assay
2.6. DNA Extraction, Bisulfite Treatment, and Quantitative Methylation-Specific PCR (qMSP)
2.7. RNA Extraction and Real-Time Quantitative PCR (RT-qCR)
2.8. Western Blot
2.9. Immunofluorescence
2.10. Dot Blot
2.11. RT2 Profiler Array
2.12. Statistical Analysis
3. Results
3.1. MLo1302 Reduces Tumor Cell Viability Across TGCT Cell Lines
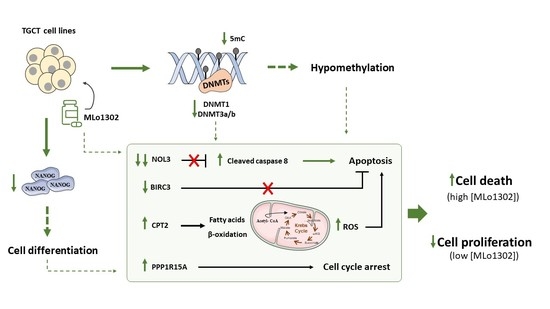
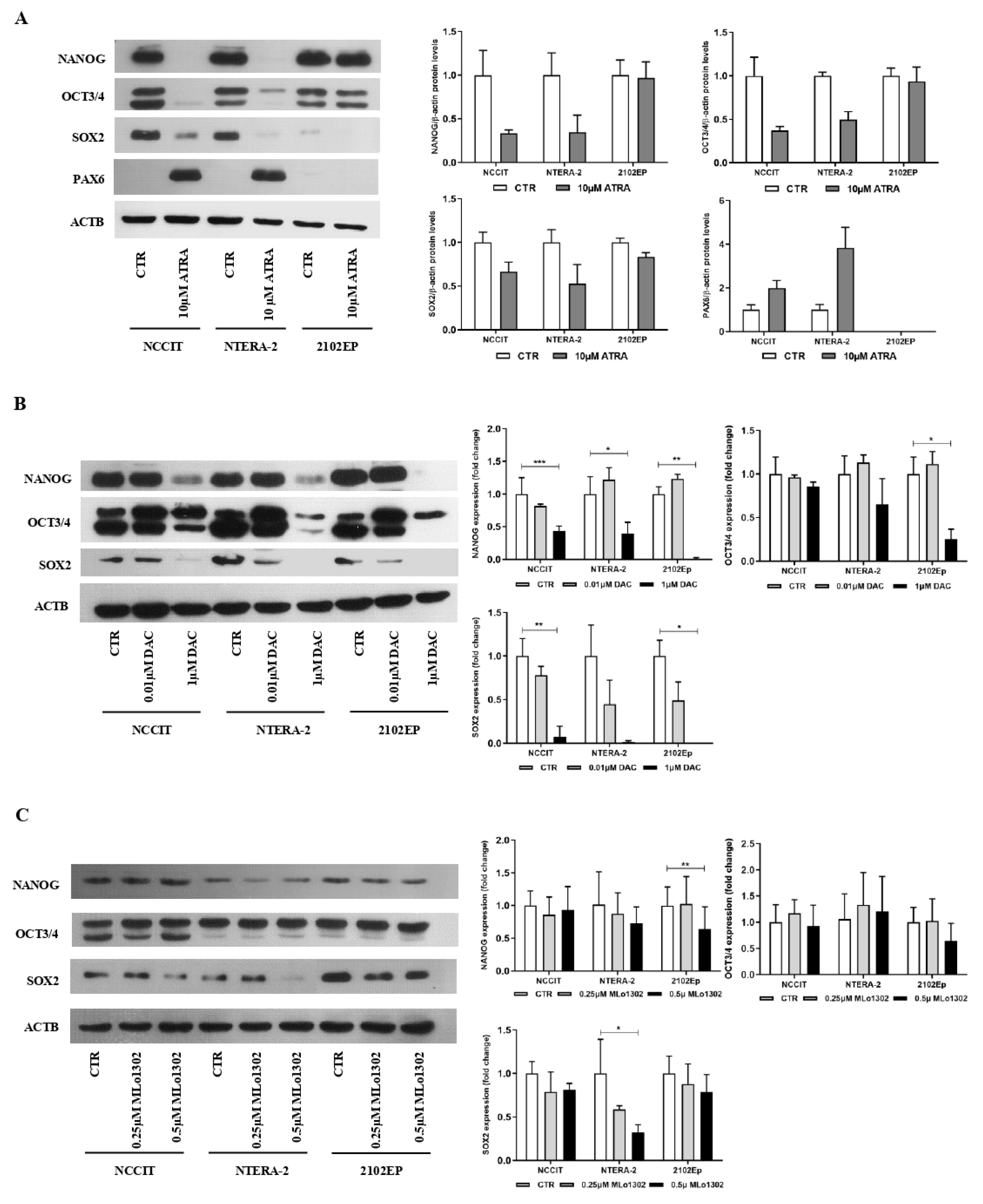
3.2. MLo1302 Treatment Results in Increased Apoptosis and Cytotoxicity, Decreased Cell Proliferation, and Reduces the Protein Expression of Pluripotency Factors
3.3. MLo1302 Effect on DNMTs, Global DNA Methylation (5 mC), and Loci-Specific Methylation Status
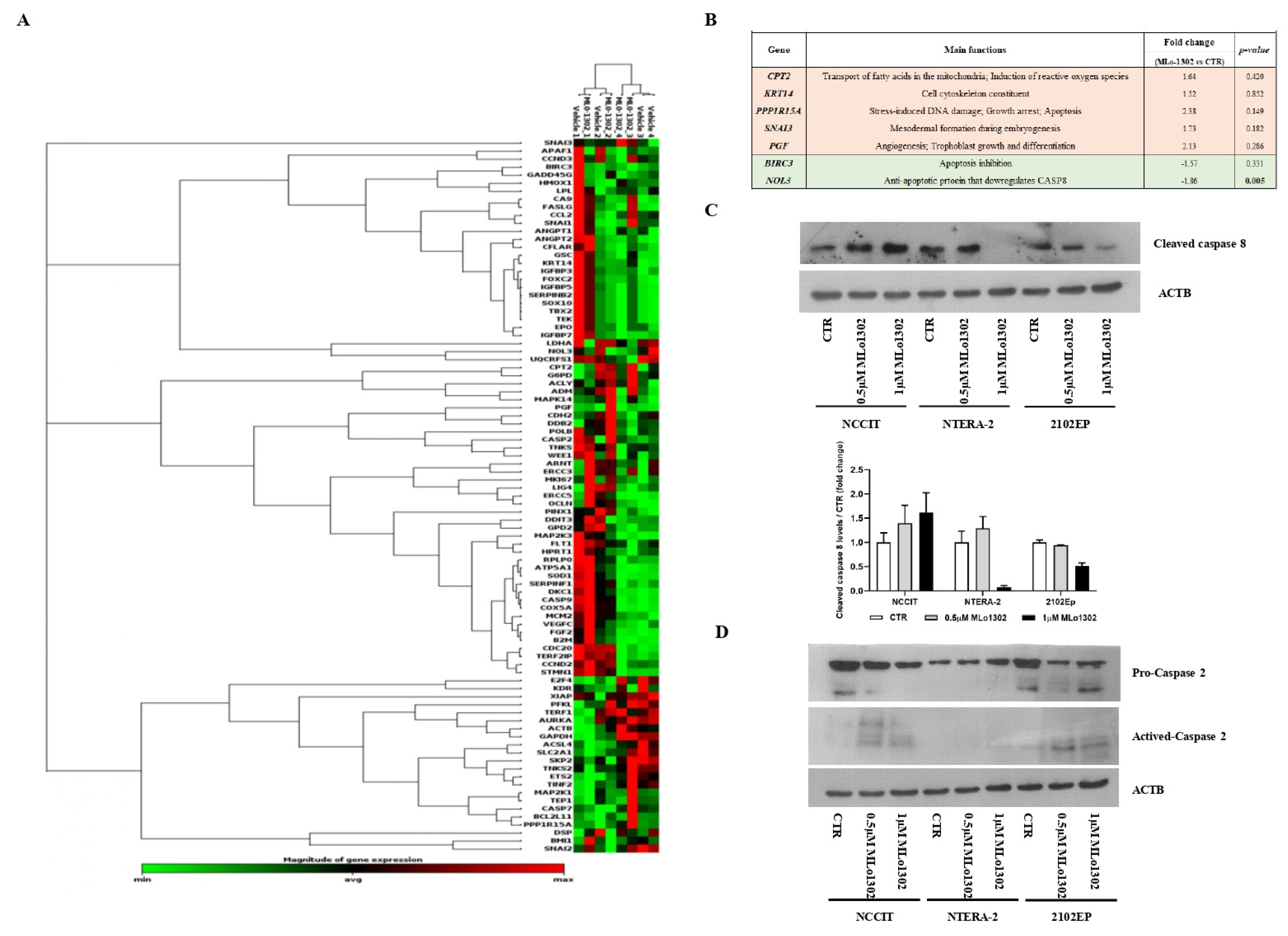
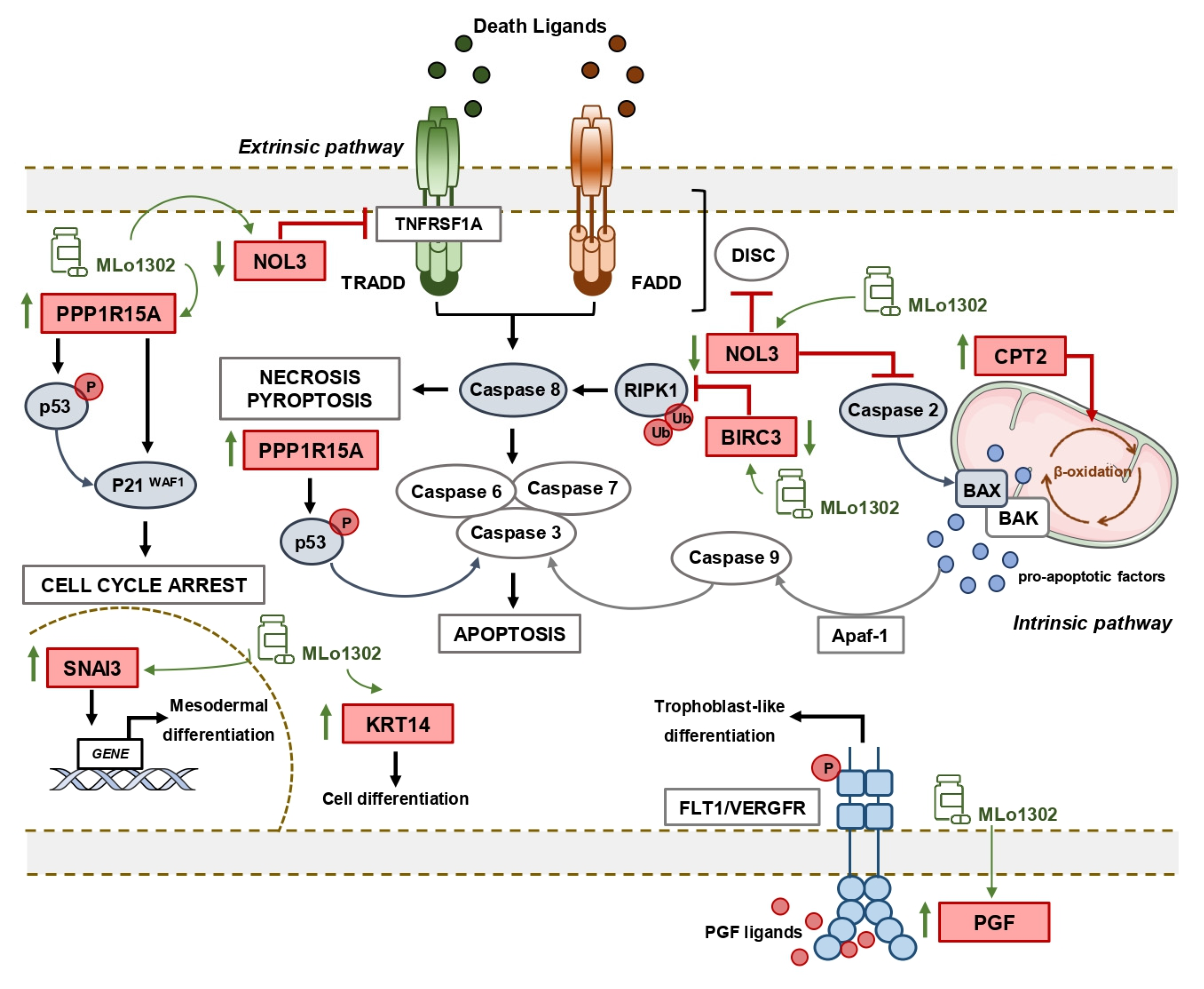
3.4. Molecular Characterization of Pathways Deregulated by MLo1302 Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, L.; Albers, P.; Berney, D.M.; Feldman, D.R.; Daugaard, G.; Gilligan, T.; Looijenga, L.H.J. Testicular cancer. Nat. Rev. Dis. Primers 2018, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Chovanec, M.; Abu Zaid, M.; Hanna, N.; El-Kouri, N.; Einhorn, L.H.; Albany, C. Long-term toxicity of cisplatin in germ-cell tumor survivors. Ann. Oncol. 2017, 28, 2670–2679. [Google Scholar] [CrossRef] [PubMed]
- Cursino-Santos, J.R.; Singh, M.; Senaldi, E.; Manwani, D.; Yazdanbakhsh, K.; Lobo, C.A. Altered parasite life-cycle processes characterize babesia divergens infection in human sickle cell anemia. Haematologica 2019, 104, 2189–2199. [Google Scholar] [CrossRef] [PubMed]
- Kalavska, K.; Conteduca, V.; De Giorgi, U.; Mego, M. Molecular mechanisms of resistance in testicular germ cell tumors-clinical implications. Curr. Cancer Drug Targets 2018, 18, 967–978. [Google Scholar] [CrossRef] [PubMed]
- Lobo, J.; Rodrigues, A.; Guimaraes, R.; Cantante, M.; Lopes, P.; Mauricio, J.; Oliveira, J.; Jeronimo, C.; Henrique, R. Detailed characterization of immune cell infiltrate and expression of immune checkpoint molecules pd-l1/ctla-4 and mmr proteins in testicular germ cell tumors disclose novel disease biomarkers. Cancers 2019, 11, 1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oing, C.; Skowron, M.A.; Bokemeyer, C.; Nettersheim, D. Epigenetic treatment combinations to effectively target cisplatin-resistant germ cell tumors: Past, present, and future considerations. Andrology 2019, 7, 487–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Plimack, E.R.; Kantarjian, H.M.; Issa, J.P. Decitabine and its role in the treatment of hematopoietic malignancies. Leuk Lymphoma 2007, 48, 1472–1481. [Google Scholar] [CrossRef]
- Linnekamp, J.F.; Butter, R.; Spijker, R.; Medema, J.P.; van Laarhoven, H.W.M. Clinical and biological effects of demethylating agents on solid tumours—A systematic review. Cancer Treat. Rev. 2017, 54, 10–23. [Google Scholar] [CrossRef] [Green Version]
- Beyrouthy, M.J.; Garner, K.M.; Hever, M.P.; Freemantle, S.J.; Eastman, A.; Dmitrovsky, E.; Spinella, M.J. High DNA methyltransferase 3b expression mediates 5-aza-deoxycytidine hypersensitivity in testicular germ cell tumors. Cancer Res. 2009, 69, 9360–9366. [Google Scholar] [CrossRef] [Green Version]
- Oing, C.; Verem, I.; Mansour, W.Y.; Bokemeyer, C.; Dyshlovoy, S.; Honecker, F. 5-azacitidine exerts prolonged pro-apoptotic effects and overcomes cisplatin-resistance in non-seminomatous germ cell tumor cells. Int. J. Mol. Sci. 2018, 20, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roth, B.J.; Elson, P.; Sledge, G.W., Jr.; Einhorn, L.H.; Trump, D.L. 5-azacytidine (nsc 102816) in refractory germ cell tumors. A phase ii trial of the eastern cooperative oncology group. Invest. New Drugs 1993, 11, 201–202. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, A.R.; Lobo, J.; Miranda-Goncalves, V.; Henrique, R.; Jeronimo, C. Epigenetic alterations as therapeutic targets in testicular germ cell tumours: Current and future application of ‘epidrugs’. Epigenetics 2020. [Google Scholar] [CrossRef] [PubMed]
- Pechalrieu, D.; Dauzonne, D.; Arimondo, P.B.; Lopez, M. Synthesis of novel 3-halo-3-nitroflavanones and their activities as DNA methyltransferase inhibitors in cancer cells. Eur. J. Med. Chem. 2020, 186, 111829. [Google Scholar] [CrossRef]
- Lobo, J.; Guimaraes-Teixeira, C.; Barros-Silva, D.; Miranda-Goncalves, V.; Camilo, V.; Guimaraes, R.; Cantante, M.; Braga, I.; Mauricio, J.; Oing, C.; et al. Efficacy of hdac inhibitors belinostat and panobinostat against cisplatin-sensitive and cisplatin-resistant testicular germ cell tumors. Cancers 2020, 12, 2903. [Google Scholar] [CrossRef]
- Palmer, R.D.; Murray, M.J.; Saini, H.K.; van Dongen, S.; Abreu-Goodger, C.; Muralidhar, B.; Pett, M.R.; Thornton, C.M.; Nicholson, J.C.; Enright, A.J.; et al. Malignant germ cell tumors display common microrna profiles resulting in global changes in expression of messenger rna targets. Cancer Res. 2010, 70, 2911–2923. [Google Scholar] [CrossRef] [Green Version]
- Josephson, R.; Ording, C.J.; Liu, Y.; Shin, S.; Lakshmipathy, U.; Toumadje, A.; Love, B.; Chesnut, J.D.; Andrews, P.W.; Rao, M.S.; et al. Qualification of embryonal carcinoma 2102ep as a reference for human embryonic stem cell research. Stem Cells 2007, 25, 437–446. [Google Scholar] [CrossRef] [Green Version]
- Lobo, J.; Nunes, S.P.; Gillis, A.J.M.; Barros-Silva, D.; Miranda-Goncalves, V.; Berg, A.V.D.; Cantante, M.; Guimaraes, R.; Henrique, R.; Jeronimo, C.; et al. Xist-promoter demethylation as tissue biomarker for testicular germ cell tumors and spermatogenesis quality. Cancers 2019, 11, 1385. [Google Scholar] [CrossRef] [Green Version]
- Costa, A.L.; Moreira-Barbosa, C.; Lobo, J.; Vilela-Salgueiro, B.; Cantante, M.; Guimaraes, R.; Lopes, P.; Braga, I.; Oliveira, J.; Antunes, L.; et al. DNA methylation profiling as a tool for testicular germ cell tumors subtyping. Epigenomics 2018, 10, 1511–1523. [Google Scholar] [CrossRef]
- Lobo, J.; Guimaraes, R.; Miranda-Goncalves, V.; Monteiro-Reis, S.; Cantante, M.; Antunes, L.; Braga, I.; Mauricio, J.; Looijenga, L.H.; Jeronimo, C.; et al. Differential expression of DNA methyltransferases and demethylases among the various testicular germ cell tumor subtypes. Epigenomics 2020, 12, 1579–1592. [Google Scholar] [CrossRef]
- Lobo, J.; Costa, A.L.; Cantante, M.; Guimaraes, R.; Lopes, P.; Antunes, L.; Braga, I.; Oliveira, J.; Pelizzola, M.; Henrique, R.; et al. M(6)a rna modification and its writer/reader virma/ythdf3 in testicular germ cell tumors: A role in seminoma phenotype maintenance. J. Transl. Med. 2019, 17, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albany, C.; Hever-Jardine, M.P.; von Herrmann, K.M.; Yim, C.Y.; Tam, J.; Warzecha, J.M.; Shin, L.; Bock, S.E.; Curran, B.S.; Chaudhry, A.S.; et al. Refractory testicular germ cell tumors are highly sensitive to the second generation DNA methylation inhibitor guadecitabine. Oncotarget 2017, 8, 2949–2959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakrishnan, S.; Hu, Q.; Krishnan, N.; Wang, D.; Smit, E.; Granger, V.; Rak, M.; Attwood, K.; Johnson, C.; Morrison, C.; et al. Decitabine, a DNA-demethylating agent, promotes differentiation via notch1 signaling and alters immune-related pathways in muscle-invasive bladder cancer. Cell Death Dis. 2017, 8, 3217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honecker, F.; Rohlfing, T.; Harder, S.; Braig, M.; Gillis, A.J.; Glaesener, S.; Barett, C.; Bokemeyer, C.; Buck, F.; Brummendorf, T.H.; et al. Proteome analysis of the effects of all-trans retinoic acid on human germ cell tumor cell lines. J. Proteom. 2014, 96, 300–313. [Google Scholar] [CrossRef] [PubMed]
- Wongtrakoongate, P.; Li, J.; Andrews, P.W. Dnmt3b inhibits the re-expression of genes associated with induced pluripotency. Exp. Cell Res. 2014, 321, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Lambrot, R.; Kimmins, S. Histone methylation is a critical regulator of the abnormal expression of pou5f1 and rassf1a in testis cancer cell lines. Int. J. Androl 2011, 34, 110–123. [Google Scholar] [CrossRef]
- Chowdhury, B.; McGovern, A.; Cui, Y.; Choudhury, S.R.; Cho, I.H.; Cooper, B.; Chevassut, T.; Lossie, A.C.; Irudayaraj, J. The hypomethylating agent decitabine causes a paradoxical increase in 5-hydroxymethylcytosine in human leukemia cells. Sci. Rep. 2015, 5, 9281. [Google Scholar] [CrossRef] [Green Version]
- Mak, P.Y.; Mak, D.H.; Ruvolo, V.; Jacamo, R.; Kornblau, S.M.; Kantarjian, H.; Andreeff, M.; Carter, B.Z. Apoptosis repressor with caspase recruitment domain modulates second mitochondrial-derived activator of caspases mimetic-induced cell death through birc2/map3k14 signalling in acute myeloid leukaemia. Br. J. Haematol. 2014, 167, 376–384. [Google Scholar] [CrossRef] [Green Version]
- Abmayr, S.; Crawford, R.W.; Chamberlain, J.S. Characterization of arc, apoptosis repressor interacting with card, in normal and dystrophin-deficient skeletal muscle. Hum. Mol. Genet. 2004, 13, 213–221. [Google Scholar] [CrossRef]
- Brown, Z.J.; Fu, Q.; Ma, C.; Kruhlak, M.; Zhang, H.; Luo, J.; Heinrich, B.; Yu, S.J.; Zhang, Q.; Wilson, A.; et al. Carnitine palmitoyltransferase gene upregulation by linoleic acid induces cd4(+) t cell apoptosis promoting hcc development. Cell Death Dis. 2018, 9, 620. [Google Scholar] [CrossRef]
- Katoh, M.; Katoh, M. Identification and characterization of human snail3 (snai3) gene in silico. Int. J. Mol. Med. 2003, 11, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Red-Horse, K.; Zhou, Y.; Genbacev, O.; Prakobphol, A.; Foulk, R.; McMaster, M.; Fisher, S.J. Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J. Clin. Investig. 2004, 114, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Brush, M.H.; Weiser, D.C.; Shenolikar, S. Growth arrest and DNA damage-inducible protein gadd34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol. Cell Biol. 2003, 23, 1292–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Berglund, A.; Kenchappa, R.S.; Forsyth, P.A.; Mule, J.J.; Etame, A.B. Birc3 is a novel driver of therapeutic resistance in glioblastoma. Sci. Rep. 2016, 6, 21710. [Google Scholar] [CrossRef] [Green Version]
- Curreri, S.A.; Fung, C.; Beard, C.J. Secondary malignant neoplasms in testicular cancer survivors. Urol. Oncol. 2015, 33, 392–398. [Google Scholar] [CrossRef]
- De Vries, G.; Rosas-Plaza, X.; van Vugt, M.; Gietema, J.A.; de Jong, S. Testicular cancer: Determinants of cisplatin sensitivity and novel therapeutic opportunities. Cancer Treat. Rev. 2020, 88, 102054. [Google Scholar] [CrossRef]
- Singh, R.; Fazal, Z.; Freemantle, S.J.; Spinella, M.J. Mechanisms of cisplatin sensitivity and resistance in testicular germ cell tumors. Cancer Drug Resist. 2019, 2, 580–594. [Google Scholar] [CrossRef] [Green Version]
- Oing, C.; Seidel, C.; Bokemeyer, C. Therapeutic approaches for refractory germ cell cancer. Expert Rev. Anticancer Ther 2018, 18, 389–397. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Biswal, B.K.; Beyrouthy, M.J.; Hever-Jardine, M.P.; Armstrong, D.; Tomlinson, C.R.; Christensen, B.C.; Marsit, C.J.; Spinella, M.J. Acute hypersensitivity of pluripotent testicular cancer-derived embryonal carcinoma to low-dose 5-aza deoxycytidine is associated with global DNA damage-associated p53 activation, anti-pluripotency and DNA demethylation. PLoS ONE 2012, 7, e53003. [Google Scholar] [CrossRef]
- Albany, C.; Fazal, Z.; Singh, R.; Bikorimana, E.; Adra, N.; Hanna, N.H.; Einhorn, L.H.; Perkins, S.M.; Sandusky, G.E.; Christensen, B.C.; et al. A phase 1 study of combined guadecitabine and cisplatin in platinum refractory germ cell cancer. Cancer Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, M.; Chabot, G.G.; Raynal, N.J.; Momparler, L.F.; Hurtubise, A.; Bernstein, M.L.; Momparler, R.L. Importance of dose-schedule of 5-aza-2′-deoxycytidine for epigenetic therapy of cancer. BMC Cancer 2008, 8, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabbour, E.; Issa, J.P.; Garcia-Manero, G.; Kantarjian, H. Evolution of decitabine development: Accomplishments, ongoing investigations, and future strategies. Cancer 2008, 112, 2341–2351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karahoca, M.; Momparler, R.L. Pharmacokinetic and pharmacodynamic analysis of 5-aza-2′-deoxycytidine (decitabine) in the design of its dose-schedule for cancer therapy. Clin. Epigenetics 2013, 5, 3. [Google Scholar] [CrossRef] [Green Version]
- Williams, L.A.; Mills, L.; Hooten, A.J.; Langer, E.; Roesler, M.; Frazier, A.L.; Krailo, M.; Nelson, H.H.; Bestrashniy, J.; Amatruda, J.F.; et al. Differences in DNA methylation profiles by histologic subtype of paediatric germ cell tumours: A report from the children’s oncology group. Br. J. Cancer 2018, 119, 864–872. [Google Scholar] [CrossRef] [Green Version]
- Juttermann, R.; Li, E.; Jaenisch, R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc. Natl. Acad. Sci. USA 1994, 91, 11797–11801. [Google Scholar] [CrossRef] [Green Version]
- Foo, R.S.; Chan, L.K.; Kitsis, R.N.; Bennett, M.R. Ubiquitination and degradation of the anti-apoptotic protein arc by mdm2. J. Biol. Chem. 2007, 282, 5529–5535. [Google Scholar] [CrossRef]
- Gustafsson, A.B.; Tsai, J.G.; Logue, S.E.; Crow, M.T.; Gottlieb, R.A. Apoptosis repressor with caspase recruitment domain protects against cell death by interfering with bax activation. J. Biol. Chem. 2004, 279, 21233–21238. [Google Scholar] [CrossRef] [Green Version]
- Jo, D.G.; Jun, J.I.; Chang, J.W.; Hong, Y.M.; Song, S.; Cho, D.H.; Shim, S.M.; Lee, H.J.; Cho, C.; Kim, D.H.; et al. Calcium binding of arc mediates regulation of caspase 8 and cell death. Mol. Cell Biol. 2004, 24, 9763–9770. [Google Scholar] [CrossRef] [Green Version]
- Kung, G.; Dai, P.; Deng, L.; Kitsis, R.N. A novel role for the apoptosis inhibitor arc in suppressing tnfalpha-induced regulated necrosis. Cell Death Differ. 2014, 21, 634–644. [Google Scholar] [CrossRef]
- Fuchs, Y.; Steller, H. Live to die another way: Modes of programmed cell death and the signals emanating from dying cells. Nat. Rev. Mol. Cell Biol. 2015, 16, 329–344. [Google Scholar] [CrossRef]
- Schroeder, A.; Warnken, U.; Roth, D.; Klika, K.D.; Vobis, D.; Barnert, A.; Bujupi, F.; Oberacker, T.; Schnolzer, M.; Nicolay, J.P.; et al. Targeting thioredoxin-1 by dimethyl fumarate induces ripoptosome-mediated cell death. Sci. Rep. 2017, 7, 43168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yagi, A.; Hasegawa, Y.; Xiao, H.; Haneda, M.; Kojima, E.; Nishikimi, A.; Hasegawa, T.; Shimokata, K.; Isobe, K. Gadd34 induces p53 phosphorylation and p21/waf1 transcription. J. Cell Biochem. 2003, 90, 1242–1249. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Nishio, N.; Ito, S.; Tanaka, Y.; Isobe, K. Negative regulation of gadd34 on myofibroblasts during cutaneous wound healing. Biomed. Res. Int. 2014, 2014, 137049. [Google Scholar] [CrossRef] [Green Version]
- Alam, H.; Sehgal, L.; Kundu, S.T.; Dalal, S.N.; Vaidya, M.M. Novel function of keratins 5 and 14 in proliferation and differentiation of stratified epithelial cells. Mol. Biol. Cell 2011, 22, 4068–4078. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M.; Katoh, M. Comparative genomics on snai1, snai2, and snai3 orthologs. Oncol. Rep. 2005, 14, 1083–1086. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.; Torry, R.J.; Torry, D.S. Deferential regulation of placenta growth factor (plgf)-mediated signal transduction in human primary term trophoblast and endothelial cells. Placenta 2004, 25, 379–386. [Google Scholar] [CrossRef]
- Lobo, J.; Jeronimo, C.; Henrique, R. Targeting the immune system and epigenetic landscape of urological tumors. Int. J. Mol. Sci. 2020, 21, 829. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lobo, J.; Cardoso, A.R.; Miranda-Gonçalves, V.; Looijenga, L.H.J.; Lopez, M.; Arimondo, P.B.; Henrique, R.; Jerónimo, C. Targeting Germ Cell Tumors with the Newly Synthesized Flavanone-Derived Compound MLo1302 Efficiently Reduces Tumor Cell Viability and Induces Apoptosis and Cell Cycle Arrest. Pharmaceutics 2021, 13, 73. https://doi.org/10.3390/pharmaceutics13010073
Lobo J, Cardoso AR, Miranda-Gonçalves V, Looijenga LHJ, Lopez M, Arimondo PB, Henrique R, Jerónimo C. Targeting Germ Cell Tumors with the Newly Synthesized Flavanone-Derived Compound MLo1302 Efficiently Reduces Tumor Cell Viability and Induces Apoptosis and Cell Cycle Arrest. Pharmaceutics. 2021; 13(1):73. https://doi.org/10.3390/pharmaceutics13010073
Chicago/Turabian StyleLobo, João, Ana Rita Cardoso, Vera Miranda-Gonçalves, Leendert H. J. Looijenga, Marie Lopez, Paola B. Arimondo, Rui Henrique, and Carmen Jerónimo. 2021. "Targeting Germ Cell Tumors with the Newly Synthesized Flavanone-Derived Compound MLo1302 Efficiently Reduces Tumor Cell Viability and Induces Apoptosis and Cell Cycle Arrest" Pharmaceutics 13, no. 1: 73. https://doi.org/10.3390/pharmaceutics13010073