Anticancer Potential of Green Synthesized Silver Nanoparticles of the Soft Coral Cladiella pachyclados Supported by Network Pharmacology and In Silico Analyses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Marine Soft Coral
2.2. Preparation of the Organic Extract
2.3. LC-HRESIMS Chemical Profiling
2.4. Preparation of Silver Nanoparticles
2.5. Characterization of Silver Nanoparticles
2.5.1. UV Spectroscopy
2.5.2. X-ray Diffraction (XRD) Studies
2.5.3. Fourier Transform Infrared Spectroscopy (FTIR)
2.5.4. Transmission Electron Microscopy Analysis (TEM)
2.5.5. Scanning Electron Microscope (SEM)
2.6. Antiproliferative Assay
2.7. In Silico and Network Pharmacology Study
2.7.1. In Silico ADME Profiling
2.7.2. Anti-Breast Cancer Activity Predictions
2.7.3. Target Proteins of Breast Cancer
2.7.4. Determination of the Potential Protein Targets of the Annotated Compounds
2.7.5. Molecular Dynamic Simulation and Binding Free Energy Calculation
2.7.6. Networks Construction and Functional Enrichment Analysis
2.8. Statistical Analysis
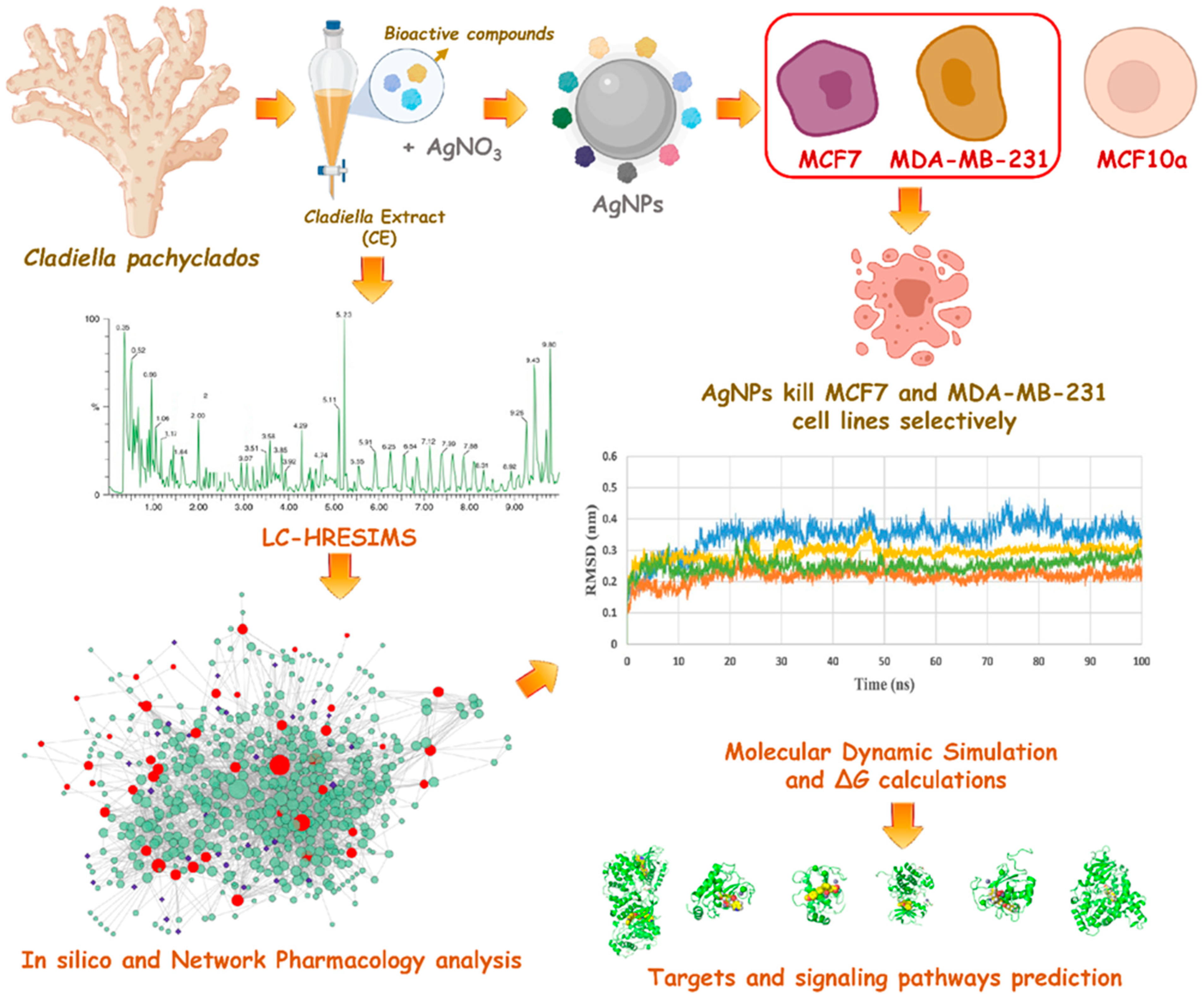
3. Results
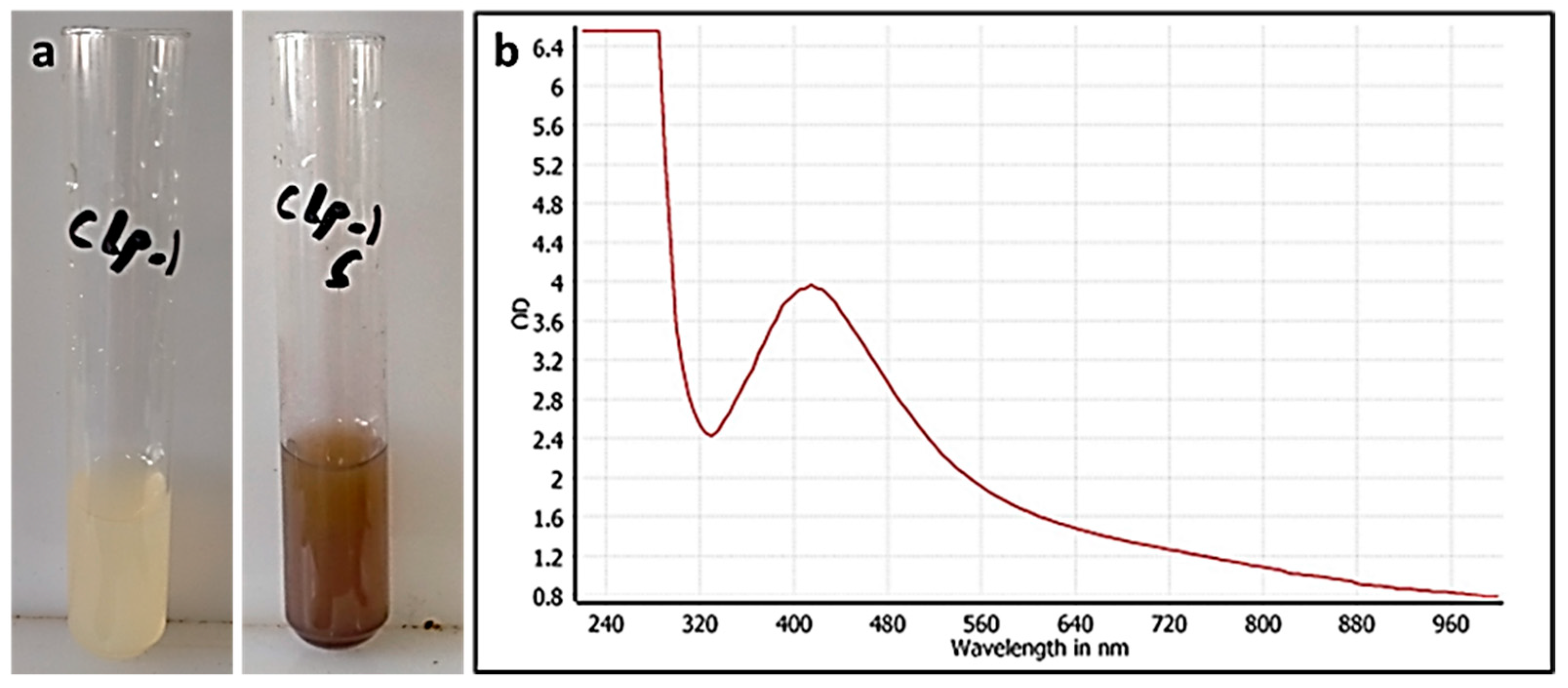
3.1. CE-Mediated Green Biosynthesis of Silver Nanoparticles
3.2. Characterization of the Prepared AgNPs
3.2.1. UV Spectroscopy
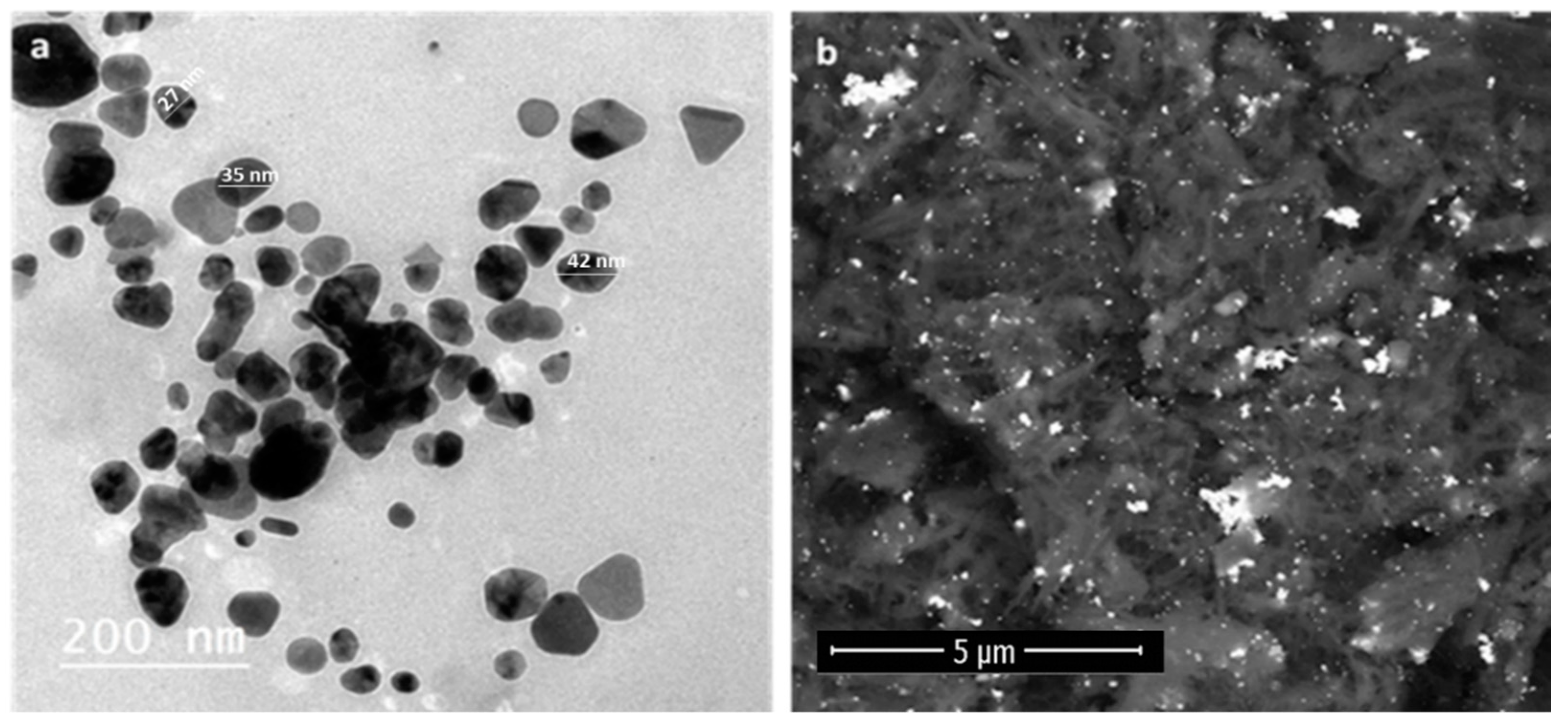
3.2.2. Electron Microscopy
3.2.3. X-ray Powder Diffraction (XRD)
3.2.4. Fourier Transform Infrared Spectroscopy Analysis (FTIR)
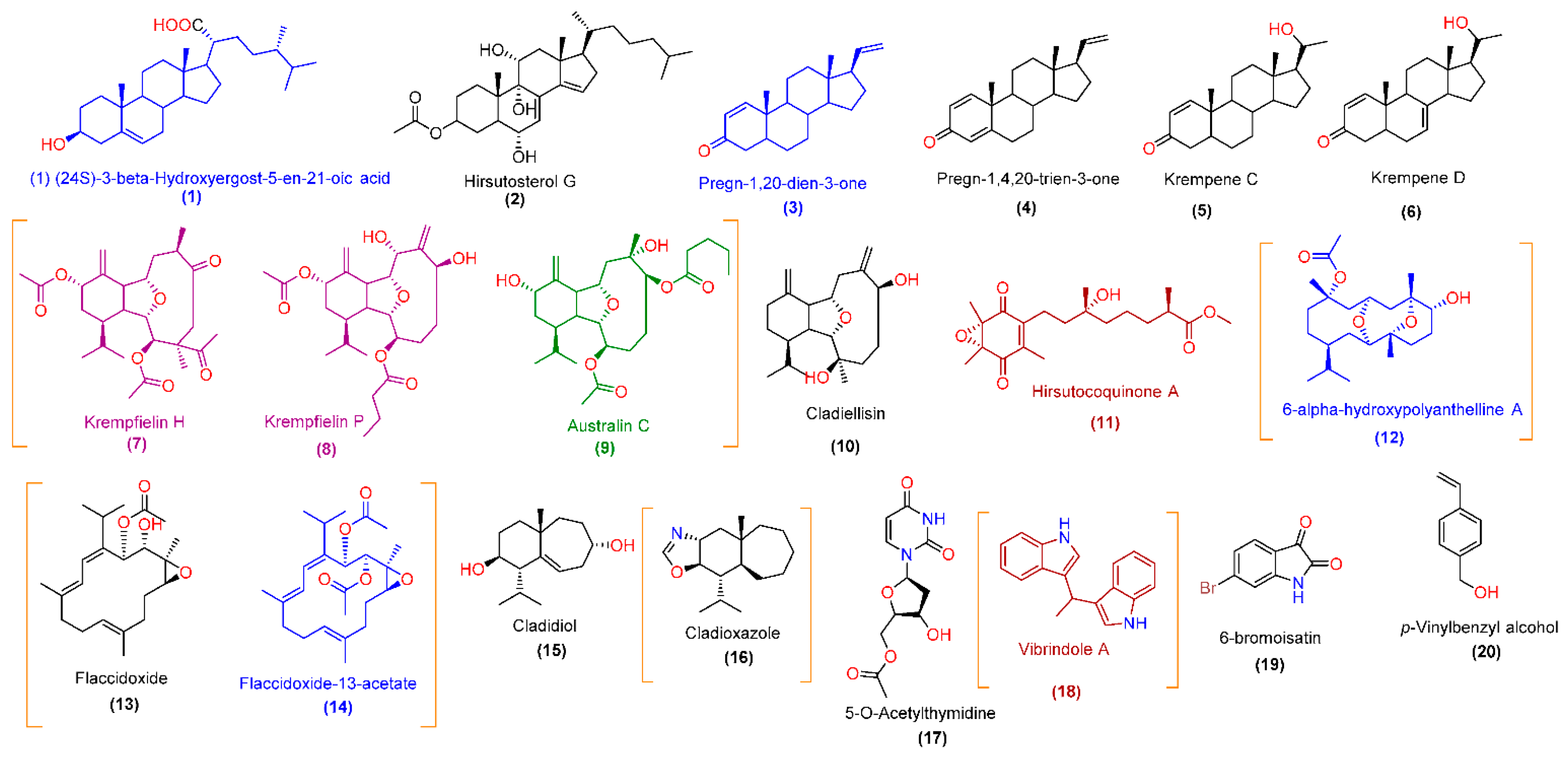
3.3. LC-HRESIMS-Assisted Chemical Profiling of CE
3.4. In Silico Investigation
3.4.1. CE-Derived Compounds with Proposed Antiproliferative Activity against BC Cell Lines
3.4.2. Protein Targets Associated with BC and the Interactions between Them (PPI Network)
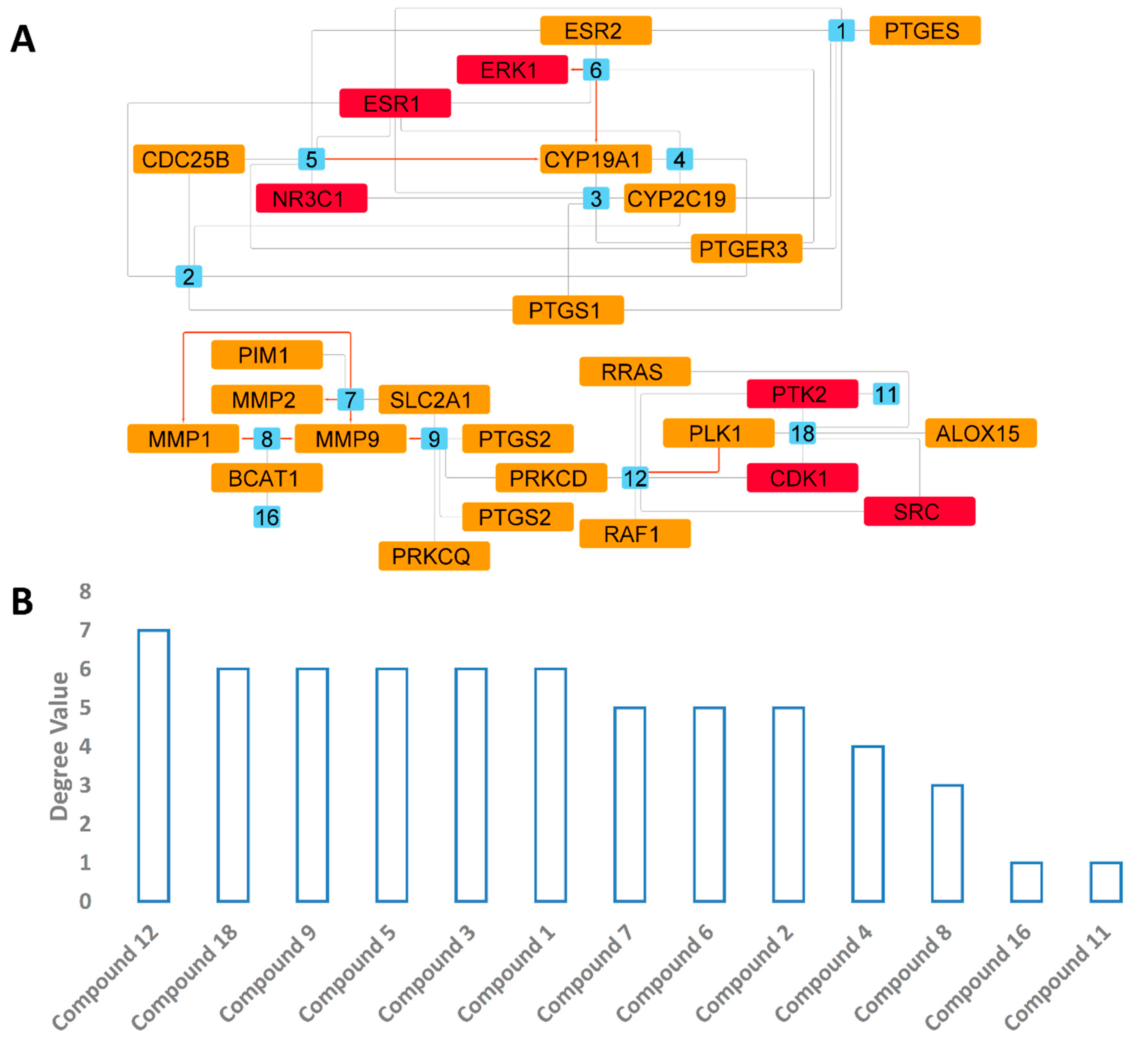
3.4.3. Predicted Targets for the Active Chemical Compounds in CE
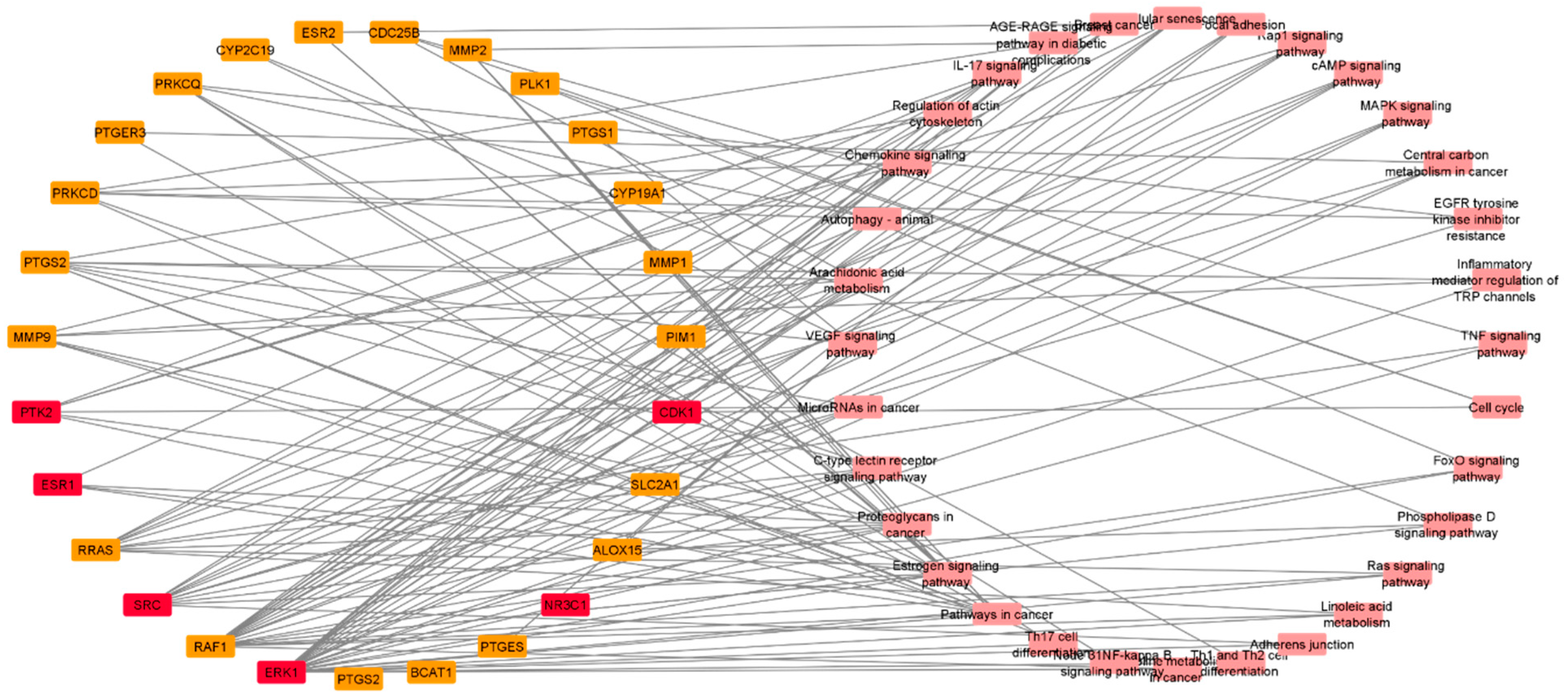
3.4.4. PPI and CPI Networks of the Predicted Targets and KEGG Enrichment Analysis
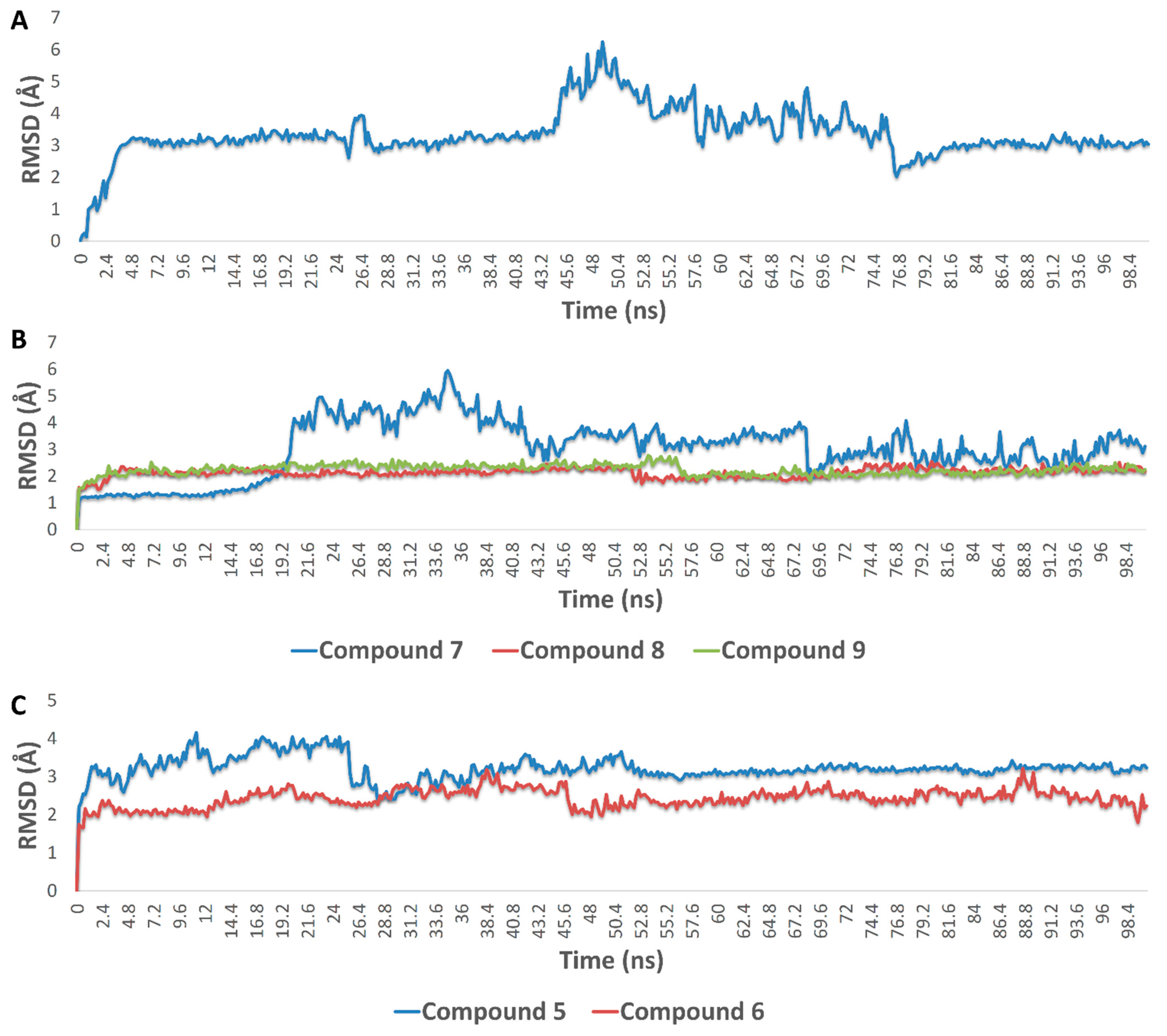
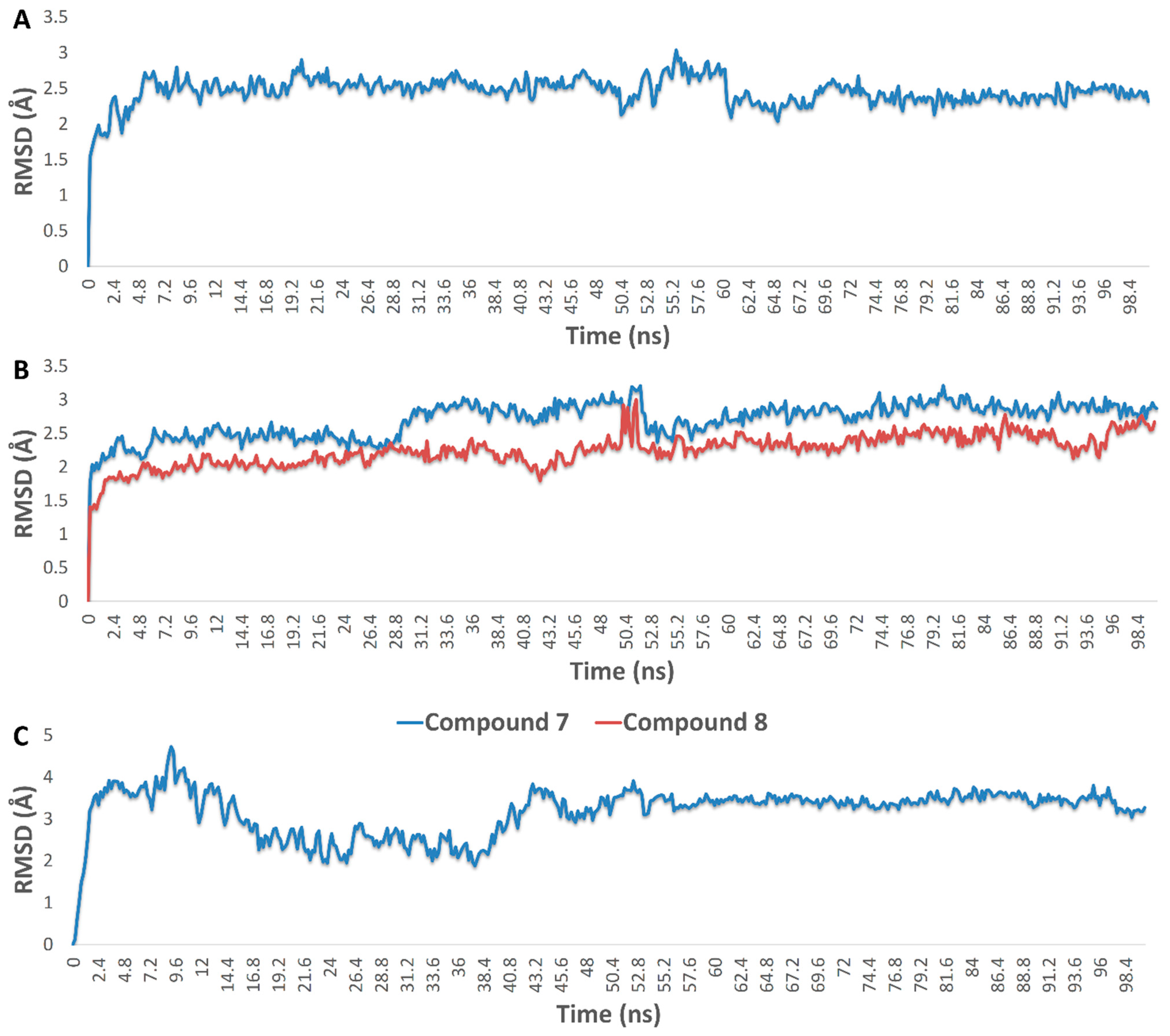
3.4.5. Molecular Modeling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Marco, B.A.; Rechelo, B.S.; Tótoli, E.G.; Kogawa, A.C.; Salgado, H.R.N. Evolution of green chemistry and its multidimensional impacts: A review. Saudi Pharm. J. 2019, 27, 1–8. [Google Scholar] [CrossRef]
- Hurst, G.A. Systems thinking approaches for international green chemistry education. Curr. Opin. Green Sustain. Chem. 2020, 21, 93–97. [Google Scholar] [CrossRef]
- Menges, N. The Role of Green Solvents and Catalysts at the Future of Drug Design and of Synthesis. Green Chem. 2017, 23, 254–257. [Google Scholar]
- Zhang, G.; Keita, B.; Biboum, R.N.; Miserque, F.; Berthet, P.; Dolbecq, A.; Mialane, P.; Catala, L.; Nadjo, L. Synthesis of various crystalline gold nanostructures in water: The polyoxometalate β-[H4PMo12O40]3− as the reducing and stabilizing agent. J. Mat. Chem. 2009, 19, 8639–8644. [Google Scholar] [CrossRef]
- Abid, J.P.; Wark, A.W.; Brevet, P.F.; Girault, H.H. Preparation of silver nanoparticles in solution from a silver salt by laser irradiation. Chem. Commun. 2002, 7, 792–793. [Google Scholar] [CrossRef]
- Kemp, M.M.; Kumar, A.; Mousa, S.; Park, T.-J.; Ajayan, P.; Kubotera, N.; Mousa, S.; Linhardt, R.J. Synthesis of gold and silver nanoparticles stabilized with glycosaminoglycans having distinctive biological activities. Biomacromolecules 2009, 10, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Maliszewska, I.; Szewczyk, K.; Waszak, K. Biological synthesis of silver nanoparticles. J. Phys. Conf. Ser. 2009, 146, 012025. [Google Scholar] [CrossRef]
- Camas, M.; Celik, F.; Sazak Camas, A.; Ozalp, H.B. Biosynthesis of gold nanoparticles using marine bacteria and Box–Behnken design optimization. Part. Sci. Technol. 2019, 37, 31–38. [Google Scholar] [CrossRef]
- Ghosh, V. Marine Bioresources as Potential Source for Synthesis of Nanoparticles. Encycl. Mar. Biotechnol. 2020, 3, 1521–1534. [Google Scholar]
- Sathiyanarayanan, G.; Vignesh, V.; Saibaba, G.; Vinothkanna, A.; Dineshkumar, K.; Viswanathan, M.B.; Selvin, J. Synthesis of carbohydrate polymer encrusted gold nanoparticles using bacterial exopolysaccharide: A novel and greener approach. RSC Adv. 2014, 4, 22817–22827. [Google Scholar] [CrossRef]
- Rabeea, M.A.; Owaid, M.N.; Aziz, A.A.; Jameel, M.S.; Dheyab, M.A. Mycosynthesis of gold nanoparticles using the extract of Flammulina velutipes, Physalacriaceae, and their efficacy for decolorization of methylene blue. J. Environ. Chem. Eng. 2020, 8, 103841. [Google Scholar] [CrossRef]
- Hadrup, N.; Lam, H.R. Oral toxicity of silver ions, silver nanoparticles and colloidal silver—A review. Regul. Toxicol. Pharmacol. 2014, 68, 1–7. [Google Scholar] [CrossRef]
- Khalifa, S.A.; Elias, N.; Farag, M.A.; Chen, L.; Saeed, A.; Hegazy, M.E.F.; El-Seedi, H.R. Marine natural products: A source of novel anticancer drugs. Mar. Drugs 2019, 17, 491. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.-H.; Lu, M.-C.; Chang, Y.-C.; Su, Y.-D.; Chen, Y.-H.; Lin, N.-C.; Fang, L.-S.; Wu, Y.-C.; Sung, P.-J. Discovery of new eunicellin-based diterpenoids from a Formosan soft coral Cladiella sp. Mar. Drugs 2013, 11, 4585–4593. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.H.; Wang, G.H.; Liaw, C.C.; Lee, M.F.; Wang, S.H.; Cheng, D.L.; Chou, T.H. Extracts from Cladiella australis, Clavularia viridis and Klyxum simplex (soft corals) are capable of inhibiting the growth of human oral squamous cell carcinoma cells. Mar. Drugs 2008, 6, 595–606. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, A.F.; Wu, M.H.; Wu, Y.C.; Dai, C.F.; Sheu, J.H. Metabolites with cytotoxic activity from the Formosan soft coral Cladiella australis. J. Chin. Chem. Soc. 2006, 53, 489–494. [Google Scholar] [CrossRef]
- Beer, C.; Foldbjerg, R.; Hayashi, Y.; Sutherland, D.S.; Autrup, H. Toxicity of silver nanoparticles—nanoparticle or silver ion? Toxicol. Lett. 2012, 208, 286–292. [Google Scholar] [CrossRef]
- Alhadrami, H.A.; Sayed, A.M.; El-Gendy, A.O.; Shamikh, Y.I.; Gaber, Y.; Bakeer, W.; Sheirf, N.H.; Attia, E.Z.; Shaban, G.M.; Khalifa, B.A.; et al. A metabolomic approach to target antimalarial metabolites in the Artemisia annua fungal endophytes. Sci. Rep. 2021, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Abdelhafez, O.H.; Ali, T.F.S.; Fahim, J.R.; Desoukey, S.Y.; Ahmed, S.; Behery, F.A.; Kamel, M.S.; Gulder, T.A.M.; Abdelmohsen, U.R. Anti-inflammatory potential of green synthesized silver nanoparticles of the soft coral Nephthea sp. supported by metabolomics analysis and docking studies. Inter. J. Nanomed. 2020, 15, 5345. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [Green Version]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Al-Jarf, R.; de Sá, A.G.; Pires, D.E.; Ascher, D.B. pdCSM-cancer: Using Graph-Based Signatures to Identify Small Molecules with Anticancer Properties. J. Chem. Inform. Model. 2021, 61, 3314–3322. [Google Scholar] [CrossRef] [PubMed]
- Lagunin, A.A.; Dubovskaja, V.I.; Rudik, A.; Pogodin, P.V.; Druzhilovskiy, D.; Gloriozova, T.A.; Filimonov, D.; Sastry, N.G.; Poroikov, V.V. CLC-Pred: A freely available web-service for in silico prediction of human cell line cytotoxicity for drug-like compounds. PLoS ONE 2018, 13, e0191838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rebhan, M.; Chalifa-Caspi, V.; Prilusky, J.; Lancet, D. GeneCards: A novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics (Oxf. Engl.) 1998, 14, 656–664. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, S.; Li, F.; Zhou, Y.; Zhang, Y.; Wang, Z.; Zhang, R.; Zhu, J.; Ren, Y.; Tan, Y.; et al. Therapeutic target database 2020: Enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020, 48, D1031–D1041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Wang, J.C.; Chu, P.Y.; Chen, C.M.; Lin, J.H. idTarget: A web server for identifying protein targets of small chemical molecules with robust scoring functions and a divide-and-conquer docking approach. Nucleic Acids Res. 2012, 40, W393–W399. [Google Scholar] [CrossRef] [Green Version]
- Alhadrami, H.A.; Orfali, R.; Hamed, A.A.; Ghoneim, M.M.; Hassan, H.M.; Hassane, A.S.I.; Rateb, M.E.; Sayed, A.M.; Gamaleldin, N.M. Flavonoid-Coated Gold Nanoparticles as Efficient Antibiotics against Gram-Negative Bacteria—Evidence from In Silico-Supported In Vitro Studies. Antibiotics 2021, 10, 968. [Google Scholar] [CrossRef]
- Alhadrami, H.A.; Sayed, A.M.; Hassan, H.M.; Youssif, K.A.; Gaber, Y.; Moatasim, Y.; Kutkat, O.; Mostafa, A.; Ali, M.A.; Rateb, M.E.; et al. Cnicin as an Anti-SARS-CoV-2: An Integrated In Silico and In Vitro Approach for the Rapid Identification of Potential COVID-19 Therapeutics. Antibiotics 2021, 10, 542. [Google Scholar] [CrossRef]
- Alhadrami, H.A.; Sayed, A.M.; Melebari, S.A.; Khogeer, A.A.; Abdulaal, W.H.; Al-Fageeh, M.B.; Algahtani, M.; Rateb, M.E. Targeting allosteric sites of human aromatase: A comprehensive in-silico and in-vitro workflow to find potential plant-based anti-breast cancer therapeutics. J. Enz. Inhib. Med. Chem. 2021, 36, 1334–1345. [Google Scholar] [CrossRef]
- Mering, C.V.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alves, C.; Diederich, M. Marine Natural Products as Anticancer Agents. Mar. Drugs 2021, 19, 447. [Google Scholar] [CrossRef] [PubMed]
- Saide, A.; Damiano, S.; Ciarcia, R.; Lauritano, C. Promising Activities of Marine Natural Products against Hematopoietic Malignancies. Biomedicines 2021, 9, 645. [Google Scholar] [CrossRef]
- Rosi, N.L.; Giljohann, D.A.; Thaxton, C.S.; Lytton-Jean, A.K.; Han, M.S.; Mirkin, C.A. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science 2006, 312, 1027–1030. [Google Scholar] [CrossRef]
- Thomas, M.; Klibanov, A.M. Conjugation to gold nanoparticles enhances polyethylenimine’s transfer of plasmid DNA into mammalian cells. Proc. Natl. Acad. Sci. USA 2003, 100, 9138–9143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, E.C.; Au, L.; Zhang, Q.; Xia, Y. The effects of size, shape, and surface functional group of gold nanostructures on their adsorption and internalization by cells. Small 2010, 6, 517–522. [Google Scholar] [CrossRef]
- Gu, H.; Ho, P.L.; Tong, E.; Wang, L.; Xu, B. Presenting vancomycin on nanoparticles to enhance antimicrobial activities. Nano Lett. 2003, 3, 1261–1263. [Google Scholar] [CrossRef]
- Kitov, P.I.; Mulvey, G.L.; Griener, T.P.; Lipinski, T.; Solomon, D.; Paszkiewicz, E.; Jacobson, J.M.; Sadowska, J.M.; Suzuki, M.; Yamamura, K.-I.; et al. In vivo supramolecular templating enhances the activity of multivalent ligands: A potential therapeutic against the Escherichia coli O157 AB5 toxins. Proc. Natl. Acad. Sci. USA 2008, 105, 16837–16842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowman, M.C.; Ballard, T.E.; Ackerson, C.J.; Feldheim, D.L.; Margolis, D.M.; Melander, C. Inhibition of HIV fusion with multivalent gold nanoparticles. J. Am. Chem. Soc. 2008, 130, 6896–6897. [Google Scholar] [CrossRef] [Green Version]
- Yavuz, M.S.; Cheng, Y.; Chen, J.; Cobley, C.M.; Zhang, Q.; Rycenga, M.; Xie, J.; Kim, C.; Song, K.H.; Schwartz, A.G.; et al. Gold nanocages covered by smart polymers for controlled release with near-infrared light. Nat. Mater. 2009, 8, 935–939. [Google Scholar] [CrossRef]
- Zhao, Y.; Tian, Y.; Cui, Y.; Liu, W.; Ma, W.; Jiang, X. Small molecule-capped gold nanoparticles as potent antibacterial agents that target gram-negative bacteria. J. Am. Chem. Soc. 2010, 132, 12349–12356. [Google Scholar] [CrossRef]
- Desai, R.; Mankad, V.; Gupta, S.K.; Jha, P.K. Size distribution of silver nanoparticles: UV-visible spectroscopic assessment. Nanosci. Nanotech. Lett. 2012, 4, 30–34. [Google Scholar] [CrossRef]
- Wang, H.; Qiao, X.; Chen, J.; Ding, S. Preparation of silver nanoparticles by chemical reduction method. Colloids Surfaces A Physicochem. Eng. Asp. 2005, 256, 111–115. [Google Scholar] [CrossRef]
- Chen, B.W.; Uvarani, C.; Huang, C.Y.; Hwang, T.L.; Dai, C.F.; Sheu, J.H. New anti-inflammatory tocopherol-derived metabolites from the Taiwanese soft coral Cladiella hirsuta. Bioorg. Med. Chem. Lett. 2015, 25, 92–95. [Google Scholar] [CrossRef]
- Tai, C.J.; Su, J.H.; Huang, C.Y.; Huang, M.S.; Wen, Z.H.; Dai, C.F.; Sheu, J.H. Cytotoxic and anti-inflammatory eunicellin-based diterpenoids from the soft coral Cladiella krempfi. Mar. Drugs 2013, 11, 788–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.W.; Chang, S.M.; Huang, C.Y.; Su, J.H.; Wen, Z.H.; Wu, Y.C.; Sheu, J.H. Hirsutosterols A–G, polyoxygenated steroids from a Formosan soft coral Cladiella hirsuta. Org. Biomol. Chem. 2011, 9, 3272–3278. [Google Scholar] [CrossRef] [PubMed]
- Mol, V.L.; Raveendran, T.V.; Parameswaran, P.S.; Kunnath, R.J.; Rajamohanan, P.R. (–)-6 α-Hydroxy polyanthellin A—A novel antifouling diterpenoid from the Indian soft coral Cladiella pachyclados (Hickson). Can. J. Chem. 2011, 89, 57–60. [Google Scholar] [CrossRef]
- Neoh, C.A.; Wu, W.T.; Dai, G.F.; Su, J.H.; Liu, C.I.; Su, T.R.; Wu, Y.J. Flaccidoxide-13-acetate extracted from the soft coral cladiella kashmani reduces human bladder cancer cell migration and invasion through reducing activation of the FAK/PI3K/AKT/mTOR signaling pathway. Molecules 2018, 23, 58. [Google Scholar] [CrossRef] [Green Version]
- Radhika, P. Chemical constituents and biological activities of the soft corals of genus Cladiella: A review. Biochem. System. Ecol. 2006, 34, 781–789. [Google Scholar] [CrossRef]
- Li, G.; Dickschat, J.S.; Guo, Y.W. Diving into the world of marine 2, 11-cyclized cembranoids: A summary of new compounds and their biological activities. Nat. Prod. Rep. 2020, 37, 1367–1383. [Google Scholar] [CrossRef]
- Bell, R.; Carmeli, S.; Sar, N. Vibrindole A, a metabolite of the marine bacterium, Vibrio parahaemolyticus, isolated from the toxic mucus of the boxfish Ostracion cubicus. J. Nat. Prod. 1994, 57, 1587–1590. [Google Scholar] [CrossRef]
- Corral, P.; Amoozegar, M.A.; Ventosa, A. Halophiles and their biomolecules: Recent advances and future applications in biomedicine. Mar. Drugs 2020, 18, 33. [Google Scholar] [CrossRef] [Green Version]
- Esmaeelian, B.; Benkendorff, K.; Johnston, M.R.; Abbott, C.A. Purified brominated indole derivatives from Dicathais orbita induce apoptosis and cell cycle arrest in colorectal cancer cell lines. Mar. Drugs 2013, 11, 3802–3822. [Google Scholar] [CrossRef] [PubMed]
- Esmaeelian, B.; Abbott, C.A.; Le Leu, R.K.; Benkendorff, K. 6-bromoisatin found in muricid mollusc extracts inhibits colon cancer cell proliferation and induces apoptosis, preventing early stage tumor formation in a colorectal cancer rodent model. Mar. Drugs 2014, 12, 17–35. [Google Scholar] [CrossRef] [PubMed]
- El-Hawary, S.S.; Sayed, A.M.; Mohammed, R.; Hassan, H.M.; Rateb, M.E.; Amin, E.; Mohammed, T.A.; El-Mesery, M.; Bin Muhsinah, A.; Alsayari, A.; et al. Bioactive brominated oxindole alkaloids from the Red Sea sponge Callyspongia siphonella. Mar. Drugs 2019, 17, 465. [Google Scholar] [CrossRef] [Green Version]
- Sayed, A.M.; Alhadrami, H.A.; El-Hawary, S.S.; Mohammed, R.; Hassan, H.M.; Rateb, M.E.; Abdelmohsen, U.R.; Bakeer, W. Discovery of two brominated oxindole alkaloids as Staphylococcal DNA gyrase and pyruvate kinase inhibitors via inverse virtual screening. Microorganisms 2020, 8, 293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.V.; Ansari, M.H.D.; Rosenkranz, D.; Maharjan, R.S.; Kriegel, F.L.; Gandhi, K.; Kanase, A.; Singh, R.; Laux, P.; Luch, A. Artificial intelligence and machine learning in computational nanotoxicology: Unlocking and empowering nanomedicine. Adv. Healthc. Mater. 2020, 9, 1901862. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.V.; Rosenkranz, D.; Ansari, M.H.D.; Singh, R.; Kanase, A.; Singh, S.P.; Johnston, B.; Tentschert, J.; Laux, P.; Luch, A. Artificial Intelligence and Machine Learning Empower Advanced Biomedical Material Design to Toxicity Prediction. Adv. Intell. Syst. 2020, 2, 2000084. [Google Scholar] [CrossRef]
- Singh, A.V.; Maharjan, R.S.; Kanase, A.; Siewert, K.; Rosenkranz, D.; Singh, R.; Laux, P.; Luch, A. Machine-Learning-Based Approach to Decode the Influence of Nanomaterial Properties on Their Interaction with Cells. ACS Appl. Mater. Interfaces 2020, 13, 1943–1955. [Google Scholar] [CrossRef] [PubMed]
- Abdelhameed, R.F.; Habib, E.S.; Eltahawy, N.A.; Hassanean, H.A.; Ibrahim, A.K.; Fahim, A.R.; Sayed, A.M.; Hendawy, O.M.; Abdelmohsen, U.R.; Ahmed, S.A. New glucose-6-phosphate dehydrogenase inhibitor from the Red Sea sponge Echinoclathria sp. Tetrahedron Lett. 2021, 72, 152986. [Google Scholar] [CrossRef]
- Musa, A.; Shady, N.; Ahmed, S.; Alnusaire, T.; Sayed, A.; Alowaiesh, B.; Sabouni, I.; Al-Sanea, M.; Mostafa, E.; Youssif, K.; et al. Antiulcer Potential of Olea europea L. cv. Arbequina Leaf Extract Supported by Metabolic Profiling and Molecular Docking. Antioxidants 2021, 10, 644. [Google Scholar] [CrossRef]
- Alzarea, S.; Elmaidomy, A.; Saber, H.; Musa, A.; Al-Sanea, M.; Mostafa, E.; Hendawy, O.; Youssif, K.; Alanazi, A.; Alharbi, M.; et al. Potential Anticancer Lipoxygenase Inhibitors from the Red Sea-Derived Brown Algae Sargassum cinereum: An In-Silico-Supported In-Vitro Study. Antibiotics 2021, 10, 416. [Google Scholar] [CrossRef]
- El-Hawwary, S.S.; Abd Almaksoud, H.M.; Saber, F.R.; Elimam, H.; Sayed, A.M.; El Raey, M.A.; Abdelmohsen, U.R. Green-synthesized zinc oxide nanoparticles, anti-Alzheimer potential and the metabolic profiling of Sabal blackburniana grown in Egypt supported by molecular modelling. RSC Adv. 2021, 11, 18009–18025. [Google Scholar] [CrossRef]
- Gamaleldin, N.M.; Bakeer, W.; Sayed, A.M.; Shamikh, Y.I.; El-Gendy, A.O.; Hassan, H.M.; Horn, H.; Abdelmohsen, U.R.; Hozzein, W.N. Exploration of chemical diversity and antitrypanosomal activity of some red sea-derived actinomycetes using the OSMAC approach supported by LC-MS-based metabolomics and molecular modelling. Antibiotics 2020, 9, 629. [Google Scholar] [CrossRef]
- Elhawary, S.S.; Sayed, A.M.; Issa, M.Y.; Ebrahim, H.S.; Alaaeldin, R.; Abd El-Kader, E.M.; Abdelmohsen, U.R. Anti-Alzheimer Chemical Constituents of Morus macroura Miq.: Chemical Profiling, In silico, and In vitro Investigations. Food Funct. 2021, 12, 8078–8089. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.-Y.; Liu, J.-C.; Liang, S.; Meng, X.-H.; Greenbaum, J.; Xiao, H.-M.; Tan, L.-J.; Deng, H.-W. Drug Repurposing for COVID-19 Treatment by Integrating Network Pharmacology and Transcriptomics. Pharmaceutics 2021, 13, 545. [Google Scholar] [CrossRef]
- Ju, Y.; Liang, H.; Du, K.; Guo, Z.; Meng, D. Isolation of triterpenoids and phytosterones from Achyranthes bidentata Bl. to treat breast cancer based on network pharmacology. Nat. Prod. Res. 2020, 25, 1–4. [Google Scholar] [CrossRef]
- Vitali, F.; Cohen, L.D.; Demartini, A.; Amato, A.; Eterno, V.; Zambelli, A.; Bellazzi, R. A network-based data integration approach to support drug repurposing and multi-target therapies in triple negative breast cancer. PLoS ONE 2016, 11, e0162407. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, S.; Sumantran, V.N.; Ranganathan, M.; Madheswaran, S. Microarray and pattern miner analysis of AXL and VIM gene networks in MDA-MB-231 cells. Mol. Med. Rep. 2018, 18, 4147–4155. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.S. Targeting Src in breast cancer. Ann. Oncol. 2008, 19, 1379–1386. [Google Scholar] [CrossRef]
- Du, Y.; Zhang, J.; Meng, Y.; Huang, M.; Yan, W.; Wu, Z. MicroRNA-143 targets MAPK3 to regulate the proliferation and bone metastasis of human breast cancer cells. AMB Express 2020, 10, 1–8. [Google Scholar] [CrossRef]
- Mendes, O.; Kim, H.T.; Lungu, G.; Stoica, G. MMP2 role in breast cancer brain metastasis development and its regulation by TIMP2 and ERK1/2. Clin. Exp. Metastasis 2007, 24, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Di, J.; Huang, H.; Qu, D.; Tang, J.; Cao, W.; Lu, Z.; Cheng, Q.; Yang, J.; Bai, J.; Zheng, J.; et al. Rap2B promotes proliferation, migration and invasion of human breast cancer through calcium-related ERK1/2 signaling pathway. Sci. Rep. 2015, 5, 1–11. [Google Scholar] [CrossRef]
- Gee, J.M.; Robertson, J.F.; Ellis, I.O.; Nicholson, R.I. Phosphorylation of ERK1/2 mitogen-activated protein kinase is associated with poor response to anti-hormonal therapy and decreased patient survival in clinical breast cancer. Int. J. Cancer 2001, 95, 247–254. [Google Scholar] [CrossRef]
- Lev, D.C.; Kim, L.S.; Melnikova, V.; Ruiz, M.; Ananthaswamy, H.N.; Price, J.E. Dual blockade of EGFR and ERK1/2 phosphorylation potentiates growth inhibition of breast cancer cells. Br. J. Cancer 2004, 91, 795–802. [Google Scholar] [CrossRef] [Green Version]
- Sirianni, R.; Chimento, A.; De Luca, A.; Casaburi, I.; Rizza, P.; Onofrio, A.; Iacopetta, D.; Puoci, F.; Andò, S.; Pezzi, V.; et al. Oleuropein and hydroxytyrosol inhibit MCF-7 breast cancer cell proliferation interfering with ERK1/2 activation. Mol. Nutr. Food Res. 2010, 54, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Illiano, M.; Sapio, L.; Salzillo, A.; Capasso, L.; Caiafa, I.; Chiosi, E.; Spina, A.; Naviglio, S. Forskolin improves sensitivity to doxorubicin of triple negative breast cancer cells via Protein Kinase A-mediated ERK1/2 inhibition. Biochem. Pharmacol. 2018, 152, 104–113. [Google Scholar] [CrossRef]
- Jezierska, A.; Motyl, T. Matrix metalloproteinase-2 involvement in breast cancer progression: A mini-review. Med. Sci. Monit. 2009, 15, RA32–RA40. [Google Scholar] [PubMed]
- Bartsch, J.E.; Staren, E.D.; Appert, H.E. Matrix metalloproteinase expression in breast cancer. J. Surg. Res. 2003, 110, 383–392. [Google Scholar] [CrossRef]
- Radisky, E.S.; Radisky, D.C. Matrix metalloproteinase-induced epithelial-mesenchymal transition in breast cancer. J. Mammary Gland. Biol. Neoplasia 2010, 15, 201–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arévalo, B.; ben Hassine, A.; Valverde, A.; Serafín, V.; Montero-Calle, A.; Raouafi, N.; Camps, J.; Arenas, M.; Barderas, R.; Yáñez-Sedeño, P.; et al. Electrochemical immunoplatform to assist in the diagnosis and classification of breast cancer through the determination of matrix-metalloproteinase-9. Talanta 2021, 225, 122054. [Google Scholar] [CrossRef]
- Kalavska, K.; Cierna, Z.; Karaba, M.; Minarik, G.; Benca, J.; Sedlackova, T.; Kolekova, D.; Mrvova, I.; Pindak, D.; Mego, M.; et al. Prognostic role of matrix metalloproteinase 9 in early breast cancer. Oncol. Lett. 2021, 21, 1. [Google Scholar]
- Balkhi, S.; Mashayekhi, F.; Salehzadeh, A.; Saedi, H.S. Matrix metalloproteinase (MMP)-1 and MMP-3 gene variations affect MMP-1 and-3 serum concentration and associates with breast cancer. Mol. Biol. Rep. 2020, 47, 9637–9644. [Google Scholar] [CrossRef]
- Mondal, S.; Adhikari, N.; Banerjee, S.; Amin, S.A.; Jha, T. Matrix metalloproteinase-9 (MMP-9) and its inhibitors in cancer: A minireview. Eur. J. Med. Chem. 2020, 194, 112260. [Google Scholar] [CrossRef]
- Ratre, P.; Mishra, K.; Dubey, A.; Vyas, A.; Jain, A.; Thareja, S. Aromatase inhibitors for the treatment of breast cancer: A journey from the scratch. Anticancer. Agents Med. Chem. 2020, 20, 1994–2004. [Google Scholar] [CrossRef] [PubMed]
- Park, H.G.; Kim, J.H.; Dancer, A.N.; Kothapalli, K.S.; Brenna, J.T. The aromatase inhibitor letrozole restores FADS2 function in ER+ MCF7 human breast cancer cells. Prostaglandins Leukot. Essent. Fat. Acids 2021, 171, 102312. [Google Scholar] [CrossRef]
- Jayaraman, S.; Hou, X.; Kuffel, M.J.; Suman, V.J.; Hoskin, T.L.; Reinicke, K.E.; Monroe, D.G.; Kalari, K.R.; Tang, X.; Goetz, M.P. Antitumor activity of Z-endoxifen in aromatase inhibitor-sensitive and aromatase inhibitor-resistant estrogen receptor-positive breast cancer. Breast Cancer Res. 2020, 22, 1–12. [Google Scholar] [CrossRef]
- Rigiracciolo, D.C.; Cirillo, F.; Talia, M.; Muglia, L.; Gutkind, J.S.; Maggiolini, M.; Lappano, R. Focal adhesion kinase fine tunes multifaced signals toward breast cancer progression. Cancers 2021, 13, 645. [Google Scholar] [CrossRef]
- Murphy, J.M.; Rodriguez, Y.A.; Jeong, K.; Ahn, E.Y.E.; Lim, S.T.S. Targeting focal adhesion kinase in cancer cells and the tumor microenvironment. Exp. Mol. Med. 2020, 52, 877–886. [Google Scholar] [CrossRef]
- Chan, K.T.; Cortesio, C.L.; Huttenlocher, A. FAK alters invadopodia and focal adhesion composition and dynamics to regulate breast cancer invasion. J. Cell Biol. 2009, 185, 357–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, S.J.; Yang, I.J.; Tettey, C.O.; Kim, K.M.; Shin, H.M. In-vitro anticancer activity of green synthesized silver nanoparticles on MCF-7 human breast cancer cells. Mater. Sci. Eng. C 2016, 68, 430–435. [Google Scholar] [CrossRef]
- Firdhouse, J.; Lalitha, P. Apoptotic efficacy of biogenic silver nanoparticles on human breast cancer MCF-7 cell lines. Prog. Biomater. 2015, 4, 113–121. [Google Scholar]
- Azizi, M.; Ghourchian, H.; Yazdian, F.; Bagherifam, S.; Bekhradnia, S.; Nyström, B. Anti-cancerous effect of albumin coated silver nanoparticles on MDA-MB 231 human breast cancer cell line. Sci. Rep. 2017, 7, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Yang, K. Exploring the pharmacological mechanism of Yanghe Decoction on HER2-positive breast cancer by a network pharmacology approach. J. Ethnopharmacol. 2017, 199, 68–85. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shen, T.; Zhou, X.; Tang, X.; Gao, R.; Xu, L.; Wang, L.; Zhou, Z.; Lin, J.; Hu, Y. Network pharmacology based virtual screening of active constituents of Prunella vulgaris L. and the molecular mechanism against breast cancer. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Wang, N.; Yang, B.; Zhang, X.; Wang, S.; Zheng, Y.; Li, X.; Liu, S.; Pan, H.; Li, Y.; Huang, Z.; et al. Network pharmacology-based validation of caveolin-1 as a key mediator of Ai Du Qing inhibition of drug resistance in breast cancer. Front. Pharmacol. 2018, 9, 1106. [Google Scholar] [CrossRef]
- Sakle, N.S.; More, S.A.; Mokale, S.N. A network pharmacology-based approach to explore potential targets of Caesalpinia pulcherima: An updated prototype in drug discovery. Sci. Rep. 2020, 10, 1–16. [Google Scholar] [CrossRef]
- Teodoro, J.S.; Simões, A.M.; Duarte, F.V.; Rolo, A.P.; Murdoch, R.C.; Hussain, S.M.; Palmeira, C.M. Assessment of the toxicity of silver nanoparticles in vitro: A mitochondrial perspective. Toxicol. Vitr. 2011, 25, 664–670. [Google Scholar] [CrossRef]
- Cunningham, B.; Engstrom, A.E.; Harper, B.J.; Harper, S.L.; Mackiewicz, M.R. Silver Nanoparticles Stable to Oxidation and Silver Ion Release Show Size-Dependent Toxicity In Vivo. Nanomaterials 2021, 11, 1516. [Google Scholar] [CrossRef]
- Jadhav, K.; Deore, S.; Dhamecha, D.; Hr, R.; Jagwani, S.; Jalalpure, S.; Bohara, R. Phytosynthesis of silver nanoparticles: Characterization, biocompatibility studies, and anticancer activity. ACS Biomater. Sci. Eng. 2018, 4, 892–899. [Google Scholar] [CrossRef] [PubMed]
- Majeed, S.; Bakhtiar NF, B.; Danish, M.; Ibrahim, M.M.; Hashim, R. Green approach for the biosynthesis of silver nanoparticles and its antibacterial and antitumor effect against osteoblast MG-63 and breast MCF-7 cancer cell lines. Sustain. Chem. Pharm. 2019, 12, 100138. [Google Scholar] [CrossRef]
- Zhang, X.F.; Liu, Z.G.; Shen, W.; Gurunathan, S. Silver nanoparticles: Synthesis, characterization, properties, applications, and therapeutic approaches. Int. J. Mol. Sci. 2016, 17, 1534. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wang, L.; Chen, Q.; Chen, C. Cytotoxic potential of silver nanoparticles. Yonsei Med. J. 2014, 55, 283–291. [Google Scholar] [CrossRef] [Green Version]

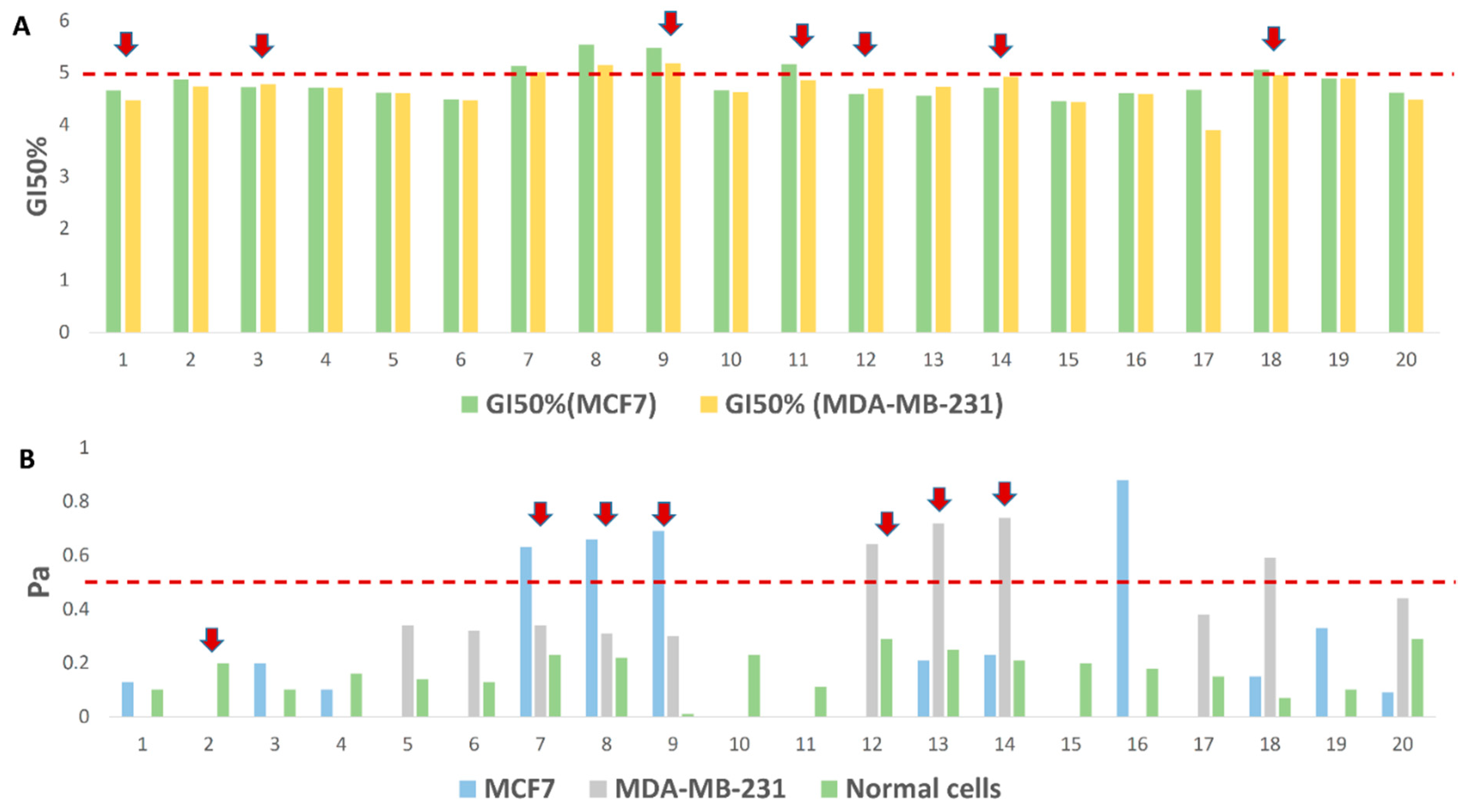


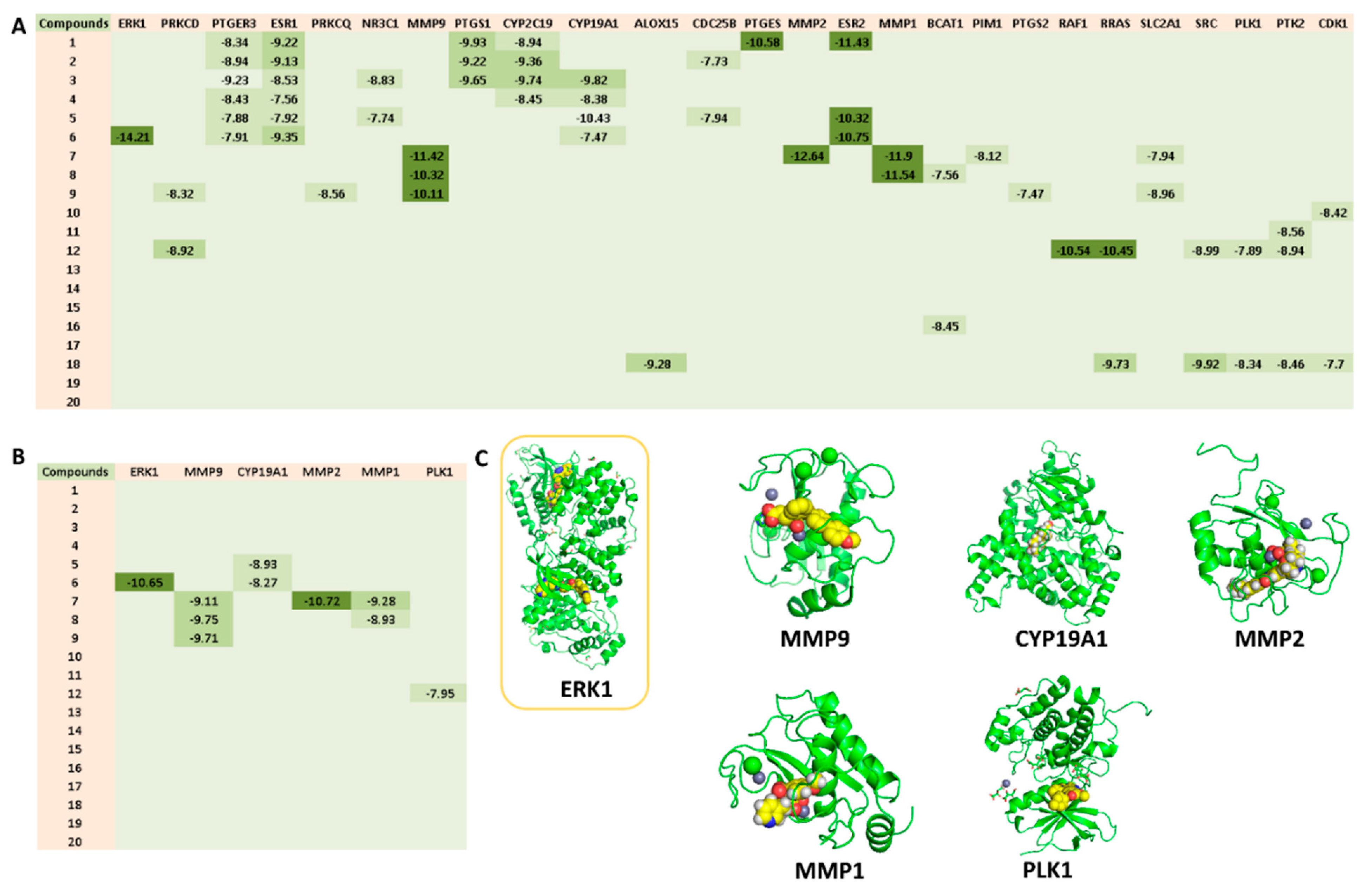
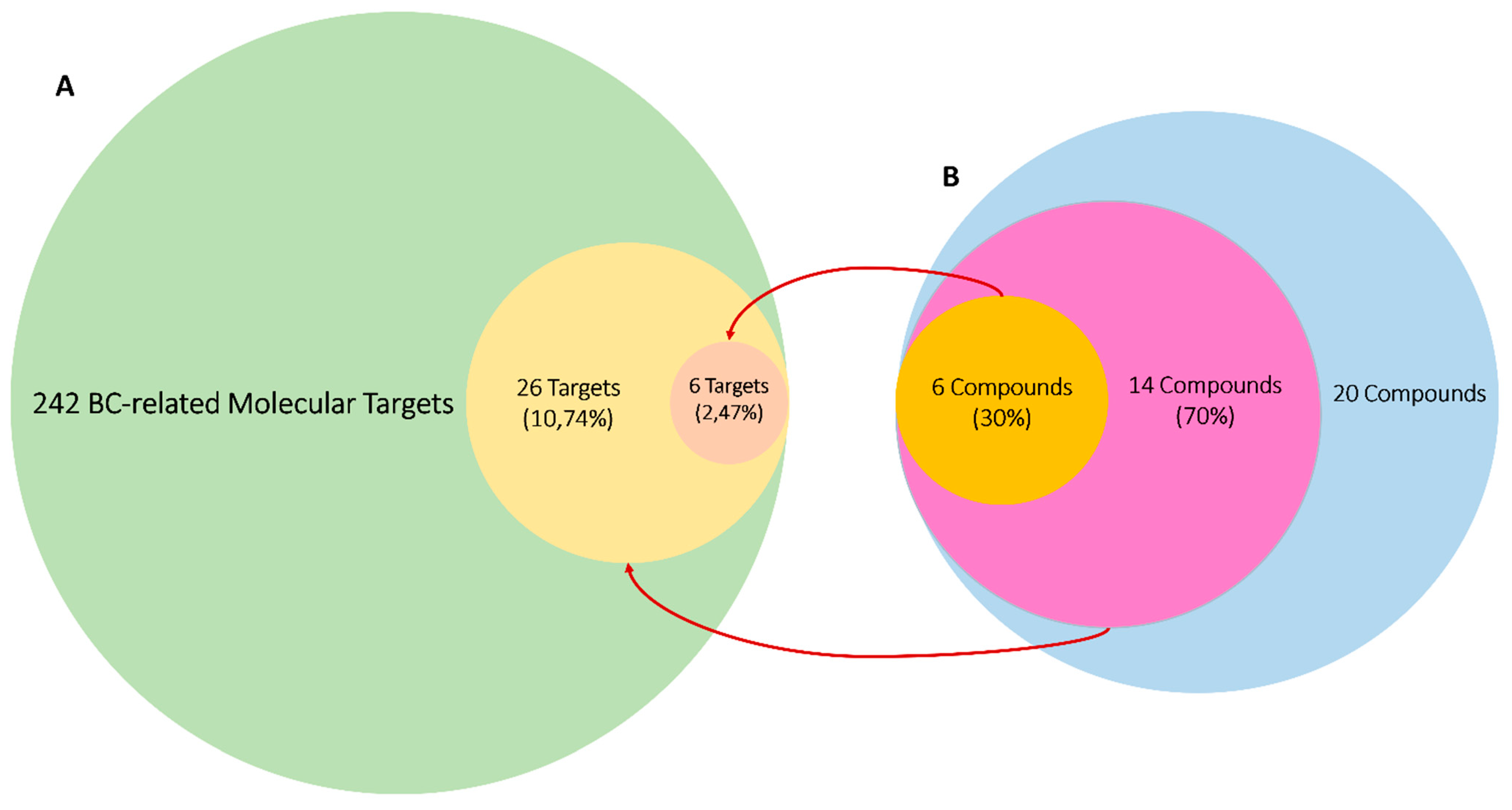
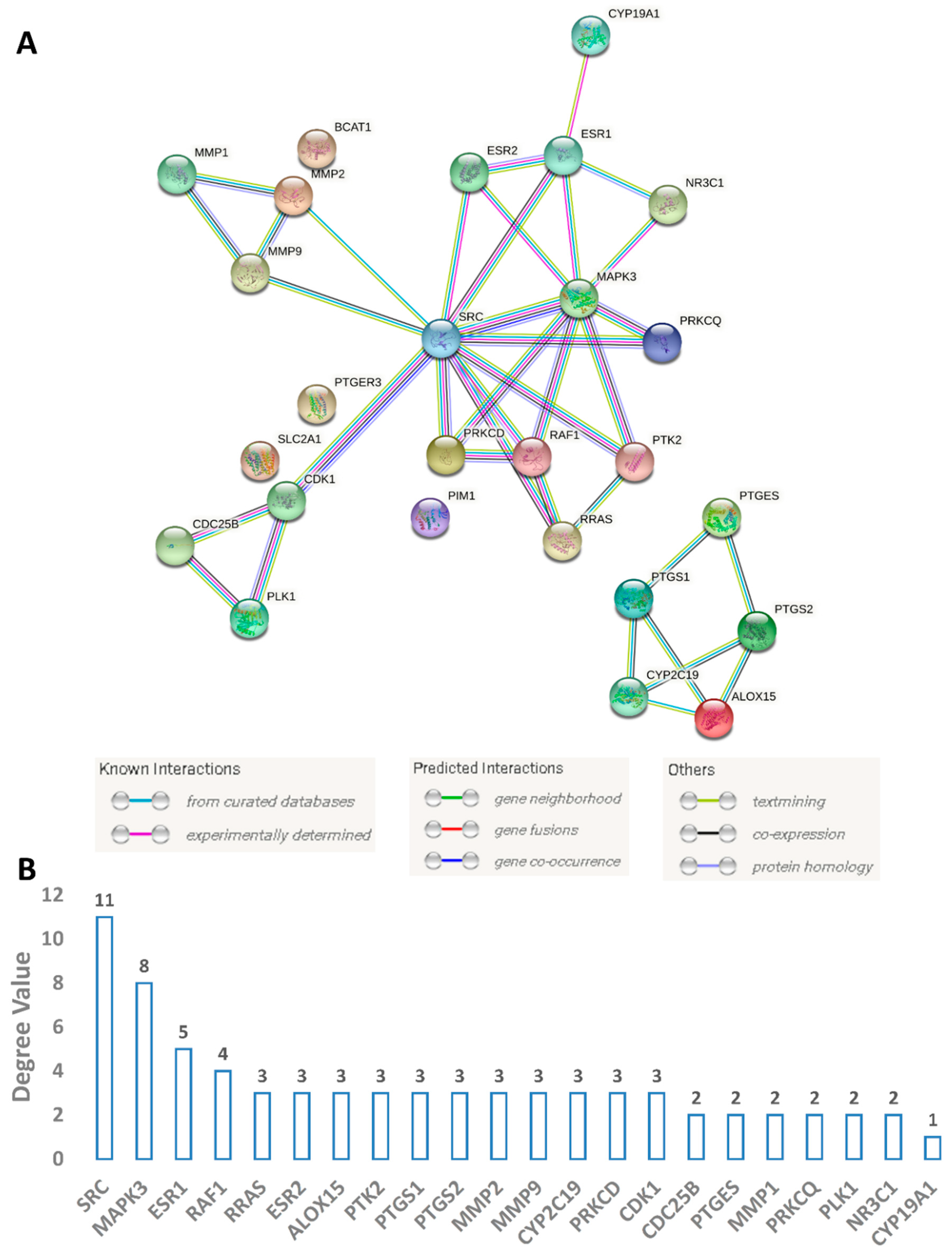



| MCF7 (SI) | MDA-MB-231 (SI) | MCF10a | |
|---|---|---|---|
| CE | 24.32 ± 0.52 c (2.95) | 9.55 ± 0.53 b (7.52) | 71.85 ± 0.5 c |
| AgNPs | 5.62 ± 0.26 b (7.34) | 1.72 ± 0.14 a (24) | 41.29 ± 0.44 b |
| Doxorubicin | 2.61 ± 0.03 a (7.7) | 1.5 ± 0.26 a (13.4) | 20.09 ± 0.72 a |
| No. | Rt | m/z | Ionization Mode | Accurate Mass | Calculated Mass | Molecular Formula | Putative Identification | Chemical Class |
|---|---|---|---|---|---|---|---|---|
| 1 | 9.11 | 431.3522 | Positive | 430.3449 | 430.3447 | C28H46O3 | (1) (24S)-3-β-Hydroxyergost-5-en-21-oic acid | Sterol |
| 2 | 9.43 | 475.3426 | Positive | 474.3353 | 474.3345 | C29H46O5 | Hirsutosterol G | Sterol |
| 3 | 9.15 | 299.2373 | Positive | 298.2395 | 298.2297 | C21H30O | Pregn-1,20-dien-3-one | Sterol |
| 4 | 9.28 | 297.2214 | Positive | 296.2141 | 296.2140 | C21H28O | Pregn-1,4,20-trien-3-one | Sterol |
| 5 | 8.88 | 315.2322 | Negative | 316.2395 | 316.2402 | C21H32O2 | Krempene C | Sterol |
| 6 | 8.95 | 313.2165 | Negative | 314.2238 | 314.2246 | C21H30O2 | Krempene D | Sterol |
| 7 | 5.43 | 463.2693 | Positive | 462.262 | 462.2618 | C26H38O7 | Krempfielin H | Diterpene (eunicellin derivative) |
| 8 | 5.67 | 451.2698 | Positive | 450.2625 | 450.2618 | C25H38O7 | Krempfielin P | Diterpene (eunicellin derivative) |
| 9 | 6.32 | 467.3005 | Positive | 466.2932 | 466.2931 | C26H42O7 | Australin C | Diterpene (eunicellin derivative) |
| 10 | 5.78 | 321.2433 | Positive | 320.236 | 320.2351 | C20H32O3 | Cladiellisin | Diterpene (eunicellin derivative) |
| 11 | 6.14 | 365.1962 | Negative | 366.2035 | 366.2042 | C20H30O6 | Hirsutocoquinone A | Diterpene (tocopherol derivative) |
| 12 | 4.72 | 381.2642 | Negative | 382.2715 | 382.2719 | C22H38O5 | 6-α-hydroxypolyanthelline A | Diterpene (eunicellin derivative) |
| 13 | 7.83 | 363.2532 | Positive | 362.2459 | 362.2457 | C22H34O4 | Flaccidoxide | Diterpene (cembrane derivative) |
| 14 | 8.17 | 405.2643 | Positive | 404.257 | 404.2563 | C24H36O5 | Flaccidoxide-13-acetate | Diterpene (cembrane derivative) |
| 15 | 3.16 | 237.1857 | Negative | 238.193 | 238.1933 | C15H26O2 | Cladidiol | Sesquiterpene |
| 16 | 3.74 | 250.2174 | Positive | 249.2101 | 249.2093 | C16H27NO | Cladioxazole | Sesquiterpene (alkaloid) |
| 17 | 3.89 | 269.0771 | Negative | 270.0844 | 270.0852 | C11H14N2O6 | 5-O-Acetylthymidine | Nucleoside |
| 18 | 3.65 | 259.1238 | Negative | 260.1311 | 260.1313 | C18H16N2 | Vibrindole A * | Indole alkaloid |
| 19 | 2.98 | 223.9344 | Negative | 224.9417 | 224.9425 | C8H4BrNO2 | 6-bromoisatin * | Indole alkaloid |
| 20 | 2.56 | 135.0813 | Positive | 134.074 | 134.0732 | C9H10O | p-Vinylbenzyl alcohol | Aromatic alcohol |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alhadrami, H.A.; Alkhatabi, H.; Abduljabbar, F.H.; Abdelmohsen, U.R.; Sayed, A.M. Anticancer Potential of Green Synthesized Silver Nanoparticles of the Soft Coral Cladiella pachyclados Supported by Network Pharmacology and In Silico Analyses. Pharmaceutics 2021, 13, 1846. https://doi.org/10.3390/pharmaceutics13111846
Alhadrami HA, Alkhatabi H, Abduljabbar FH, Abdelmohsen UR, Sayed AM. Anticancer Potential of Green Synthesized Silver Nanoparticles of the Soft Coral Cladiella pachyclados Supported by Network Pharmacology and In Silico Analyses. Pharmaceutics. 2021; 13(11):1846. https://doi.org/10.3390/pharmaceutics13111846
Chicago/Turabian StyleAlhadrami, Hani A., Heba Alkhatabi, Fahad H. Abduljabbar, Usama Ramadan Abdelmohsen, and Ahmed M. Sayed. 2021. "Anticancer Potential of Green Synthesized Silver Nanoparticles of the Soft Coral Cladiella pachyclados Supported by Network Pharmacology and In Silico Analyses" Pharmaceutics 13, no. 11: 1846. https://doi.org/10.3390/pharmaceutics13111846
APA StyleAlhadrami, H. A., Alkhatabi, H., Abduljabbar, F. H., Abdelmohsen, U. R., & Sayed, A. M. (2021). Anticancer Potential of Green Synthesized Silver Nanoparticles of the Soft Coral Cladiella pachyclados Supported by Network Pharmacology and In Silico Analyses. Pharmaceutics, 13(11), 1846. https://doi.org/10.3390/pharmaceutics13111846









