Polyelectrolyte Encapsulation and Confinement within Protein Cage-Inspired Nanocompartments
Abstract
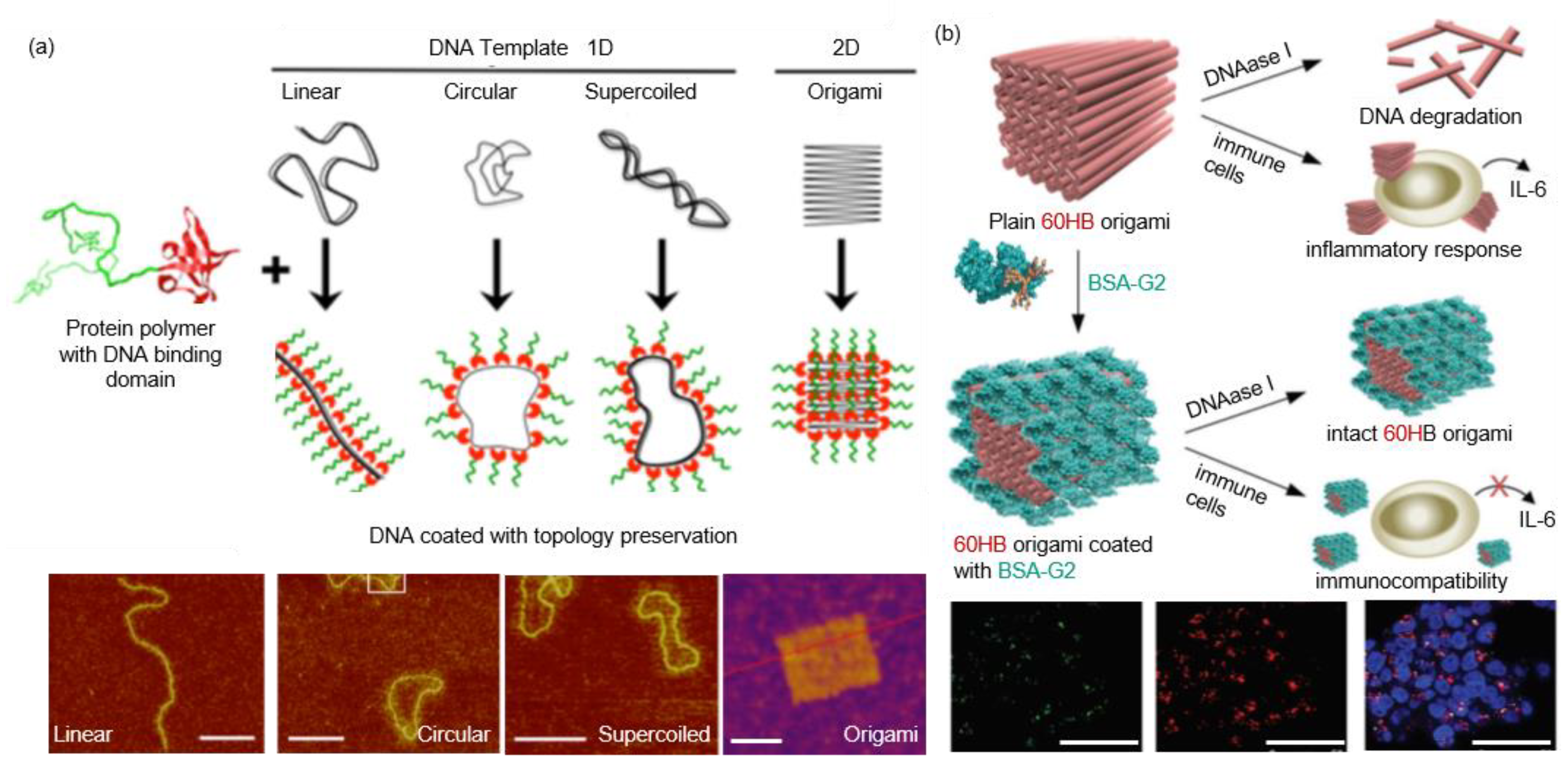
1. Introduction
2. Native Structures of Protein Cages
2.1. BMV
2.2. CCMV
2.3. RCNMV
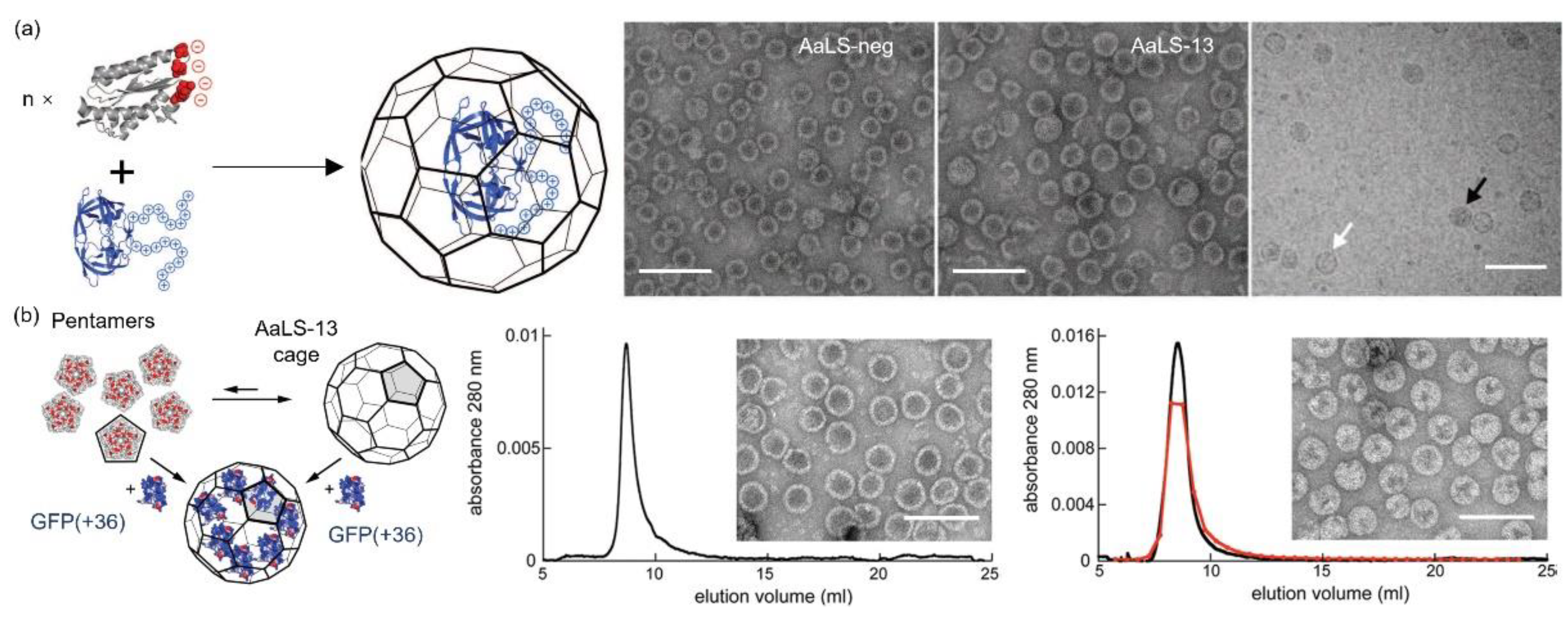
2.4. AaLS
3. Protein Cages Encapsulating Noncognate Polyelectrolytes
3.1. BMV
3.2. CCMV
3.3. RCNMV
3.4. AaLS
3.5. Other Protein Cages
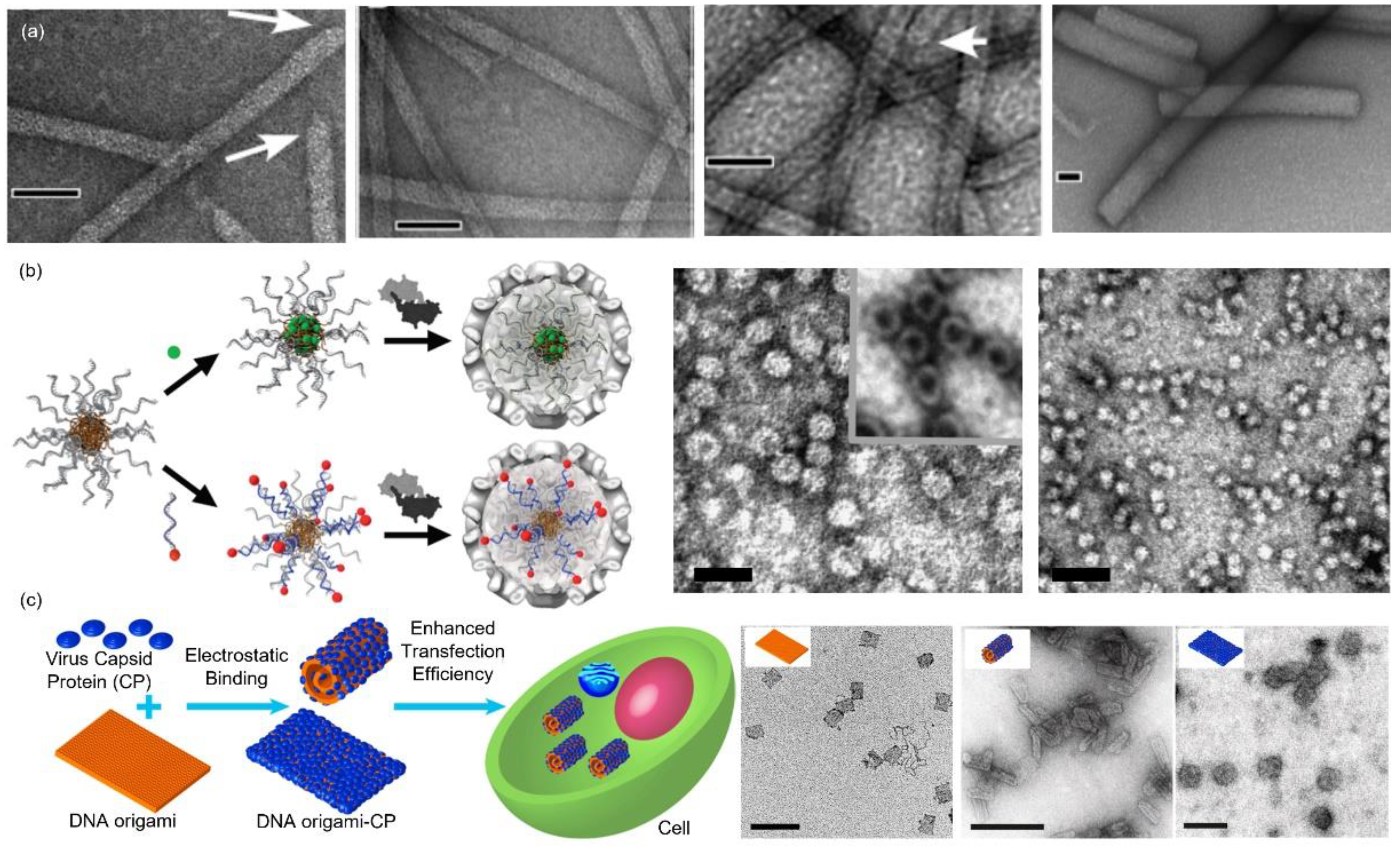
4. Virus-Inspired Core-Shell Structure Formed by Protein (Shell) and Polyelectrolyte (Core)
4.1. Nucleic Acids
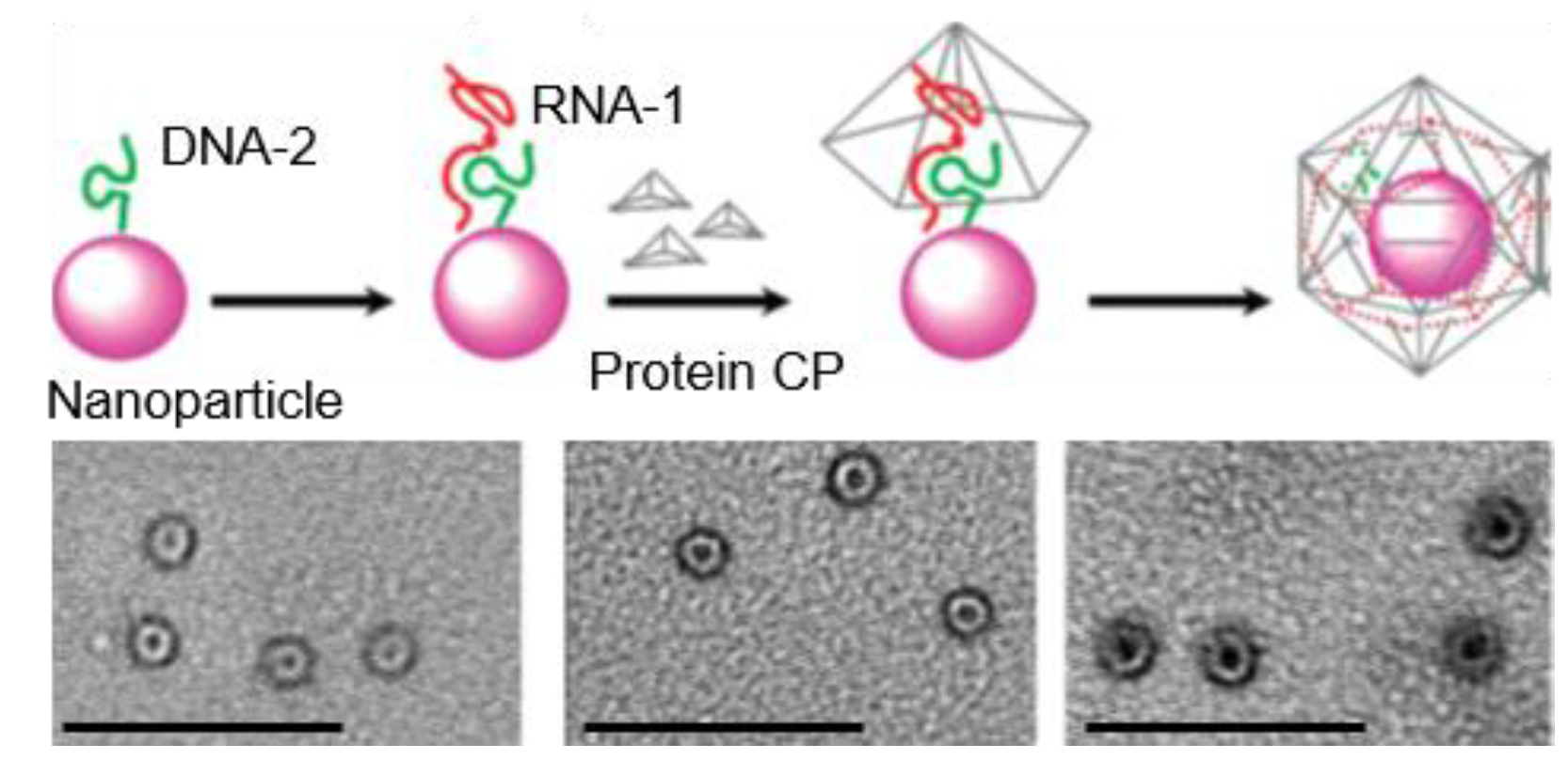
4.2. Nanoparticles
4.3. Organic Polymers
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yeates, T.O.; Crowley, C.S.; Tanaka, S. Bacterial Microcompartment Organelles: Protein Shell Structure and Evolution. Annu. Rev. Biophys. 2010, 39, 185–205. [Google Scholar] [CrossRef]
- Gabashvili, A.N.; Chmelyuk, N.S.; Efremova, M.V.; Malinovskaya, J.A.; Semkina, A.S.; Abakumov, M.A. Encapsulins—Bacterial Protein Nanocompartments: Structure, Properties, and Application. Biomolecules 2020, 10, 966. [Google Scholar] [CrossRef]
- Tanaka, S.; Sawaya, M.R.; Yeates, T.O. Structure and Mechanisms of a Protein-Based Organelle in Escherichia coli. Science 2010, 327, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Siu, K.; Chen, R.P.; Sun, Q.; Chen, L.; Tsai, S.; Chen, W. Synthetic Scaffolds for Pathway Enhancement. Curr. Opin. Biotechnol. 2015, 36, 98–106. [Google Scholar] [CrossRef]
- Uchida, M.; Klem, M.T.; Allen, M.; Suci, P.; Flenniken, M.; Gillitzer, E.; Varpness, Z.; Liepold, L.O.; Young, M.; Douglas, T. Biological Containers: Protein Cages as Multifunctional Nanoplatforms. Adv. Mater. 2007, 19, 1025–1042. [Google Scholar] [CrossRef]
- Jutz, G.; van Rijn, P.; Miranda, B.S.; Böker, A. Ferritin: A Versatile Building Block for Bionanotechnology. Chem. Rev. 2015, 115, 1653–1701. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Meining, W.; Fischer, M.; Bacher, A.; Ladenstein, R. X-Ray Structure Analysis and Crystallographic Refinement of Lumazine Synthase from the Hyperthermophile Aquifex Aeolicus at 1.6 Å Resolution Determinants of Thermostability Revealed from Structural Comparisons. J. Mol. Biol. 2001, 306, 1099–1114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Konarev, P.V.; Petoukhov, M.V.; Svergun, D.I.; Xing, L.; Cheng, R.H.; Haase, I.; Fischer, M.; Bacher, A.; Ladenstein, R.; et al. Multiple Assembly States of Lumazine Synthase: A Model Relating Catalytic Function and Molecular Assembly. J. Mol. Biol. 2006, 362, 753–770. [Google Scholar] [CrossRef]
- Azuma, Y.; Edwardson, T.G.W.; Hilvert, D. Tailoring Lumazine Synthase Assemblies for Bionanotechnology. Chem. Soc. Rev. 2018, 47, 3543–3557. [Google Scholar] [CrossRef] [PubMed]
- Aumiller, W.M.; Uchida, M.; Douglas, T. Protein Cage Assembly across Multiple Length Scales. Chem. Soc. Rev. 2018, 47, 3433–3469. [Google Scholar] [CrossRef] [PubMed]
- Hagan, M.F. A Theory for Viral Capsid Assembly around Electrostatic Cores. J. Chem. Phys. 2009, 130, 114902. [Google Scholar] [CrossRef]
- Perlmutter, J.D.; Hagan, M.F. Mechanisms of Virus Assembly. Annu. Rev. Phys. Chem. 2015, 66, 217–239. [Google Scholar] [CrossRef]
- Kegel, W.K.; van der Schoot, P. Competing Hydrophobic and Screened-Coulomb Interactions in Hepatitis B Virus Capsid Assembly. Biophys. J. 2004, 86, 3905–3913. [Google Scholar] [CrossRef]
- Ceres, P.; Zlotnick, A. Weak Protein-Protein Interactions are Sufficient to Drive Assembly of Hepatitis B Virus Capsids. Biochemistry 2002, 41, 11525–11531. [Google Scholar] [CrossRef]
- Kegel, W.K.; van der Schoot, P. Physical Regulation of the Self-Assembly of Tobacco Mosaic Virus Coat Protein. Biophys. J. 2006, 91, 1501–1512. [Google Scholar] [CrossRef]
- Kivenson, A.; Hagan, M.F. Mechanisms of Capsid Assembly around a Polymer. Biophys. J. 2010, 99, 619–628. [Google Scholar] [CrossRef]
- Garmann, R.F.; Comas-Garcia, M.; Knobler, C.M.; Gelbart, W.M. Physical Principles in the Self-Assembly of a Simple Spherical Virus. Acc. Chem. Res. 2016, 49, 48–55. [Google Scholar] [CrossRef]
- Rother, M.; Nussbaumer, M.G.; Renggli, K.; Bruns, N. Protein Cages and Synthetic Polymers: A Fruitful Symbiosis for Drug Delivery Applications, Bionanotechnology and Materials Science. Chem. Soc. Rev. 2016, 45, 6213–6249. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Garcia, A.; Kraft, D.J.; Janssen, A.F.J.; Bomans, P.H.H.; Sommerdijk, N.A.J.M.; Thies-Weesie, D.M.E.; Favretto, M.E.; Brock, R.; de Wolf, F.A.; Werten, M.W.T.; et al. Design and Self-Assembly of Simple Coat Proteins for Artificial Viruses. Nat. Nanotechnol. 2014, 9, 698–702. [Google Scholar] [CrossRef]
- Elsaesser, A.; Howard, C.V. Toxicology of Nanoparticles. Adv. Drug Deliv. Rev. 2012, 64, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Douglas, T.; Young, M. Host-Guest Encapsulation of Materials by Assembled Virus Protein Cages. Nature 1998, 393, 152–155. [Google Scholar] [CrossRef]
- Maassen, S.J.; van der Ham, A.M.; Cornelissen, J.J.L.M. Combining Protein Cages and Polymers: From Understanding Self-Assembly to Functional Materials. ACS Macro Lett. 2016, 5, 987–994. [Google Scholar] [CrossRef]
- Korpi, A.; Anaya-Plaza, E.; Välimäki, S.; Kostiainen, M. Highly Ordered Protein Cage Assemblies: A Toolkit for New Materials. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, 12, e1578. [Google Scholar] [CrossRef]
- Caspar, D.L.; Klug, A. Physical Principles in the Construction of Regular Viruses. Cold Spring Harb. Symp. Quant. Biol. 1962, 27, 1–24. [Google Scholar] [CrossRef]
- Johnson, J.E.; Speir, J.A. Quasi-Equivalent Viruses a Paradigm for Protein Assemblies. J. Mol. Biol. 1997, 269, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Lucas, R.W.; Larson, S.B.; Mcpherson, A. The Crystallographic Structure of Brome Mosaic Virus. J. Mol. Biol. 2002, 317, 95–108. [Google Scholar] [CrossRef]
- Larson, S.B.; Lucas, R.W.; McPherson, A. Crystallographic Structure of the T=1 Particle of Brome Mosaic Virus. J. Mol. Biol. 2005, 346, 815–831. [Google Scholar] [CrossRef]
- Speir, J.A.; Munshi, S.; Wang, G.; Baker, T.S.; Johnson, J.E. Structures of the Native and Swollen Forms of Cowpea Chlorotic Mottle Virus Determined by X-Ray Crystallography and Cryo-Electron Microscopy. Structure 1995, 3, 63–78. [Google Scholar] [CrossRef]
- Sikkema, F.D.; Comellas-Aragonès, M.; Fokkink, R.G.; Verduin, B.J.M.; Cornelissen, J.J.L.M.; Nolte, R.J.M. Monodisperse Polymer-Virus Hybrid Nanoparticles. Org. Biomol. Chem. 2007, 5, 54–57. [Google Scholar] [CrossRef]
- Basnayake, V.R.; Sit, T.L.; Lommel, S.A. The Genomic RNA Packaging Scheme of Red Clover Necrotic Mosaic Virus. Virology 2006, 345, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M.B.; Guenther, R.H.; Tama, F.; Sit, T.L.; Brooks, C.L.; Mikhailov, A.M.; Orlova, E.V.; Baker, T.S.; Lommel, S.A. Removal of Divalent Cations Induces Structural Transitions in Red Clover Necrotic Mosaic Virus, Revealing a Potential Mechanism for RNA Release. J. Virol. 2006, 80, 10395–10406. [Google Scholar] [CrossRef]
- Cuillel, M.; Berthet-Colominas, C.; Timmins, P.A.; Zulauf, M. Reassembly of Brome Mosaic Virus from Dissociated Virus: A Neutron Scattering Study. Eur. Biophys. J. 1987, 15, 169–176. [Google Scholar] [CrossRef]
- Bancroft, J.B.; Hiebert, E.; Bracker, C.E. The Effects of Various Polyanions on Shell Formation of Some Spherical Viruses. Virology 1969, 39, 924–930. [Google Scholar] [CrossRef]
- Bancroft, J.B.; Bracker, C.E.; Wagner, G.W. Structures Derived from Cowpea Chlorotic Mottle and Brome Mosaic Virus Protein. Virology 1969, 38, 324–335. [Google Scholar] [CrossRef]
- Chen, C.; Daniel, M.C.; Quinkert, Z.T.; De, M.; Stein, B.; Bowman, V.D.; Chipman, P.R.; Rotello, V.M.; Kao, C.C.; Dragnea, B. Nanoparticle-Templated Assembly of Viral Protein Cages. Nano Lett. 2006, 6, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Dufort, C.; Daniel, M.; Murali, A.; Chen, C.; Gopinath, K.; Stein, B.; De, M.; Rotello, V.M.; Holzenburg, A.; et al. Core-Controlled Polymorphism in Virus-like Particles. Proc. Natl. Acad. Sci. USA 2007, 104, 1354–1359. [Google Scholar] [CrossRef]
- Cheng, Z.; Guillermo, R.L.; Irina, B.; Tsvetkova, M.F.; Hagan, B.D. Defects and Chirality in the Nanoparticle-Directed Assembly of Spherocylindrical Shells of Virus Coat Proteins. ACS Nano 2018, 12, 5323–5332. [Google Scholar] [CrossRef]
- Daniel, M.; Tsvetkova, I.B.; Quinkert, Z.T.; Murali, A.; De, M.; Rotello, V.M.; Kao, C.C.; Dragnea, B. Role of Surface Charge Density in Nanoparticle-Templated Assembly of Bromovirus Protein Cages. ACS Nano 2010, 4, 3853–3860. [Google Scholar] [CrossRef][Green Version]
- Tsvetkova, I.; Chen, C.; Rana, S.; Kao, C.C.; Rotello, V.M.; Dragnea, B. Pathway Switching in Templated Virus-like Particle Assembly. Soft Matter 2012, 8, 4571–4577. [Google Scholar] [CrossRef]
- Huang, X.; Bronstein, L.M.; Retrum, J.; Dufort, C.; Tsvetkova, I.; Aniagyei, S.; Stein, B.; Stucky, G.; McKenna, B.; Remmes, N.; et al. Self-Assembled Virus-like Particles with Magnetic Cores. Nano Lett. 2007, 7, 2407–2416. [Google Scholar] [CrossRef]
- Malyutin, A.G.; Cheng, H.; Sanchez-Felix, O.R.; Carlson, K.; Stein, B.D.; Konarev, P.V.; Svergun, D.I.; Dragnea, B.; Bronstein, L.M. Coat Protein-Dependent Behavior of Poly(Ethylene Glycol) Tails in Iron Oxide Core Virus-like Nanoparticles. ACS Appl. Mater. Interfaces 2015, 7, 12089–12098. [Google Scholar] [CrossRef]
- Dixit, S.K.; Goicochea, N.L.; Daniel, M.C.; Murali, A.; Bronstein, L.; De, M.; Stein, B.; Rotello, V.M.; Kao, C.C.; Dragnea, B. Quantum Dot Encapsulation in Viral Capsids. Nano Lett. 2006, 6, 1993–1999. [Google Scholar] [CrossRef] [PubMed]
- Dragnea, B.; Chen, C.; Kwak, E.; Stein, B.; Kao, C.C. Gold Nanoparticles as Spectroscopic Enhancers for in Vitro Studies on Single Viruses. J. Am. Chem. Soc. 2003, 125, 6374–6375. [Google Scholar] [CrossRef]
- Bancroft, J.B.; Hiebert, E. Formation of an Infectious Nucleoprotein from Protein and Nucleic Acid Isolated from a Small Spherical Virus. Virology 1967, 32, 354–356. [Google Scholar] [CrossRef]
- Tama, F.; Brooks, C.L. The Mechanism and Pathway of PH Induced Swelling in Cowpea Chlorotic Mottle Virus. J. Mol. Biol. 2002, 318, 733–747. [Google Scholar] [CrossRef]
- Zlotnick, A.; Aldrich, R.; Johnson, J.M.; Ceres, P.; Young, M.J. Mechanism of Capsid Assembly for an Icosahedral Plant Virus. Virology 2000, 277, 450–456. [Google Scholar] [CrossRef]
- Johnson, J.M.; Willits, D.A.; Young, M.J.; Zlotnick, A. Interaction with Capsid Protein Alters RNA Structure and the Pathway for in Vitro Assembly of Cowpea Chlorotic Mottle Virus. J. Mol. Biol. 2004, 335, 455–464. [Google Scholar] [CrossRef]
- Mukherjee, S.; Pfeifer, C.M.; Johnson, J.M.; Liu, J.; Zlotnick, A. Redirecting the Coat Protein of a Spherical Virus to Assemble into Tubular Nanostructures. J. Am. Chem. Soc. 2006, 128, 2538–2539. [Google Scholar] [CrossRef]
- De Ruiter, M.V.; van der Hee, R.M.; Driessen, A.J.M.; Keurhorst, E.D.; Hamid, M.; Cornelissen, J.J.L.M. Polymorphic Assembly of Virus-Capsid Proteins around DNA and the Cellular Uptake of the Resulting Particles. J. Control. Release 2019, 307, 342–354. [Google Scholar] [CrossRef]
- Kwak, M.; Minten, I.J.; Anaya, D.; Musser, A.J.; Brasch, M.; Nolte, R.J.M.; Mu, K.; Cornelissen, J.J.L.M.; Herrmann, A. Virus-like Particles Templated by DNA Micelles: A General Method for Loading Virus Nanocarriers. J. Am. Chem. Soc. 2010, 132, 7834–7835. [Google Scholar] [CrossRef]
- Alemdaroglu, F.E.; Alemdaroglu, N.C.; Langguth, P.; Herrmann, A. DNA Block Copolymer Micelles-a Combinatorial Tool for Cancer Nanotechnology. Adv. Mater. 2008, 20, 899–902. [Google Scholar] [CrossRef]
- Brasch, M.; Putri, R.M.; de Ruiter, M.V.; Luque, D.; Koay, M.S.T.; Castón, J.R.; Cornelissen, J.J.L.M. Assembling Enzymatic Cascade Pathways inside Virus-Based Nanocages Using Dual-Tasking Nucleic Acid Tags. J. Am. Chem. Soc. 2017, 139, 1512–1519. [Google Scholar] [CrossRef] [PubMed]
- Mikkilä, J.; Eskelinen, A.P.; Niemelä, E.H.; Linko, V.; Frilander, M.J.; Törmä, P.; Kostiainen, M.A. Virus-Encapsulated DNA Origami Nanostructures for Cellular Delivery. Nano Lett. 2014, 14, 2196–2200. [Google Scholar] [CrossRef]
- Comellas-Aragonès, M.; de la Escosura, A.; Dirks, A.J.; van der Ham, A.; Fusté-Cuñé, A.; Cornelissen, J.J.L.M.; Nolte, R.J.M. Controlled Integration of Polymers into Viral Capsids. Biomacromolecules 2009, 10, 3141–3147. [Google Scholar] [CrossRef]
- Maassen, S.J.; Huskens, J.; Cornelissen, J.J.L.M. Elucidating the Thermodynamic Driving Forces of Polyanion-Templated Virus-like Particle Assembly. J. Phys. Chem. B 2019, 123, 9733–9741. [Google Scholar] [CrossRef] [PubMed]
- Chevreuil, M.; Law-Hine, D.; Chen, J.; Bressanelli, S.; Combet, S.; Constantin, D.; Degrouard, J.; Möller, J.; Zeghal, M.; Tresset, G. Nonequilibrium Self-Assembly Dynamics of Icosahedral Viral Capsids Packaging Genome or Polyelectrolyte. Nat. Commun. 2018, 9, 3071. [Google Scholar] [CrossRef] [PubMed]
- Baigl, D.; Seery, T.A.P.; Williams, C.E. Preparation and Characterization of Hydrosoluble, Partially Charged Poly(Styrenesulfonate)s of Various Controlled Charge Fractions and Chain Lengths. Macromolecules 2002, 35, 2318–2326. [Google Scholar] [CrossRef]
- Hu, Y.; Zandi, R.; Anavitarte, A.; Knobler, C.M.; Gelbart, W.M. Packaging of a Polymer by a Viral Capsid: The Interplay between Polymer Length and Capsid Size. Biophys. J. 2008, 94, 1428–1436. [Google Scholar] [CrossRef]
- Cadena-Nava, R.D.; Hu, Y.; Garmann, R.F.; Ng, B.; Zelikin, A.N.; Knobler, C.M.; Gelbart, W.M. Exploiting Fluorescent Polymers to Probe the Self-Assembly of Virus-like Particles. J. Phys. Chem. B 2011, 115, 2386–2391. [Google Scholar] [CrossRef]
- Ng, B.C.; Chan, S.T.; Lin, J.; Tolbert, S.H. Using Polymer Conformation to Control Architecture in Semiconducting Polymer/Viral Capsid Assemblies. ACS Nano 2011, 5, 7730–7738. [Google Scholar] [CrossRef]
- Välimäki, S.; Liu, Q.; Schoonen, L.; Vervoort, D.F.M.; Nonappa; Linko, V.; Nolte, R.J.M.; van Hest, J.C.M.; Kostiainen, M.A. Engineered Protein Cages for Selective Heparin Encapsulation. J. Mater. Chem. B 2021, 9, 1272–1276. [Google Scholar] [CrossRef] [PubMed]
- Brasch, M.; de la Escosura, A.; Ma, Y.; Uetrecht, C.; Heck, A.J.R.; Torres, T.; Cornelissen, J.J.L.M. Encapsulation of Phthalocyanine Supramolecular Stacks into Virus-like Particles. J. Am. Chem. Soc. 2011, 133, 6878–6881. [Google Scholar] [CrossRef] [PubMed]
- Sinn, S.; Yang, L.; Biedermann, F.; Wang, D.; Kübel, C.; Cornelissen, J.J.L.M.; Cola, L.D. Templated Formation of Luminescent Virus-like Particles by Tailor-Made Pt(II) Amphiphiles. J. Am. Chem. Soc. 2018, 140, 2355–2362. [Google Scholar] [CrossRef]
- Luque, D.; de la Escosura, A.; Snijder, J.; Brasch, M.; Burnley, R.J.; Koay, M.S.T.; Carrascosa, J.L.; Wuite, G.J.L.; Roos, W.H.; Heck, A.J.R.; et al. Self-Assembly and Characterization of Small and Monodisperse Dye Nanospheres in a Protein Cage. Chem. Sci. 2014, 5, 575–581. [Google Scholar] [CrossRef]
- Setaro, F.; Brasch, M.; Hahn, U.; Koay, M.S.T.; Cornelissen, J.J.L.M.; de la Escosura, A.; Torres, T. Generation-Dependent Templated Self-Assembly of Biohybrid Protein Nanoparticles around Photosensitizer Dendrimers. Nano Lett. 2015, 15, 1245–1251. [Google Scholar] [CrossRef]
- Galindo, J.; Brasch, M.; Anaya-Plaza, E.; de la Escosura, A.; Velders, A.H.; Reinhoudt, D.N.; Torres, T.; Koay, M.S.T.; Cornelissen, J.J.L.M. Self-Assembly Triggered by Self-Assembly: Optically Active, Paramagnetic Micelles Encapsulated in Protein Cage Nanoparticles. J. Inorg. Biochem. 2014, 136, 140–146. [Google Scholar] [CrossRef]
- Aniagyei, S.E.; Kennedy, C.J.; Stein, B.; Willits, D.A.; Douglas, T.; Young, M.J.; De, M.; Rotello, V.M.; Srisathiyanarayanan, D.; Kao, C.C.; et al. Synergistic Effects of Mutations and Nanoparticle Templating in the Self-Assembly of Cowpea Chlorotic Mottle Virus Capsids. Nano Lett. 2009, 9, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Verwegen, M.; de Ruiter, M.V.; Maassen, S.J.; Traulsen, C.H.; Cornelissen, J.J.L.M. Protein Cages as Containers for Gold Nanoparticles. J. Phys. Chem. B 2016, 120, 6352–6357. [Google Scholar] [CrossRef]
- Basnayake, V.R.; Sit, T.L.; Lommel, S.A. The Red Clover Necrotic Mosaic Virus Origin of Assembly Is Delimited to the RNA-2 Trans-Activator. Virology 2009, 384, 169–178. [Google Scholar] [CrossRef]
- Loo, L.; Guenther, R.H.; Basnayake, V.R.; Lommel, S.A.; Franzen, S. Controlled Encapsidation of Gold Nanoparticles by a Viral Protein Shell. J. Am. Chem. Soc. 2006, 128, 4502–4503. [Google Scholar] [CrossRef]
- Loo, L.; Guenther, R.H.; Lommel, S.A.; Franzen, S. Encapsidation of Nanoparticles by Red Clover Necrotic Mosaic Virus. J. Am. Chem. Soc. 2007, 129, 11111–11117. [Google Scholar] [CrossRef] [PubMed]
- Loo, L.; Guenther, R.H.; Lommel, S.A.; Franzen, S. Infusion of Dye Molecules into Red Clover Necrotic Mosaic Virus. Chem. Commun. 2008, 1, 88–90. [Google Scholar] [CrossRef] [PubMed]
- Lockney, D.M.; Guenther, R.N.; Loo, L.; Overton, W.; Antonelli, R.; Clark, J.; Hu, M.; Luft, C.; Lommel, S.A.; Franzen, S. The Red Clover Necrotic Mosaic Virus Capsid as a Multifunctional Cell Targeting Plant Viral Nanoparticle. Bioconjug. Chem. 2011, 22, 67–73. [Google Scholar] [CrossRef]
- Azuma, Y.; Zschoche, R.; Hilvert, D. The C-Terminal Peptide of Aquifex Aeolicus Riboflavin Synthase Directs Encapsulation of Native and Foreign Guests by a Cage-Forming Lumazine Synthase. J. Biol. Chem. 2017, 292, 10321–10327. [Google Scholar] [CrossRef]
- Seebeck, F.P.; Woycechowsky, K.J.; Zhuang, W.; Rabe, J.P.; Hilvert, D. A Simple Tagging System for Protein Encapsulation. J. Am. Chem. Soc. 2006, 128, 4516–4517. [Google Scholar] [CrossRef]
- Wörsdörfer, B.; Woycechowsky, K.J.; Hilvert, D. Directed Evolution of a Protein Container. Science 2011, 331, 589–592. [Google Scholar] [CrossRef]
- Wörsdörfer, B.; Pianowski, Z.; Hilvert, D. Efficient in Vitro Encapsulation of Protein Cargo by an Engineered Protein Container. J. Am. Chem. Soc. 2012, 134, 909–911. [Google Scholar] [CrossRef]
- Azuma, Y.; Zschoche, R.; Tinzl, M.; Hilvert, D. Quantitative Packaging of Active Enzymes into a Protein Cage. Angew. Chem. Int. Ed. 2016, 55, 1531–1534. [Google Scholar] [CrossRef]
- Frey, R.; Mantri, S.; Rocca, M.; Hilvert, D. Bottom-up Construction of a Primordial Carboxysome Mimic. J. Am. Chem. Soc. 2016, 138, 10072–10075. [Google Scholar] [CrossRef]
- Azuma, Y.; Bader, D.L.V.; Hilvert, D. Substrate Sorting by a Supercharged Nanoreactor. J. Am. Chem. Soc. 2018, 140, 860–863. [Google Scholar] [CrossRef]
- Beck, T.; Tetter, S.; Künzle, M.; Hilvert, D. Construction of Matryoshka-Type Structures from Supercharged Protein Nanocages. Angew. Chem. Int. Ed. 2015, 127, 951–954. [Google Scholar] [CrossRef]
- Sasaki, E.; Dragoman, R.M.; Mantri, S.; Dirin, D.N.; Kovalenko, M.V.; Hilvert, D. Self-Assembly of Proteinaceous Shells around Positively Charged Gold Nanomaterials Enhances Colloidal Stability in High-Ionic-Strength Buffers. ChemBioChem 2020, 21, 74–79. [Google Scholar] [CrossRef]
- Lilavivat, S.; Sardar, D.; Jana, S.; Thomas, G.C.; Woycechowsky, K.J. In Vivo Encapsulation of Nucleic Acids Using an Engineered Nonviral Protein Capsid. J. Am. Chem. Soc. 2012, 134, 13152–13155. [Google Scholar] [CrossRef]
- Fu, J.; Woycechowsky, K.J. Guest Sequence Can Influence RNA Encapsulation by an Engineered Cationic Protein Capsid. Biochemistry 2020, 59, 1517–1526. [Google Scholar] [CrossRef]
- Azuma, Y.; Edwardson, T.G.W.; Terasaka, N.; Hilvert, D. Modular Protein Cages for Size-Selective RNA Packaging in Vivo. J. Am. Chem. Soc. 2018, 140, 566–569. [Google Scholar] [CrossRef]
- Terasaka, N.; Azuma, Y.; Hilverta, D. Laboratory Evolution of Virus-Like Nucleocapsids from Nonviral Protein Cages. Proc. Natl. Acad. Sci. USA 2018, 115, 5432–5437. [Google Scholar] [CrossRef]
- Tetter, S.; Terasaka, N.; Steinauer, A.; Bingham, R.J.; Clark, S.; Scott, A.J.P.; Patel, N.; Leibundgut, M.; Wroblewski, E.; Ban, N.; et al. Evolution of A Virus-Like Architecture and Packaging Mechanism in a Repurposed Bacterial Protein. Science 2021, 372, 1220–1224. [Google Scholar] [CrossRef]
- Sana, B.; Johnson, E.; Magueres, P.L.; Criswell, A.; Cascio, D.; Lim, S. The Role of Nonconserved Residues of Archaeoglobus Fulgidus Ferritin on Its Unique Structure and Biophysical Properties. J. Biol. Chem. 2013, 288, 32663–32672. [Google Scholar] [CrossRef]
- Swift, J.; Butts, C.A.; Cheung-Lau, J.; Yerubandi, V.; Dmochowski, I.J. Efficient Self-Assembly of Archaeoglobus fulgidus Ferritin around Metallic Cores. Langmuir 2009, 25, 5219–5225. [Google Scholar] [CrossRef]
- Tetter, S.; Hilvert, D. Enzyme Encapsulation by a Ferritin Cage. Angew. Chem. Int. Ed. 2017, 56, 15129–15132. [Google Scholar] [CrossRef]
- Bigotti, M.G.; Clarke, A.R. Chaperonins: The Hunt for the Group II Mechanism. Arch. Biochem. Biophys. 2008, 474, 331–339. [Google Scholar] [CrossRef]
- Nussbaumer, M.G.; Bisig, C.; Bruns, N. Using the Dendritic Polymer PAMAM to Form Gold Nanoparticles in the Protein Cage Thermosome. Chem. Commun. 2016, 52, 10537–10539. [Google Scholar] [CrossRef]
- Nussbaumer, M.G.; Duskey, J.T.; Rother, M.; Renggli, K.; Chami, M.; Bruns, N. Chaperonin-Dendrimer Conjugates for siRNA Delivery. Adv. Sci. 2016, 3, 1600046. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.L.; Heuser, J.E.; Chugani, D.C.; Rome, L.H. Vaults. III. Vault Ribonucleoprotein Particles Open into Flower-like Structures with Octagonal Symmetry. J. Cell Biol. 1991, 112, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Slesina, M.; Inman, E.M.; Moore, A.E.; Goldhaber, J.I.; Rome, L.H. Movement of Vault Particles Visualized by GFP-Tagged Major Vault Protein. Cell Tissue Res. 2006, 324, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Rome, L.H.; Kickhoefer, V.A. Development of the Vault Particle as a Platform Technology. ACS Nano 2013, 7, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Ng, B.C.; Yu, M.; Gopal, A.; Rome, L.H.; Monbouquette, H.G.; Tolbert, S.H. Encapsulation of Semiconducting Polymers in Vault Protein Cages. Nano Lett. 2008, 8, 3503–3509. [Google Scholar] [CrossRef] [PubMed]
- Glasgow, J.E.; Capehart, S.L.; Francis, M.B.; Tullman-Ercek, D. Osmolyte-Mediated Encapsulation of Proteins inside MS2 Viral Capsids. ACS Nano 2012, 6, 8658–8664. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, J.D.; Brown, S.D.; Lau, J.L.; Finn, M.G. RNA-Directed Packaging of Enzymes within Virus-like Particles. Angew. Chem. Int. Ed. 2010, 122, 9842–9845. [Google Scholar] [CrossRef]
- Das, S.; Zhao, L.; Elofson, K.; Finn, M.G. Enzyme Stabilization by Virus-Like Particles. Biochemistry 2020, 59, 2870–2881. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, G.L.; Lajoie, M.J.; Gustafson, H.H.; Sellers, D.L.; Nattermann, U.; Ellis, D.; Bale, J.B.; Ke, S.; Lenz, G.H.; Yehdego, A.; et al. Evolution of A Designed Protein Assembly Encapsulating its Own RNA Genome. Nature 2017, 552, 415–420. [Google Scholar] [CrossRef]
- Edwardson, T.G.W.; Mori, T.; Hilvert, D. Rational Engineering of a Designed Protein Cage for siRNA Delivery. J. Am. Chem. Soc. 2018, 140, 10439–10442. [Google Scholar] [CrossRef]
- Edwardson, T.G.W.; Tetter, S.; Hilvert, D. Two-Tier Supramolecular Encapsulation of Small Molecules in a Protein Cage. Nat. Commun. 2020, 11, 5410. [Google Scholar] [CrossRef]
- Stupka, I.; Heddle, J.G. Artificial Protein Cages—Inspiration, Construction, and Observation. Curr. Opin. Struct. Biol. 2020, 64, 66–73. [Google Scholar] [CrossRef]
- Niemeyer, C.M. Biotechnology Meets Materials Science. Angew. Chem. Int. Ed. 2001, 40, 4128–4158. [Google Scholar] [CrossRef]
- Heddle, J.G.; Chakraborti, S.; Iwasaki, K. Natural and Artificial Protein Cages: Design, Structure and Therapeutic Applications. Curr. Opin. Struct. Biol. 2017, 43, 148–155. [Google Scholar] [CrossRef]
- Giacca, M.; Zacchigna, S. Virus-Mediated Gene Delivery for Human Gene Therapy. J. Control. Release 2012, 161, 377–388. [Google Scholar] [CrossRef]
- Ibraheem, D.; Elaissari, A.; Fessi, H. Gene Therapy and DNA Delivery Systems. Int. J. Pharm. 2014, 459, 70–83. [Google Scholar] [CrossRef]
- Yang, Y.J.; Holmberg, A.L.; Olsen, B.D. Artificially Engineered Protein Polymers. Annu. Rev. Chem. Biomol. Eng. 2017, 8, 549–577. [Google Scholar] [CrossRef]
- Keller, A.; Linko, V. Challenges and Perspectives of DNA Nanostructures in Biomedicine. Angew. Chem. Int. Ed. 2020, 59, 15818–15833. [Google Scholar] [CrossRef]
- Vazquez, E.; Roldán, M.; Diez-Gil, C.; Unzueta, U.; Domingo-Espín, J.; Cedano, J.; Conchillo, O.; Ratera, I.; Veciana, J.; Daura, X.; et al. Protein Nanodisk Assembling and Intracellular Trafficking Powered by an Arginine-Rich (R9) Peptide. Nanomedicine 2010, 5, 259–268. [Google Scholar] [CrossRef]
- Unzueta, U.; Saccardo, P.; Domingo-Espín, J.; Cedano, J.; Conchillo-Solé, O.; García-Fruitós, E.; Céspedes, M.V.; Corchero, J.L.; Daura, X.; Mangues, R.; et al. Sheltering DNA in Self-Organizing, Protein-Only Nano-Shells as Artificial Viruses for Gene Delivery. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 535–541. [Google Scholar] [CrossRef]
- Hernandez-Ggarcia, A.; Werten, M.W.T.; Stuart, M.C.; de Wolf, F.A.; de Vries, R. Coating of Single DNA Molecules by Genetically Engineered Protein Diblock Copolymers. Small 2012, 8, 3491–3501. [Google Scholar] [CrossRef]
- Hernandez-Garcia, A.; Estrich, N.A.; Werten, M.W.T.; van der Maarel, J.R.C.; Labean, T.H.; de Wolf, F.A.; Stuart, M.A.C.; de Vries, R. Precise Coating of a Wide Range of DNA Templates by a Protein Polymer with a DNA Binding Domain. ACS Nano 2017, 11, 144–152. [Google Scholar] [CrossRef]
- Kostiainen, M.A.; Hardy, J.G.; Smith, D.K. High-Affinity Multivalent DNA Binding by Using Low-Molecular-Weight Dendrons. Angew. Chem. Int. Ed. 2005, 44, 2556–2559. [Google Scholar] [CrossRef]
- Kostiainen, M.A.; Szilvay, G.R.; Smith, D.K.; Linder, M.B.; Ikkala, O. Multivalent Dendrons for High-Affinity Adhesion of Proteins to DNA. Angew. Chem. Int. Ed. 2006, 45, 3538–3542. [Google Scholar] [CrossRef]
- Kostiainen, M.A.; Szilvay, G.R.; Lehtinen, J.; Smith, D.K.; Linder, M.B.; Urtti, A.; Ikkala, O. Precisely Defined Protein-Polymer Conjugates: Construction of Synthetic DNA Binding Domains on Proteins by Using Multivalent Dendrons. ACS Nano 2007, 1, 103–113. [Google Scholar] [CrossRef]
- Auvinen, H.; Zhang, H.; Nonappa; Kopilow, A.; Niemelä, E.H.; Nummelin, S.; Correia, A.; Santos, H.A.; Linko, V.; Kostiainen, M.A. Protein Coating of DNA Nanostructures for Enhanced Stability and Immunocompatibility. Adv. Healthc. Mater. 2017, 6, 1700692. [Google Scholar] [CrossRef]
- Daraee, H.; Eatemadi, A.; Abbasi, E.; Aval, S.F.; Kouhi, M.; Akbarzadeh, A. Application of Gold Nanoparticles in Biomedical and Drug Delivery. Artif. Cells Nanomed. Biotechnol. 2016, 44, 410–422. [Google Scholar] [CrossRef]
- Dulińska-Litewka, J.; Łazarczyk, A.; Hałubiec, P.; Szafrański, O.; Karnas, K.; Karewicz, A. Superparamagnetic Iron Oxide Nanoparticles-Current and Prospective Medical Applications. Materials 2019, 12, 617. [Google Scholar] [CrossRef]
- Lynch, I.; Dawson, K.A. Protein-Nanoparticle Interactions. Nanotoday 2008, 3, 40–47. [Google Scholar] [CrossRef]
- Nel, A.E.; Mädler, L.; Velegol, D.; Xia, T.; Hoek, E.M.V.; Somasundaran, P.; Klaessig, F.; Castranova, V.; Thompson, M. Understanding Biophysicochemical Interactions at the Nano-Bio Interface. Nat. Mater. 2009, 8, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Cedervall, T.; Lynch, I.; Lindman, S.; Berggård, T.; Thulin, E.; Nilsson, H.; Dawson, K.A.; Linse, S. Understanding the Nanoparticle-Protein Corona Using Methods to Quantify Exchange Rates and Affinities of Proteins for Nanoparticles. Proc. Natl. Acad. Sci. USA 2007, 104, 2050–2055. [Google Scholar] [CrossRef] [PubMed]
- Cedervall, T.; Lynch, I.; Foy, M.; Berggård, T.; Donnelly, S.C.; Cagney, G.; Linse, S.; Dawson, K.A. Detailed Identification of Plasma Proteins Adsorbed on Copolymer Nanoparticles. Angew. Chem. Int. Ed. 2007, 46, 5754–5756. [Google Scholar] [CrossRef]
- Rocker, C.; Po, M.; Zhang, F.; Parak, W.J.; Nienhaus, G.U. A Quantitative Fluorescence Study of Protein Monolayer Formation on Colloidal Nanoparticles. Nat. Nanotechnol. 2009, 4, 577–580. [Google Scholar] [CrossRef] [PubMed]
- Casals, E.; Pfaller, T.; Duschl, A.; Oostingh, G.J.; Puntes, V. Time Evolution of the Nanoparticle Protein Corona. ACS Nano 2010, 4, 3623–3632. [Google Scholar] [CrossRef]
- Ding, F.; Radic, S.; Chen, R.; Chen, P.; Geitner, N.K.; Brown, J.M.; Ke, P.C. Direct Observation of a Single Nanoparticle-Ubiquitin Corona Formation. Nanoscale 2013, 5, 9162–9169. [Google Scholar] [CrossRef] [PubMed]
- Paoli, S.H.D.; Diduch, L.L.; Tegegn, T.Z.; Orecna, M.; Strader, M.B.; Karnaukhova, E.; Bonevich, J.E.; Holada, K.; Simak, J. The Effect of Protein Corona Composition on the Interaction of Carbon Nanotubes with Human Blood Platelets. Biomaterials 2014, 35, 6182–6194. [Google Scholar] [CrossRef]
- Treuel, L.; Brandholt, S.; Maffre, P.; Wiegele, S.; Shang, L.; Nienhaus, G.U. Impact of Protein Modification on the Protein Corona on Nanoparticles and Nanoparticle-Cell Interactions. ACS Nano 2014, 8, 503–513. [Google Scholar] [CrossRef]
- Ding, H.; Ma, Y. Computer Simulation of the Role of Protein Corona in Cellular Delivery of Nanoparticles. Biomaterials 2014, 35, 8703–8710. [Google Scholar] [CrossRef]
- Duan, G.; Kang, S.; Tian, X.; Garate, J.A.; Zhao, L.; Ge, C.; Zhou, R. Protein Corona Mitigates the Cytotoxicity of Graphene Oxide by Reducing Its Physical Interaction with Cell Membrane. Nanoscale 2015, 7, 15214–15224. [Google Scholar] [CrossRef] [PubMed]
- Duracher, D.; Veyret, R.; Ela, A.; Pichot, C. Adsorption of Bovine Serum Albumin Protein onto Amino-Containing Thermosensitive Core-Shell Latexes. Polym. Int. 2004, 53, 618–626. [Google Scholar] [CrossRef]
- Yan, X.; Kong, J.; Yang, C.; Fu, G. Facile Synthesis of Hairy Core-Shell Structured Magnetic Polymer Submicrospheres and Their Adsorption of Bovine Serum Albumin. J. Colloid Interface Sci. 2015, 445, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Gref, R.; Lück, M.; Quellec, P.; Marchand, M.; Dellacherie, E.; Harnisch, S.; Blunk, T.; Müller, R.H. ‘Stealth’ Corona-Core Nanoparticles Surface Modified by Polyethylene Glycol (PEG): Influences of the Corona (PEG Chain Length and Surface Density) and of the Core Composition on Phagocytic Uptake and Plasma Protein Adsorption. Colloids Surf. B Biointerfaces 2000, 18, 301–313. [Google Scholar] [CrossRef]
- Gossmanna, R.; Fahrländera, E.; Hummel, M.; Mulac, D.; Brockmeyer, J.; Langer, K. Comparative Examination of Adsorption of Serum Proteins on HSA- and PLGA-Based Nanoparticles Using SDS–PAGE and LC–MS. Eur. J. Pharm. Biopharm. 2015, 93, 80–87. [Google Scholar] [CrossRef]
- Meka, V.S.; Sing, M.K.G.; Pichika, M.R.; Nali, S.R.; Kolapalli, V.R.M.; Kesharwani, P. A Comprehensive Review on Polyelectrolyte Complexes. Drug Discov. Today 2017, 22, 1697–1706. [Google Scholar] [CrossRef]
- Siyawamwaya, M.; Choonara, Y.E.; Bijukumar, D.; Kumar, P.; du Toit, L.C.; Pillay, V. A Review: Overview of Novel Polyelectrolyte Complexes as Prospective Drug Bioavailability Enhancers. Int. J. Polym. Mater. Polym. Biomater. 2015, 64, 955–968. [Google Scholar] [CrossRef]
- Vllasaliu, D.; Fowler, R.; Stolnik, S. PEGylated Nanomedicines: Recent Progress and Remaining Concerns. Expert Opin. Drug Deliv. 2014, 11, 139–154. [Google Scholar] [CrossRef]
- Huang, P.S.; Boyken, S.E.; Baker, D. The Coming of Age of de Novo Protein Design. Nature 2016, 537, 320–327. [Google Scholar] [CrossRef]
- Li, L.; Fang, C.J.; Ryan, J.C.; Niemi, E.C.; Lebrón, J.A.; Björkman, P.J.; Arase, H.; Torti, F.M.; Torti, S.V.; Nakamura, M.C.; et al. Binding and Uptake of H-Ferritin Are Mediated by Human Transferrin Receptor-1. Proc. Natl. Acad. Sci. USA 2010, 107, 3505–3510. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Shaukat, A.; Kyllönen, D.; Kostiainen, M.A. Polyelectrolyte Encapsulation and Confinement within Protein Cage-Inspired Nanocompartments. Pharmaceutics 2021, 13, 1551. https://doi.org/10.3390/pharmaceutics13101551
Liu Q, Shaukat A, Kyllönen D, Kostiainen MA. Polyelectrolyte Encapsulation and Confinement within Protein Cage-Inspired Nanocompartments. Pharmaceutics. 2021; 13(10):1551. https://doi.org/10.3390/pharmaceutics13101551
Chicago/Turabian StyleLiu, Qing, Ahmed Shaukat, Daniella Kyllönen, and Mauri A. Kostiainen. 2021. "Polyelectrolyte Encapsulation and Confinement within Protein Cage-Inspired Nanocompartments" Pharmaceutics 13, no. 10: 1551. https://doi.org/10.3390/pharmaceutics13101551
APA StyleLiu, Q., Shaukat, A., Kyllönen, D., & Kostiainen, M. A. (2021). Polyelectrolyte Encapsulation and Confinement within Protein Cage-Inspired Nanocompartments. Pharmaceutics, 13(10), 1551. https://doi.org/10.3390/pharmaceutics13101551