Correlation between Biophysical Properties of Niosomes Elaborated with Chloroquine and Different Tensioactives and Their Transfection Efficiency
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Niosomes and Nioplexes
2.2. Plasmid Propagation
2.3. Nano DSC Studies
2.4. Rheological Studies
2.5. Morphology, Size, Dispersity and Superficial Charge
2.6. Nioplexes Membrane Packing Studies
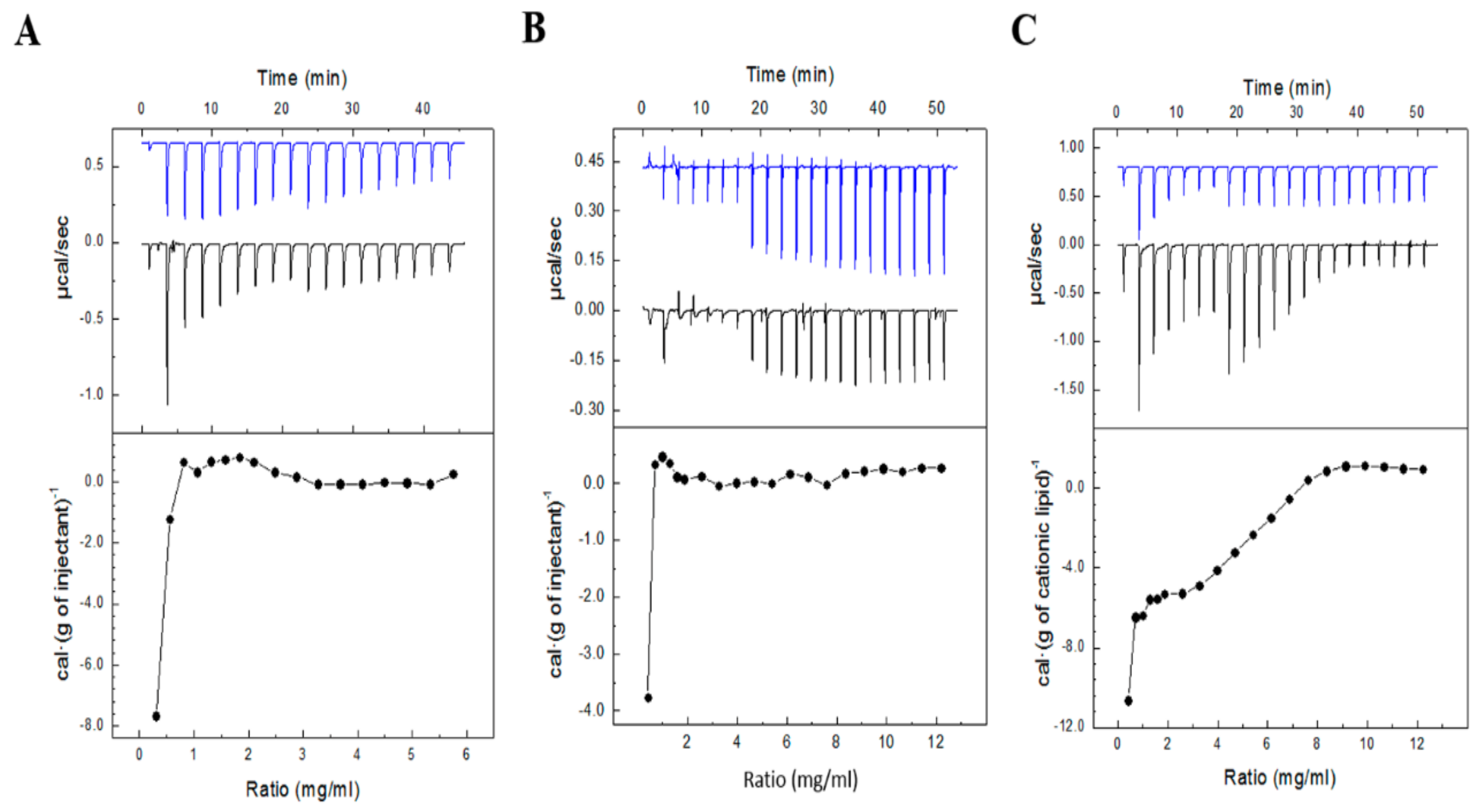
2.7. ITC Studies
2.8. DNA Release from Niosomes and Protection Capacity
2.9. Buffer Capacity Assay
2.10. Vulnerability Assay of Complexes in the Late Endosome
2.11. Cell Culture and Transfection Assays
2.12. Analysis of EGFP Expression and Cell Viability
2.13. Statistical Analysis
3. Results
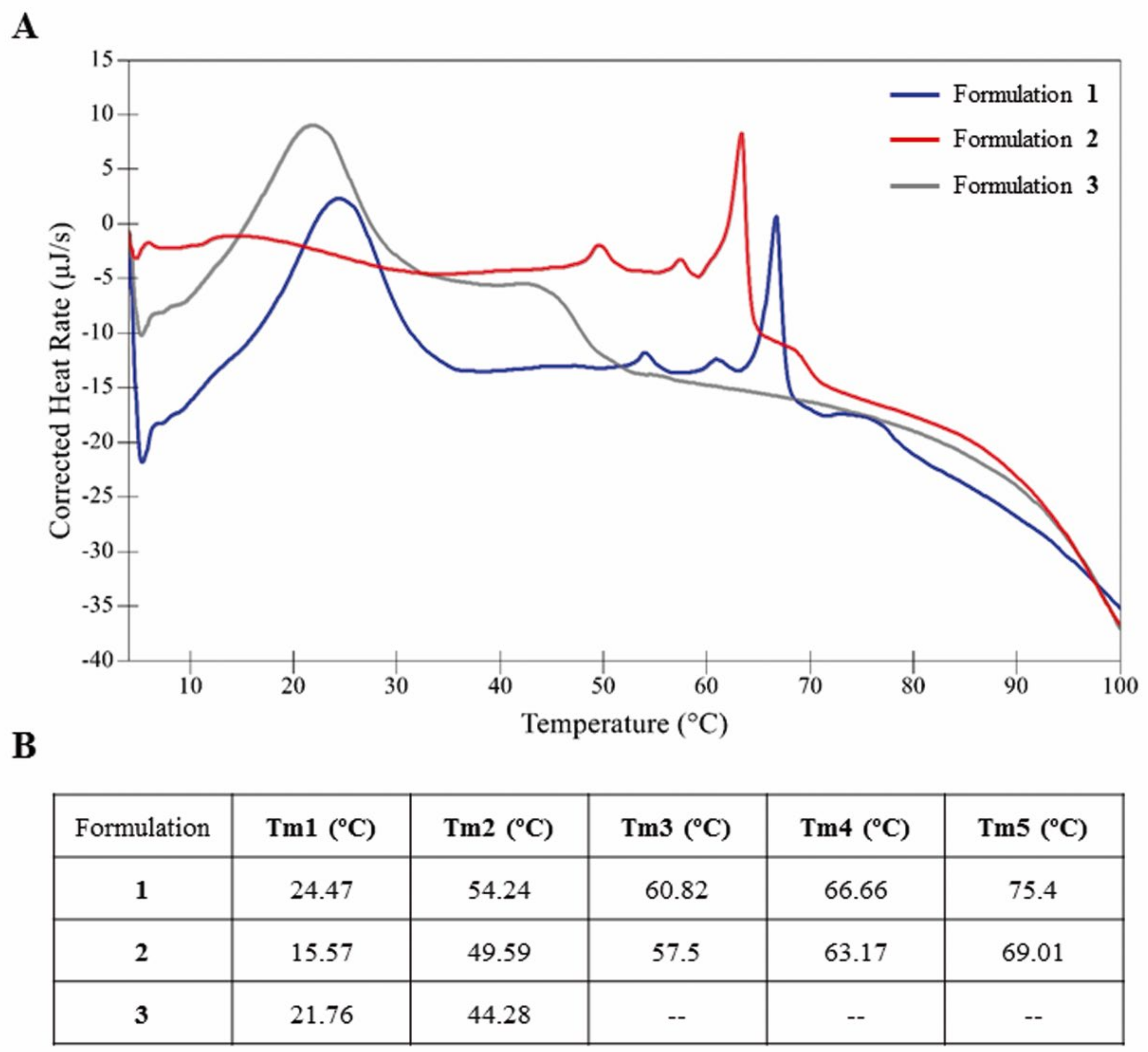
3.1. Characterization of the Thermostability of Niosomes
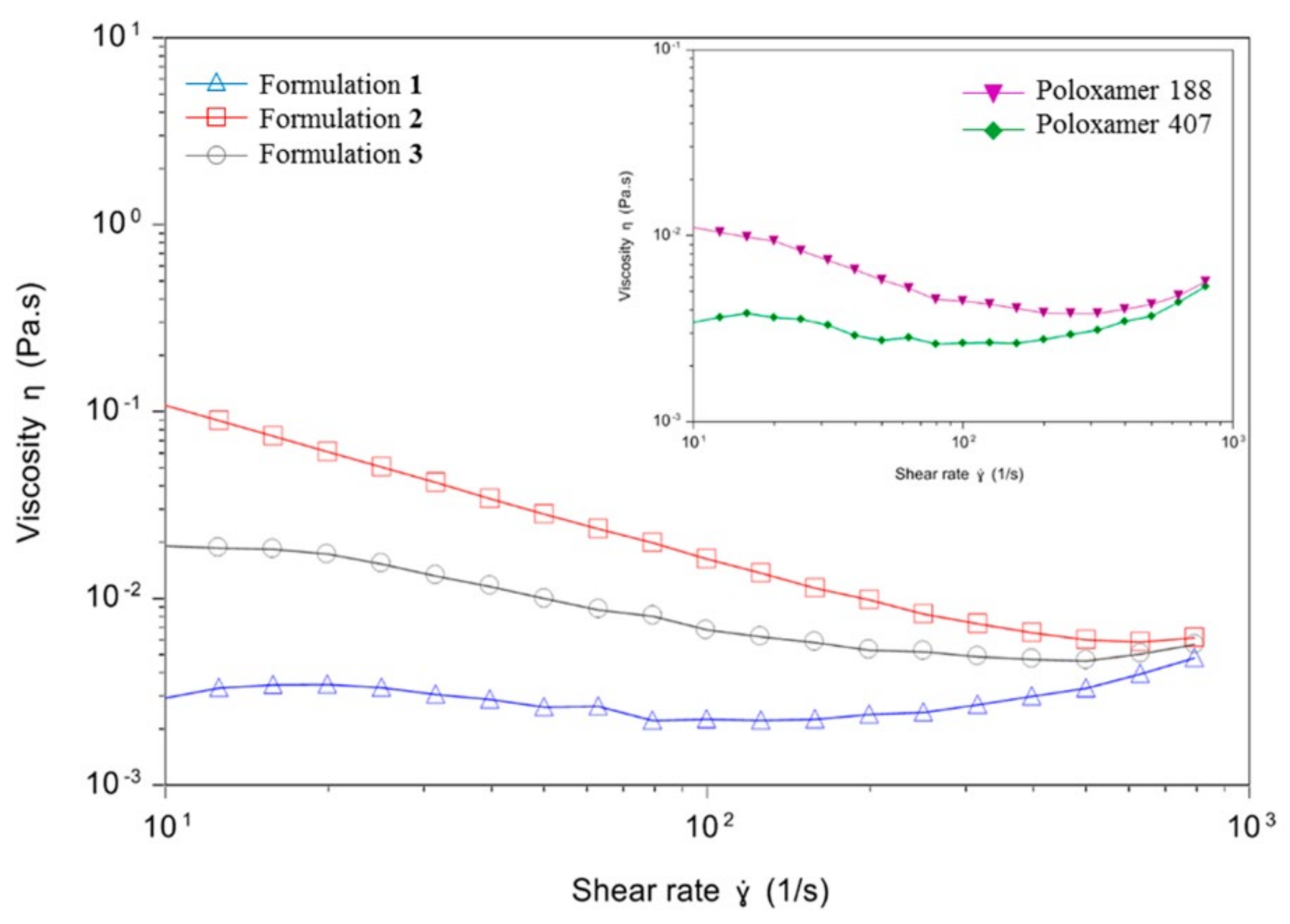
3.2. Rheological Properties of Niosomes
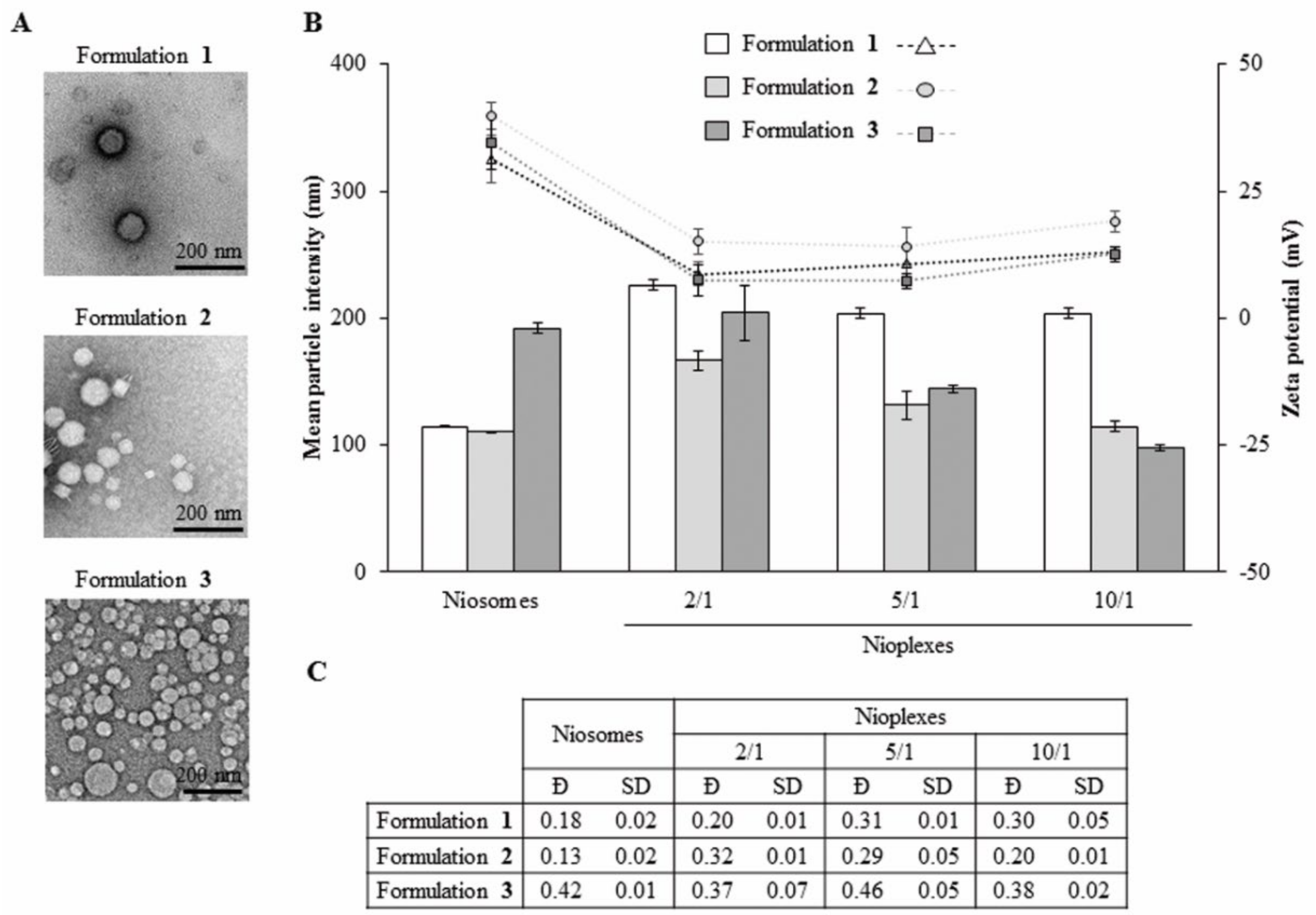
3.3. Morphology, Size, Dispersity and Superficial Charge
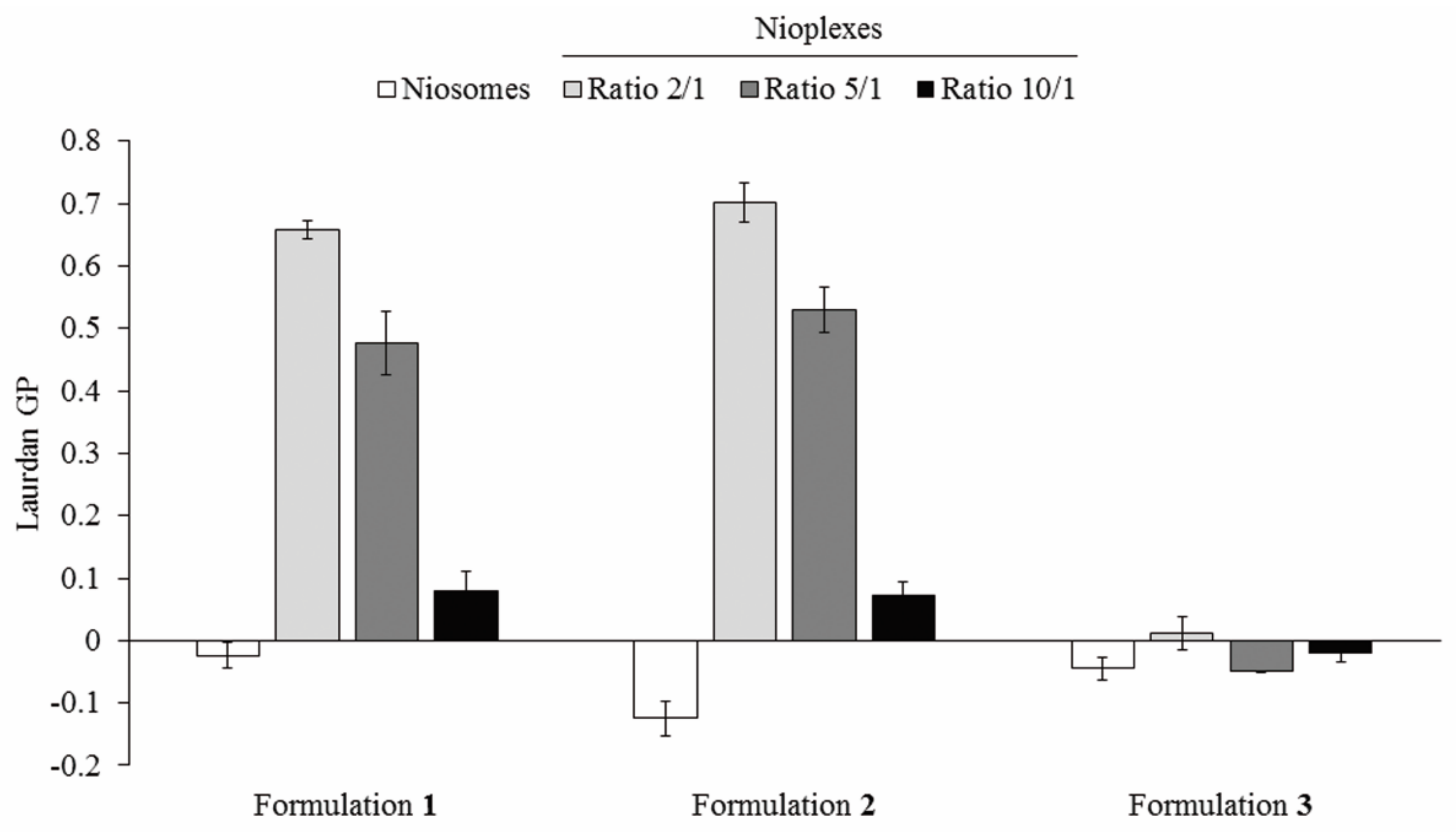
3.4. Nioplexes Membrane Packing Studies
3.5. Evaluation of Niosome-pEGFP Interactions by ITC
3.6. Buffer Capacity and Endosomal Escape of Nioplexes
3.7. Cell Viability and Transfection Efficiency of Nioplexes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- U.S. Food and Drug Administration. What Is Gene Therapy? Available online: https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/what-gene-therapy (accessed on 30 September 2020).
- Dunbar, C.E.; High, K.A.; Joung, J.K.; Kohn, D.B.; Ozawa, K.; Sadelain, M. Gene therapy comes of age. Science 2018, 359, eaan4672. [Google Scholar] [CrossRef] [Green Version]
- Ramamoorth, M.; Narvekar, A. Non viral vectors in gene therapy—An overview. J. Clin. Diagn. Res. 2015, 9, GE01–GE06. [Google Scholar] [CrossRef] [PubMed]
- Gallego, I.; Villate-Beitia, I.; Martínez-Navarrete, G.; Menéndez, M.; López-Méndez, T.; Soto-Sánchez, C.; Zárate, J.; Puras, G.; Fernández, E.; Pedraz, J.L. Non-viral vectors based on cationic niosomes and minicircle DNA technology enhance gene delivery efficiency for biomedical applications in retinal disorders. Nanomedicine 2019, 17, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Ojeda, E.; Puras, G.; Agirre, M.; Zárate, J.; Grijalvo, S.; Pons, R.; Eritja, R.; Martinez-Navarrete, G.; Soto-Sanchez, C.; Fernández, E.; et al. Niosomes based on synthetic cationic lipids for gene delivery: The influence of polar head-groups on the transfection efficiency in HEK-293, ARPE-19 and MSC-D1 cells. Org. Biomol. Chem. 2015, 13, 1068–1081. [Google Scholar] [CrossRef] [Green Version]
- Puras, G.; Mashal, M.; Zárate, J.; Agirre, M.; Ojeda, E.; Grijalvo, S.; Eritja, R.; Diaz-Tahoces, A.; Martínez Navarrete, G.; Avilés-Trigueros, M.; et al. A novel cationic niosome formulation for gene delivery to the retina. J. Control. Release 2014, 174, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Villate-Beitia, I.; Gallego, I.; Martínez-Navarrete, G.; Zárate, J.; López-Méndez, T.; Soto-Sánchez, C.; Santos-Vizcaíno, E.; Puras, G.; Fernández, E.; Pedraz, J.L. Polysorbate 20 non-ionic surfactant enhances retinal gene delivery efficiency of cationic niosomes after intravitreal and subretinal administration. Int. J. Pharm. 2018, 550, 388–397. [Google Scholar] [CrossRef]
- Zhang, H. Onivyde for the therapy of multiple solid tumors. OncoTargets Ther. 2016, 9, 3001–3007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoy, S.M. Patisiran: First global approval. Drugs 2018, 78, 1625–1631. [Google Scholar] [CrossRef]
- Sharma, O.; Sultan, A.A.; Ding, H.; Triggle, C.R. A review of the progress and challenges of developing a vaccine for COVID-19. Front. Immunol. 2020, 11, 585354. [Google Scholar] [CrossRef]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef]
- Uchegbu, I.F.; Vyas, S.P. Non-ionic surfactant based vesicles (Niosomes) in drug delivery. Int. J. Pharm. 1998, 172, 33–70. [Google Scholar] [CrossRef]
- Huang, Y.; Rao, Y.; Chen, J.; Yang, V.C.; Liang, W. Polysorbate cationic synthetic vesicle for gene delivery. J. Biomed. Mater. Res. A 2011, 96, 513–519. [Google Scholar] [CrossRef] [Green Version]
- Bartelds, R.; Nematollahi, M.H.; Pols, T.; Stuart, M.C.A.; Pardakhty, A.; Asadikaram, G.; Poolman, B. Niosomes, an alternative for liposomal delivery. PLoS ONE 2018, 13, e0194179. [Google Scholar]
- Ling Yeo, P.; Ling Lim, C.; Moi Chye, S.; Kiong Ling, A.P.; Yiah Koh, R. Niosomes: A review of their structure, properties, methods of preparation and medical applications. Asian Biomed. 2018, 11, 301–314. [Google Scholar]
- Li, M.; Du, C.; Guo, N.; Teng, Y.; Meng, X.; Sun, H.; Li, S.; Yu, P.; Galons, H. Composition design and medical application of liposomes. Eur. J. Med. Chem. 2019, 164, 640–653. [Google Scholar] [CrossRef] [PubMed]
- Al Qtaish, N.; Gallego, I.; Villate-Beitia, I.; Sainz-Ramos, M.; López-Méndez, T.B.; Grijalvo, S.; Eritja, R.; Soto-Sánchez, C.; Martínez-Navarrete, G.; Fernández, E.; et al. Niosome-based approach for in situ gene delivery to retina and brain cortex as immune-privileged tissues. Pharmaceutics 2020, 12, 198. [Google Scholar] [CrossRef] [Green Version]
- Ojeda, E.; Puras, G.; Agirre, M.; Zarate, J.; Grijalvo, S.; Eritja, R.; Martinez-Navarrete, G.; Soto-Sánchez, C.; Diaz-Tahoces, A.; Aviles-Trigueros, M.; et al. The influence of the polar head-group of synthetic cationic lipids on the transfection efficiency mediated by niosomes in rat retina and brain. Biomaterials 2016, 77, 267–279. [Google Scholar] [CrossRef] [Green Version]
- Ojeda, E.; Puras, G.; Agirre, M.; Zarate, J.; Grijalvo, S.; Eritja, R.; DiGiacomo, L.; Caracciolo, G.; Pedraz, J. The role of helper lipids in the intracellular disposition and transfection efficiency of niosome formulations for gene delivery to retinal pigment epithelial cells. Int. J. Pharm. 2016, 503, 115–126. [Google Scholar] [CrossRef] [Green Version]
- Zhi, D.; Zhang, S.; Cui, S.; Zhao, Y.; Wang, Y.; Zhao, D. The headgroup evolution of cationic lipids for gene delivery. Bioconj. Chem. 2013, 24, 487–519. [Google Scholar] [CrossRef] [PubMed]
- Carrière, M.; Tranchant, I.; Niore, P.; Byk, G.; Mignet, N.; Escriou, V.; Scherman, D.; Herscovici, J. Optimization of cationic lipid mediated gene transfer: Structure-function, physico-chemical, and cellular studies. J. Liposome Res. 2002, 12, 95–106. [Google Scholar] [CrossRef]
- Zhi, D.; Bai, Y.; Yang, J.; Cui, S.; Zhao, Y.; Chen, H.; Zhang, S. A review on cationic lipids with different linkers for gene delivery. Adv. Colloid Interface Sci. 2018, 253, 117–140. [Google Scholar] [CrossRef]
- Kim, B.; Hwang, G.; Seu, Y.; Choi, J.; Jin, K.S.; Doh, K. DOTAP/DOPE ratio and cell type determine transfection efficiency with DOTAP-liposomes. Biochim. Biophys. Acta 2015, 1848, 1996–2001. [Google Scholar] [CrossRef] [Green Version]
- Abou-Taleb, H.A.; Khallaf, R.A.; Abdel-Aleem, J.A. Intranasal niosomes of nefopam with improved bioavailability: Preparation, optimization, and in-vivo evaluation. Drug Des. Dev. Ther. 2018, 12, 3501–3516. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Mageed, H.M.; Fahmy, A.S.; Shaker, D.S.; Mohamed, S.A. Development of novel delivery system for nanoencapsulation of catalase: Formulation, characterization, and in vivo evaluation using oxidative skin injury model. Artif. Cells Nanomed. Biotechnol. 2018, 46, 362–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manosroi, A.; Wongtrakul, P.; Manosroi, J.; Sakai, H.; Sugawara, F.; Yuasa, M.; Abe, M. Characterization of Vesicles Prepared with various non-ionic surfactants mixed with cholesterol. Colloids Surf. B Biointerfaces 2003, 30, 129–138. [Google Scholar] [CrossRef]
- Junquera, E.; Aicart, E. Recent progress in gene therapy to deliver nucleic acids with multivalent cationic vectors. Adv. Colloid Interface Sci. 2016, 233, 161–175. [Google Scholar] [CrossRef]
- Grijalvo, S.; Puras, G.; Zárate, J.; Sainz-Ramos, M.; Qtaish, N.A.L.; López, T.; Mashal, M.; Attia, N.; Díaz, D.; Pons, R.; et al. Cationic niosomes as non-viral vehicles for nucleic acids: Challenges and opportunities in gene delivery. Pharmaceutics 2019, 11, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mashal, M.; Attia, N.; Puras, G.; Martínez-Navarrete, G.; Fernández, E.; Pedraz, J.L. Retinal gene delivery enhancement by lycopene incorporation into cationic niosomes based on DOTMA and polysorbate. J. Control. Release 2017, 254, 55–64. [Google Scholar] [CrossRef]
- Mashal, M.; Attia, N.; Martínez-Navarrete, G.; Soto-Sánchez, C.; Fernández, E.; Grijalvo, S.; Eritja, R.; Puras, G.; Pedraz, J.L. Gene delivery to the rat retina by non-viral vectors based on chloroquine-containing cationic niosomes. J. Control. Release 2019, 304, 181–190. [Google Scholar] [CrossRef]
- Rajera, R.; Nagpal, K.; Singh, S.K.; Mishra, D.N. Niosomes: A controlled and novel drug delivery system. Biol. Pharm. Bull. 2011, 34, 945–953. [Google Scholar] [CrossRef] [Green Version]
- Tassler, S.; Pawlowska, D.; Janich, C.; Dobner, B.; Wölk, C.; Brezesinski, G. Lysine-based amino-functionalized lipids for gene transfection: The influence of the chain composition on 2D properties. Phys. Chem. Chem. Phys. 2018, 20, 6936–6944. [Google Scholar] [CrossRef] [Green Version]
- Adler-Moore, J.; Proffitt, R.T. AmBisome: Liposomal formulation, structure, mechanism of action and pre-clinical experience. J. Antimicrob. Chemother. 2002, 49 (Suppl. S1), 21–30. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Hanning, S.; Falconer, J.; Locke, M.; Wen, J. Recent advances in non-ionic surfactant vesicles (Niosomes): Fabrication, characterization, pharmaceutical and cosmetic applications. Eur. J. Pharm. Biopharm. 2019, 144, 18–39. [Google Scholar] [CrossRef] [Green Version]
- Mahale, N.B.; Thakkar, P.D.; Mali, R.G.; Walunj, D.R.; Chaudhari, S.R. Niosomes: Novel sustained release nonionic stable vesicular systems—An overview. Adv. Colloid Interface Sci. 2012, 183–184, 46–54. [Google Scholar]
- Ojeda, E.; Agirre, M.; Villate-Beitia, I.; Mashal, M.; Puras, G.; Zarate, J.; Pedraz, J.L. Elaboration and physicochemical characterization of niosome-based nioplexes for gene delivery purposes. Methods Mol. Biol. 2016, 1445, 63–75. [Google Scholar] [PubMed]
- Grijalvo, S.; Ocampo, S.M.; Perales, J.C.; Eritja, R. Synthesis of lipid–oligonucleotide conjugates for RNA interference studies. Chem. Biodivers. 2011, 8, 287–299. [Google Scholar] [CrossRef] [Green Version]
- Parasassi, T.; De Stasio, G.; d’Ubaldo, A.; Gratton, E. Phase fluctuation in phospholipid membranes revealed by laurdan fluorescence. Biophys. J. 1990, 57, 1179–1186. [Google Scholar] [CrossRef] [Green Version]
- Agirre, M.; Zarate, J.; Puras, G.; Ojeda, E.; Pedraz, J.L. Improving transfection efficiency of ultrapure oligochitosan/DNA polyplexes by medium acidification. Drug Deliv. 2015, 22, 100–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mochizuki, S.; Kanegae, N.; Nishina, K.; Kamikawa, Y.; Koiwai, K.; Masunaga, H.; Sakurai, K. The role of the helper lipid Dioleoylphosphatidylethanolamine (DOPE) for DNA transfection cooperating with a cationic lipid bearing ethylenediamine. Biochim. Biophys. Acta (BBA) Biomembr. 2013, 1828, 412–418. [Google Scholar] [CrossRef] [Green Version]
- Agirre, M.; Ojeda, E.; Zarate, J.; Puras, G.; Grijalvo, S.; Eritja, R.; García del Caño, G.; Barrondo, S.; González-Burguera, I.; López de Jesús, M.; et al. New insights into gene delivery to human neuronal precursor NT2 cells: A comparative study between lipoplexes, nioplexes, and polyplexes. Mol. Pharm. 2015, 12, 4056–4066. [Google Scholar] [CrossRef] [Green Version]
- Erbacher, P.; Roche, A.C.; Monsigny, M.; Midoux, P. Putative role of chloroquine in gene transfer into a human hepatoma cell line by DNA/Lactosylated Polylysine complexes. Exp. Cell Res. 1996, 225, 186–194. [Google Scholar] [CrossRef]
- Pippa, N.; Demetzos, C.; Pispas, S. Drug Delivery Nanosystems: From Bioinspiration and Biomimetics to Clinical Applications, 1st ed.; Jenny Stanford Publishing: Singapore, 2019; p. 418. [Google Scholar]
- Olson, J.A.; Schwartz, J.A.; Hahka, D.; Nguyen, N.; Bunch, T.; Jensen, G.M.; Adler-Moore, J.P. Toxicity and efficacy differences between liposomal amphotericin B formulations in uninfected and aspergillus fumigatus infected mice. Med. Mycol. 2015, 53, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Azanza, J.R.; Sádada, B.; Reis, J. Liposomal formulations of amphotericin B: Differences according to the scientific evidence. Rev. Esp. Quim. 2015, 28, 275–281. [Google Scholar]
- Olson, J.A.; Adler-Moore, J.P.; Jensen, G.M.; Schwartz, J.; Dignani, M.C.; Proffitt, R.T. Comparison of the physicochemical, antifungal, and toxic properties of two liposomal amphotericin B products. Antimicrob. Agents Chemother. 2008, 52, 259–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Committee for Human Medicinal Products. Reflection Paper on the Data Requirements for Intravenous Liposomal Products Developed with Reference to an Innovator Liposomal Product; European Medicines Agency: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Bobo, D.; Robinson, K.J.; Islam, J.; Thurecht, K.J.; Corrie, S.R. Nanoparticle-based medicines: A review of FDA-approved materials and clinical trials to date. Pharm. Res. 2016, 33, 2373–2387. [Google Scholar] [CrossRef]
- Savla, R.; Browne, J.; Plassat, V.; Wasan, K.M.; Wasan, E.K. Review and analysis of FDA approved drugs using lipid-based formulations. Drug Dev. Ind. Pharm. 2017, 43, 1743–1758. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, G.P.; Sesma, J.Z.; Díez, M.A.; Díaz-Tahoces, A.; Avilés-Trigeros, M.; Grijalvo, S.; Eritja, R.; Fernández, E.; Pedraz, J.L. A novel formulation based on 2,3-Di(Tetradecyloxy)Propan-1-Amine Cationic lipid combined with polysorbate 80 for efficient gene delivery to the retina. Pharm. Res. 2014, 31, 1665–1675. [Google Scholar] [CrossRef]
- Villate-Beitia, I.; Truong, N.F.; Gallego, I.; Zárate, J.; Puras, G.; Pedraz, J.L.; Segura, T. Hyaluronic acid hydrogel scaffolds loaded with cationic niosomes for efficient non-viral gene delivery. RSC Adv. 2018, 8, 31934–31942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Yang, J.; Huang, L.; Liu, D. Effect of non-ionic surfactants on the formation of DNA/emulsion complexes and emulsion-mediated gene transfer. Pharm. Res. 1996, 13, 1642–1646. [Google Scholar] [CrossRef]
- Dong, H.; Qin, Y.; Huang, Y.; Ji, D.; Wu, F. Poloxamer 188 Rescues MPTP-induced lysosomal membrane integrity impairment in cellular and mouse models of Parkinson’s disease. Neurochem. Int. 2019, 126, 178–186. [Google Scholar] [CrossRef]
- Moloughney, J.G.; Weisleder, N. Poloxamer 188 (P188) as a membrane resealing reagent in biomedical applications. Recent Pat. Biotechnol. 2012, 6, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Attia, N.; Mashal, M.; Soto-Sánchez, C.; Martínez-Navarrete, G.; Fernández, E.; Grijalvo, S.; Eritja, R.; Puras, G.; Pedraz, J.L. Gene transfer to rat cerebral cortex mediated by polysorbate 80 and poloxamer 188 nonionic surfactant vesicles. Drug Des. Devel. Ther. 2018, 12, 3937–3949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sriadibhatla, S.; Yang, Z.; Gebhart, C.; Alakhov, V.Y.; Kabanov, A. Transcriptional activation of gene expression by pluronic block copolymers in stably and transiently transfected cells. Mol. Ther. 2006, 13, 804–813. [Google Scholar] [CrossRef] [PubMed]
- Dumortier, G.; Grossiord, J.L.; Agnely, F.; Chaumeil, J.C. A review of poloxamer 407 pharmaceutical and pharmacological characteristics. Pharm. Res. 2006, 23, 2709–2728. [Google Scholar] [CrossRef]
- Minnelli, C.; Moretti, P.; Fulgenzi, G.; Mariani, P.; Laudadio, E.; Armeni, T.; Galeazzi, R.; Mobbili, G. A poloxamer-407 modified liposome encapsulating epigallocatechin-3-gallate in the presence of magnesium: Characterization and protective effect against oxidative damage. Int. J. Pharm. 2018, 552, 225–234. [Google Scholar] [CrossRef]
- Shu, H.; Zhang, Y.; Zhang, M.; Wu, J.; Cui, M.; Liu, K.; Wang, J. Addition of free poloxamer 407 to a new gene vector P407-PEI-K12 solution forms a sustained-release in situ hypergel that enhances cell transfection and extends gene expression. Oncol. Lett. 2019, 17, 3085–3096. [Google Scholar] [CrossRef] [Green Version]
- Sorensen, M.; Sehested, M.; Jensen, P.B. pH-Dependent regulation of camptothecin-induced cytotoxicity and cleavable complex formation by the antimalarial agent chloroquine. Biochem. Pharmacol. 1997, 54, 373–380. [Google Scholar] [CrossRef]
- Allison, J.L.; O’Brien, R.L.; Hahn, F.E. DNA: Reaction with chloroquine. Science 1965, 149, 1111–1113. [Google Scholar] [CrossRef]
- Gallego, I.; Villate-Beitia, I.; Soto-Sánchez, C.; Menendez, M.; Grijalvo, S.; Eritja, R.; Martinez-Navarrete, G.; Humphreys, L.; Lopez-Mendez, T.; Puras, G.; et al. Brain Angiogenesis induced by non-viral gene therapy with potential therapeutic benefits for central nervous system diseases. Mol. Pharm. 2020, 17, 1848–1858. [Google Scholar] [CrossRef] [PubMed]
- Almulathanon, A.A.Y.; Ranucci, E.; Ferruti, P.; Garnett, M.C.; Bosquillon, C. Comparison of gene transfection and cytotoxicity mechanisms of linear Poly(Amidoamine) and branched Poly(Ethyleneimine) polyplexes. Pharm. Res. 2018, 35, 86. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zeidan, R.; Mishra, S.; Liu, A.; Pun, S.H.; Kulkarni, R.P.; Jensen, G.S.; Bellocq, N.C.; Davis, M.E. Structure-function correlation of chloroquine and analogues as transgene expression enhancers in nonviral gene delivery. J. Med. Chem. 2006, 49, 6522–6531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarro, M.; Castro, W.; Higuera-Padilla, A.R.; Sierraalta, A.; Abad, M.J.; Taylor, P.; Sánchez-Delgado, R.A. Synthesis, characterization and biological activity of trans-Platinum(II) complexes with chloroquine. J. Inorg. Biochem. 2011, 105, 1684–1691. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.; Lee, J.J.; Yang, H.; Baek, S.; Kim, S.; Kim, S.; Lee, T.; Song, D.; Park, G. Evaluation of similar quality attribute characteristics in SB5 and reference product of adalimumab. MAbs 2019, 11, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Bunjes, H.; Unruh, T. Characterization of lipid nanoparticles by differential scanning calorimetry, X-Ray and Neutron scattering. Adv. Drug Deliv. Rev. 2007, 59, 379–402. [Google Scholar] [CrossRef]
- Chiu, M.H.; Prenner, E.J. Differential scanning calorimetry: An Invaluable tool for a detailed thermodynamic characterization of macromolecules and their interactions. J. Pharm. Bioallied Sci. 2011, 3, 39–59. [Google Scholar] [PubMed]
- Vicente-Pascual, M.; Gómez-Aguado, I.; Rodríguez-Castejón, J.; Rodríguez-Gascón, A.; Muntoni, E.; Battaglia, L.; del Pozo-Rodríguez, A.; Solinís Aspiazu, M.Á. Topical administration of SLN-Based gene therapy for the treatment of corneal inflammation by De Novo IL-10 production. Pharmaceutics 2020, 12, 584. [Google Scholar] [CrossRef]
- Denn, M.M.; Morris, J.F. Rheology of non-brownian suspensions. Annu. Rev. Chem. Biomol. Eng. 2014, 5, 203–228. [Google Scholar] [CrossRef]
- Dos Santos, T.; Varela, J.; Lynch, I.; Salvati, A.; Dawson, K.A. Quantitative assessment of the comparative nanoparticle-uptake efficiency of a range of cell lines. Small 2011, 7, 3341–3349. [Google Scholar] [CrossRef] [Green Version]
- Hosseinkhani, H.; Tabata, Y. Self assembly of DNA nanoparticles with polycations for the delivery of genetic materials into cells. J. Nanosci. Nanotechnol. 2006, 6, 2320–2328. [Google Scholar] [CrossRef]
- Caracciolo, G.; Amenitsch, H. Cationic liposome/DNA complexes: From structure to interactions with cellular membranes. Eur. Biophys. J. 2012, 41, 815–829. [Google Scholar] [CrossRef]
- Paecharoenchai, O.; Niyomtham, N.; Ngawhirunpat, N.; Rojanarata, T.; Yingyongnarongkul, B.; Opanasopit, P. Cationic niosomes composed of spermine-based cationic lipids mediate high gene transfection efficiency. J. Drug Target. 2012, 20, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, H.; Lingwood, D.; Levental, I.; Sampaio, J.L.; Kalvodova, L.; Rajendran, L.; Simons, K. Order of lipid phases in model and plasma membranes. Proc. Natl. Acad. Sci. USA 2009, 106, 16645–16650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dopierała, K.; Karwowska, K.; Petelska, A.D.; Prochaska, K. Thermodynamic, viscoelastic and electrical properties of lipid membranes in the presence of astaxanthin. Biophys. Chem. 2020, 258, 106318. [Google Scholar] [CrossRef]
- Shao, X.; Wei, X.; Song, X.; Hao, L.; Cai, X.; Zhang, Z.; Peng, Q.; Lin, Y. Independent effect of polymeric nanoparticle zeta potential/surface charge, on their cytotoxicity and affinity to cells. Cell Prolif. 2015, 48, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Lam, J.K.W. Endosomal Escape Pathways for Non-Viral Nucleic Acid Delivery Systems.; IntechOpen: London, UK, 2012. [Google Scholar]
- Varkouhi, A.K.; Scholte, M.; Storm, G.; Haisma, H.J. Endosomal escape pathways for delivery of biologicals. J. Control. Release 2011, 151, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.A.; Selby, L.I.; Johnston, A.P.R.; Such, G.K. The endosomal escape of nanoparticles: Toward more efficient cellular delivery. Bioconjug. Chem. 2019, 30, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Ong, V.; Mei, V.; Cao, L.; Lee, K.; Chung, E.J. Nanomedicine for cystic fibrosis. SLAS Technol. Transl. Life Sci. Innov. 2019, 24, 169–180. [Google Scholar] [CrossRef]
- Baradaran Eftekhari, R.; Maghsoudnia, N.; Dorkoosh, F.A. Chloroquine: A brand-new scenario for an old drug. Expert Opin. Drug Deliv. 2020, 17, 275–277. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sainz-Ramos, M.; Villate-Beitia, I.; Gallego, I.; AL Qtaish, N.; Menéndez, M.; Lagartera, L.; Grijalvo, S.; Eritja, R.; Puras, G.; Pedraz, J.L. Correlation between Biophysical Properties of Niosomes Elaborated with Chloroquine and Different Tensioactives and Their Transfection Efficiency. Pharmaceutics 2021, 13, 1787. https://doi.org/10.3390/pharmaceutics13111787
Sainz-Ramos M, Villate-Beitia I, Gallego I, AL Qtaish N, Menéndez M, Lagartera L, Grijalvo S, Eritja R, Puras G, Pedraz JL. Correlation between Biophysical Properties of Niosomes Elaborated with Chloroquine and Different Tensioactives and Their Transfection Efficiency. Pharmaceutics. 2021; 13(11):1787. https://doi.org/10.3390/pharmaceutics13111787
Chicago/Turabian StyleSainz-Ramos, Myriam, Ilia Villate-Beitia, Idoia Gallego, Nuseibah AL Qtaish, Margarita Menéndez, Laura Lagartera, Santiago Grijalvo, Ramón Eritja, Gustavo Puras, and José Luis Pedraz. 2021. "Correlation between Biophysical Properties of Niosomes Elaborated with Chloroquine and Different Tensioactives and Their Transfection Efficiency" Pharmaceutics 13, no. 11: 1787. https://doi.org/10.3390/pharmaceutics13111787
APA StyleSainz-Ramos, M., Villate-Beitia, I., Gallego, I., AL Qtaish, N., Menéndez, M., Lagartera, L., Grijalvo, S., Eritja, R., Puras, G., & Pedraz, J. L. (2021). Correlation between Biophysical Properties of Niosomes Elaborated with Chloroquine and Different Tensioactives and Their Transfection Efficiency. Pharmaceutics, 13(11), 1787. https://doi.org/10.3390/pharmaceutics13111787









