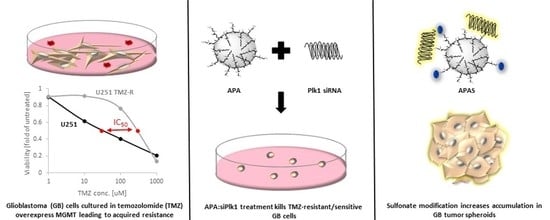
Sulfonated Amphiphilic Poly(α)glutamate Amine—A Potential siRNA Nanocarrier for the Treatment of Both Chemo-Sensitive and Chemo-Resistant Glioblastoma Tumors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.2.1. Establishment of mCherry and iRFP Stably-Expressing U251 Cells
2.2.2. Establishment of mCherry U251 Chemo-Resistant Cell Line (U251 TMZ-R)
2.3. IC50 Determination Assay
2.4. RNA Isolation and Quantitative Real-Time RT-PCR (qRT-PCR)
2.5. Animal Models
Animals Ethics Statement
2.6. Frozen OCT Tissue Fixation
2.7. Immunostaining
2.8. Survival Analysis Based on TCGA Data
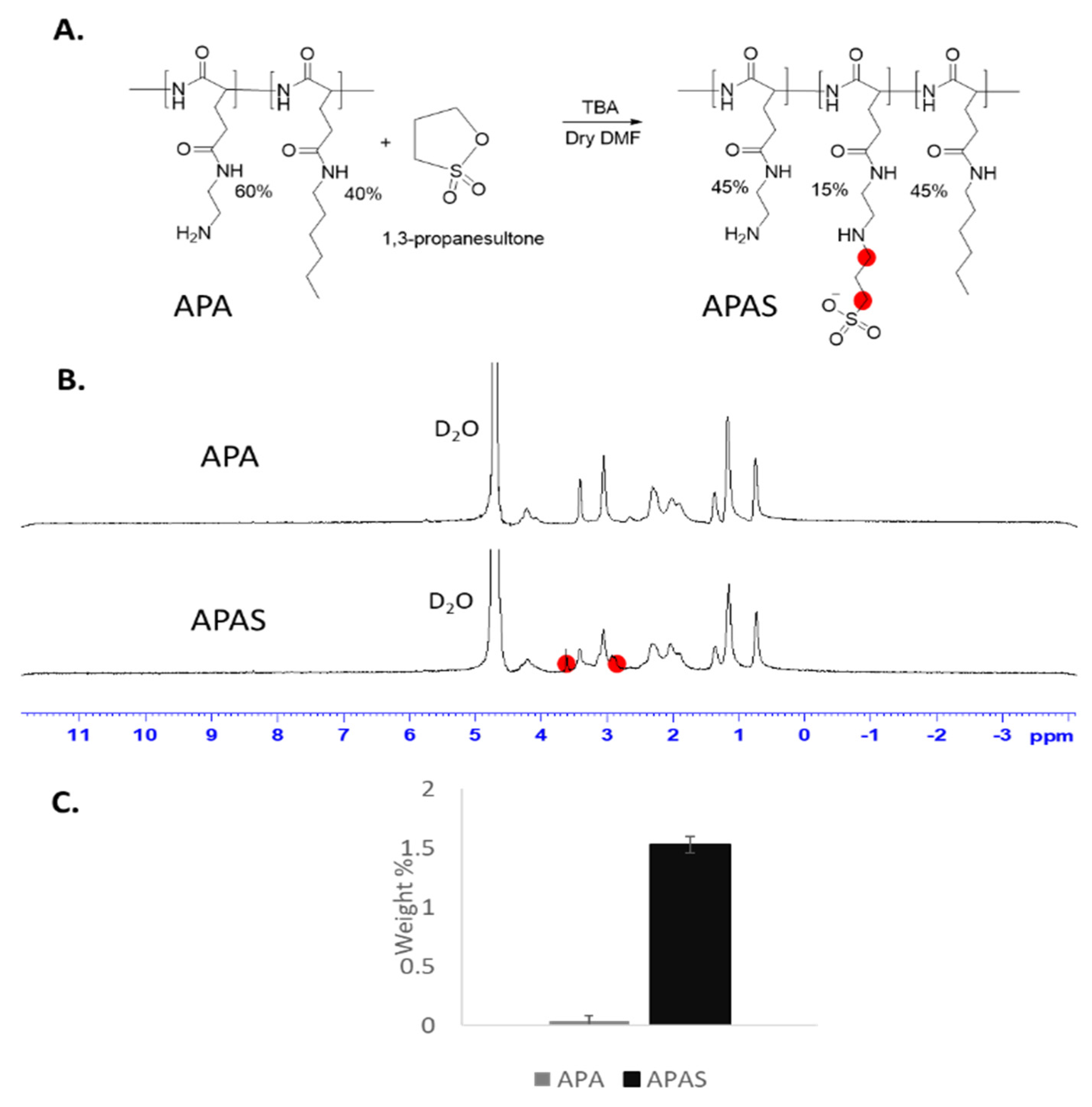
2.9. Synthesis of APA-Sulfonate (APAS)
2.10. Multi-Angle Static Light Scattering (MALS)
2.11. Scanning Electron Microscope (SEM)
2.12. Dynamic Light Scattering (DLS) and Phase Analysis Light Scattering (PALS)
2.13. Elemental Analysis (EDS)
2.14. Proliferation Assay
2.15. Multicellular Tumor Spheroids (MCTS)
2.16. SELP Expression in 3D vs. 2D Cell Culture
2.17. Infra-Red Spectroscopy
2.18. Electrophoretic Shift Assay (EMSA)
2.19. Western Blot
2.20. H-Nuclear Magnetic Resonance (NMR)
2.21. Internalization of APA/APAS:Cy5-siRNA Polyplexes into U251 Cells (2D)
2.22. Statistical Analysis
3. Results and Discussion
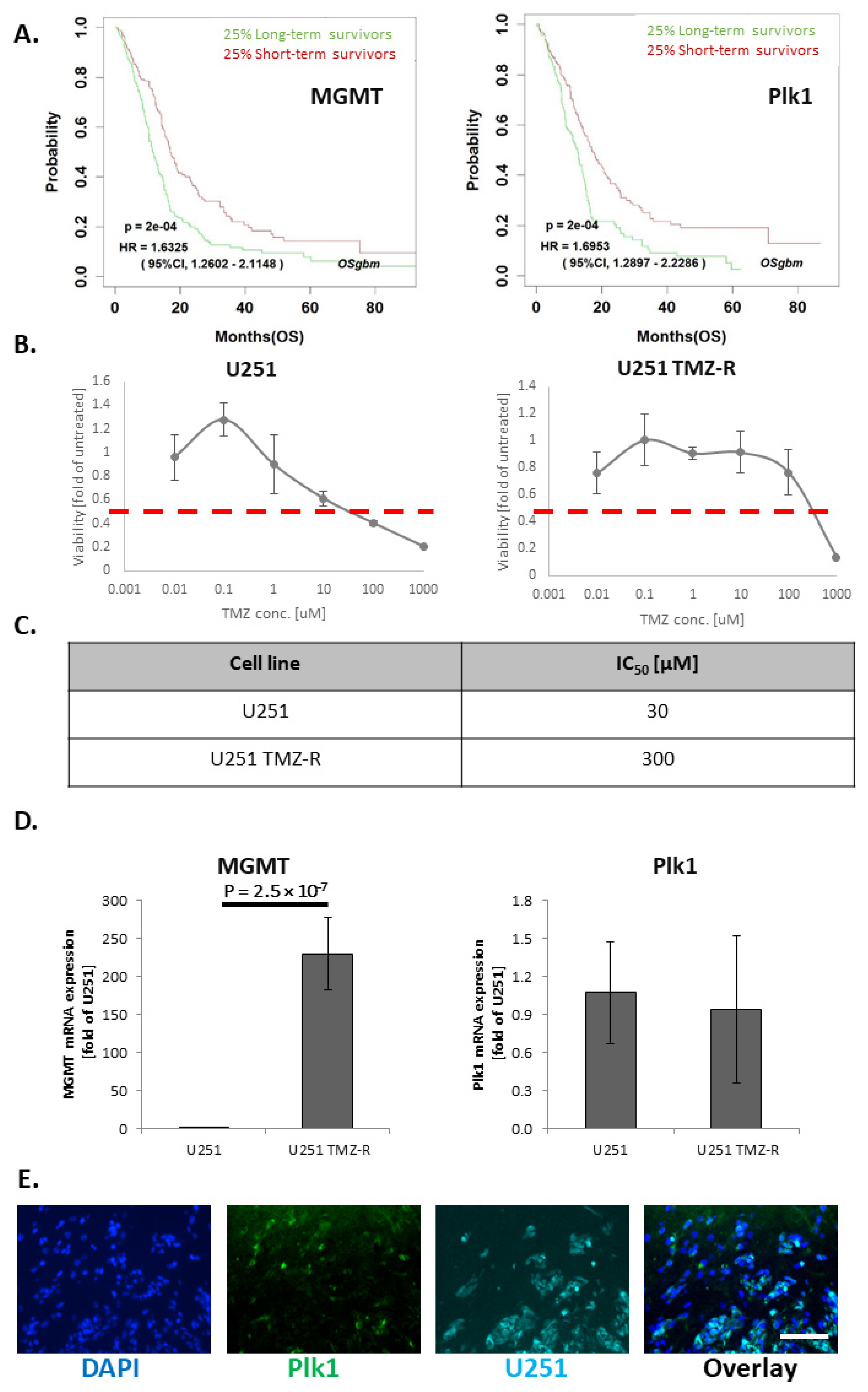
3.1. TMZ-Induced Resistance in U251 GB Cells Enhances MGMT mRNA Levels but Does Not Alter Plk1 mRNA Levels
3.2. APA:siPlk1 Complexes Efficiently Silence Plk1 Expression in Both U251 and U251 TMZ-R Cells
3.3. Treatment with APA:siPlk1 Polyplexes Reduces the Viability of Both U251 Cells and U251 TMZ-R Clone
3.4. SELP Is Expressed on the Membranes of GB Cells and Represents a Suitable Candidate for Active Targeting for the Delivery of siRNA Polyplexes
3.5. Sulfonation of Amphiphilic Poly(α)glutamate Amine (APA)
3.6. APAS forms Active Complexes with siRNA and Enables Silencing of GB-Relevant Genes
3.7. Sulfonate Modification Facilitated Internalization of Cy5-siRNA into U251 and U251 TMZ-R Spheroids
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gittleman, H.; Boscia, A.; Ostrom, Q.T.; Truitt, G.; Fritz, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. Survivorship in adults with malignant brain and other central nervous system tumor from 2000–2014. Neuro-Oncology 2018, 20 (Suppl. 7), vii6–vii16. [Google Scholar]
- Castro, M.G.; Cowen, R.; Williamson, I.K.; David, A.; Jimenez-Dalmaroni, M.J.; Yuan, X.; Bigliari, A.; Williams, J.C.; Hu, J.; Lowenstein, P.R. Current and future strategies for the treatment of malignant brain tumors. Pharmacol. Ther. 2003, 98, 71–108. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Schapira, A.H. Neurology and Clinical Neuroscience; Elsevier: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Rommelaere, J.; Raykov, Z.; Grekova, S.; Kiprijanova, I.; Geletneky, K.; Koch, U.; Aprahamian, M. Oncolytic Virotherapy for Prevention of Tumor Recurrence. U.S. Patent No. 9,861,669, 9 January 2018. [Google Scholar]
- Hegi, M.E.; Diserens, A.-C.; Gorlia, T.; Hamou, M.-F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [Green Version]
- Papachristodoulou, A.; Signorell, R.D.; Werner, B.; Brambilla, D.; Luciani, P.; Cavusoglu, M.; Grandjean, J.; Silginer, M.; Rudin, M.; Martin, E. Chemotherapy sensitization of glioblastoma by focused ultrasound-mediated delivery of therapeutic liposomes. J. Control. Release 2019, 295, 130–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sloan, A.E.; Roger, L.; Murphy, C.; Reese, J.; Lazarus, H.M.; Dropulic, B.; Gerson, S.L. A phase I study of MGMT-P140K transfected hematopoetic progenitor cells (HPC) combined with TMZ/O6BG dose escalation for newly diagnosed, MGMT unmethylated glioblastoma: Tolerance and evidence of survival benefit. Am. Soc. Clin. Oncol. 2019, 37, 2062. [Google Scholar] [CrossRef]
- Yu, W.; Zhang, L.; Wei, Q.; Shao, A. O6-methylguanine-DNA Methyltransferase (MGMT): Challenges and New Opportunities in Glioma Chemotherapy. Front. Oncol. 2020, 9, 1547. [Google Scholar] [CrossRef] [Green Version]
- Ofek, P.; Calderon, M.; Mehrabadi, F.S.; Krivitsky, A.; Ferber, S.; Tiram, G.; Yerushalmi, N.; Kredo-Russo, S.; Grossman, R.; Ram, Z.; et al. Restoring the oncosuppressor activity of microRNA-34a in glioblastoma using a polyglycerol-based polyplex. Nanomedicine 2016, 12, 2201–2214. [Google Scholar] [CrossRef] [Green Version]
- Tiram, G.; Ferber, S.; Ofek, P.; Eldar-Boock, A.; Ben-Shushan, D.; Yeini, E.; Krivitsky, A.; Blatt, R.; Almog, N.; Henkin, J.; et al. Reverting the molecular fingerprint of tumor dormancy as a therapeutic strategy for glioblastoma. FASEB J. 2018, 32, 5835–5850. [Google Scholar] [CrossRef]
- Zafir-Lavie, I.; Sherbo, S.; Goltsman, H.; Badinter, F.; Yeini, E.; Ofek, P.; Miari, R.; Tal, O.; Liran, A.; Shatil, T.; et al. Successful intracranial delivery of trastuzumab by gene-therapy for treatment of HER2-positive breast cancer brain metastases. J. Control. Release 2018, 291, 80–89. [Google Scholar] [CrossRef]
- Shatsberg, Z.; Zhang, X.; Ofek, P.; Malhotra, S.; Krivitsky, A.; Scomparin, A.; Tiram, G.; Calderón, M.; Haag, R.; Satchi-Fainaro, R. Functionalized nanogels carrying an anticancer microRNA for glioblastoma therapy. J. Control. Release 2016, 239, 159–168. [Google Scholar] [CrossRef]
- Higuchi, F.; Fink, A.L.; Kiyokawa, J.; Miller, J.J.; Koerner, M.V.; Cahill, D.P.; Wakimoto, H. PLK1 inhibition targets Myc-activated malignant glioma cells irrespective of mismatch repair deficiency–mediated acquired resistance to temozolomide. Mol. Cancer Ther. 2018, 17, 2551–2563. [Google Scholar] [CrossRef] [Green Version]
- Strebhardt, K.; Ullrich, A. Targeting polo-like kinase 1 for cancer therapy, Nature reviews. Cancer 2006, 6, 321–330. [Google Scholar] [PubMed]
- Pezuk, J.; Brassesco, M.; Morales, A.; de Oliveira, J.; Queiroz, R.d.; Machado, H.; Carlotti, C.; Neder, L.; Scrideli, C.; Tone, L. Polo-like kinase 1 inhibition causes decreased proliferation by cell cycle arrest, leading to cell death in glioblastoma. Cancer Gene Ther. 2013, 20, 499–506. [Google Scholar] [CrossRef] [Green Version]
- Lerner, R.G.; Grossauer, S.; Kadkhodaei, B.; Meyers, I.; Sidorov, M.; Koeck, K.; Hashizume, R.; Ozawa, T.; Phillips, J.J.; Berger, M.S. Targeting a Plk1-controlled polarity checkpoint in therapy-resistant glioblastoma-propagating cells. Cancer Res. 2015, 75, 5355–5366. [Google Scholar] [CrossRef] [Green Version]
- Polyak, D.; Krivitsky, A.; Scomparin, A.; Eliyahu, S.; Kalinski, H.; Avkin-Nachum, S.; Satchi-Fainaro, R. Systemic delivery of siRNA by aminated poly(α)glutamate for the treatment of solid tumors. J. Control. Release 2017, 257, 132–143. [Google Scholar] [CrossRef]
- Gibori, H.; Eliyahu, S.; Krivitsky, A.; Ben-Shushan, D.; Epshtein, Y.; Tiram, G.; Blau, R.; Ofek, P.; Lee, J.S.; Ruppin, E.; et al. Amphiphilic nanocarrier-induced modulation of PLK1 and miR-34a leads to improved therapeutic response in pancreatic cancer. Nat. Commun 2018, 9, 16. [Google Scholar] [CrossRef] [Green Version]
- Scomparin, A.; Polyak, D.; Krivitsky, A.; Satchi-Fainaro, R. Achieving successful delivery of oligonucleotides—From physico-chemical characterization to in vivo evaluation. Biotechnol. Adv. 2015, 33 Pt 3, 1294–1309. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shushan, D.; Markovsky, E.; Gibori, H.; Tiram, G.; Scomparin, A.; Satchi-Fainaro, R. Overcoming obstacles in microRNA delivery towards improved cancer therapy. Drug Deliv. Transl. Res. 2014, 4, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Tiram, G.; Scomparin, A.; Ofek, P.; Satchi-Fainaro, R. Interfering cancer with polymeric siRNA nanomedicines. J. Biomed. Nanotechnol. 2014, 10, 50–66. [Google Scholar] [CrossRef]
- Maeda, H.; Wu, J.; Sawa, T.; Matsumura, Y.; Hori, K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: A review. J. Control. Release 2000, 65, 271–284. [Google Scholar] [CrossRef]
- Markovsky, E.; Baabur-Cohen, H.; Eldar-Boock, A.; Omer, L.; Tiram, G.; Ferber, S.; Ofek, P.; Polyak, D.; Scomparin, A.; Satchi-Fainaro, R. Administration, distribution, metabolism and elimination of polymer therapeutics. J. Contr. Release 2012, 161, 446–460. [Google Scholar] [CrossRef]
- Krivitsky, A.; Polyak, D.; Scomparin, A.; Eliyahu, S.; Ori, A.; Avkin-Nachum, S.; Krivitsky, V.; Satchi-Fainaro, R. Structure–function correlation of aminated poly (α) glutamate as siRNA nanocarriers. Biomacromolecules 2016, 17, 2787–2800. [Google Scholar] [CrossRef]
- Krivitsky, A.; Krivitsky, V.; Polyak, D.; Scomparin, A.; Eliyahu, S.; Gibori, H.; Yeini, E.; Pisarevsky, E.; Blau, R.; Satchi-Fainaro, R. Molecular weight-dependent activity of aminated poly (α) glutamates as siRNA nanocarriers. Polymers 2018, 10, 548. [Google Scholar] [CrossRef] [Green Version]
- Krivitsky, A.; Polyak, D.; Scomparin, A.; Eliyahu, S.; Ofek, P.; Tiram, G.; Kalinski, H.; Avkin-Nachum, S.; Gracia, N.F.; Albertazzi, L.; et al. Amphiphilic poly(alpha)glutamate polymeric micelles for systemic administration of siRNA to tumors. Nanomedicine 2018, 14, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Yeini, E.; Ofek, P.; Albeck, N.; Ajamil, D.R.; Neufeld, L.; Eldar-Boock, A.; Kleiner, R.; Vaskovich, D.; Koshrovski-Michael, S.; Israeli, S.D. Targeting Glioblastoma: Advances in Drug Delivery and Novel Therapeutic Approaches. Adv. Ther. 2020, 4, 2000124. [Google Scholar] [CrossRef]
- Muldoon, L.L.; Soussain, C.; Jahnke, K.; Johanson, C.; Siegal, T.; Smith, Q.R.; Hall, W.A.; Hynynen, K.; Senter, P.D.; Peereboom, D.M. Chemotherapy delivery issues in central nervous system malignancy: A reality check. J. Clin. Oncol. 2007, 25, 2295–2305. [Google Scholar] [CrossRef] [Green Version]
- Lorant, D.E.; Topham, M.K.; Whatley, R.E.; McEver, R.P.; McIntyre, T.M.; Prescott, S.M.; Zimmerman, G.A. Inflammatory roles of P-selectin. J. Clin. Investig. 1993, 92, 559–570. [Google Scholar] [CrossRef] [Green Version]
- Läubli, H.; Borsig, L. Selectins Promote Tumor Metastasis; Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2010; pp. 169–177. [Google Scholar]
- Yeini, E.; Ofek, P.; Pozzi, S.; Albeck, N.; Ben-Shushan, D.; Tiram, G.; Golan, S.; Kleiner, R.; Sheinin, R.; Dangoor, S.I.; et al. P-selectin axis plays a key role in microglia immunophenotype and glioblastoma progression. Nat. Commun. 2021, 12, 1912. [Google Scholar] [CrossRef] [PubMed]
- Ferber, S.; Tiram, G. Co-targeting the tumor endothelium and P-selectin-expressing glioblastoma cells leads to a remarkable therapeutic outcome. eLife 2017, 6, e25281. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, P.P.; Moore, K.L.; McEver, R.P.; Cummings, R.D. Tyrosine sulfation of P-selectin glycoprotein ligand-1 is required for high affinity binding to P-selectin. J. Biol. Chem. 1995, 270, 22677–22680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segal, E.; Pan, H.; Ofek, P.; Udagawa, T.; Kopeckova, P.; Kopecek, J.; Satchi-Fainaro, R. Targeting angiogenesis-dependent calcified neoplasms using combined polymer therapeutics. PLoS ONE 2009, 4, e5233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santana-Magal, N.; Farhat-Younis, L.; Gutwillig, A.; Gleiberman, A.; Rasoulouniriana, D.; Tal, L.; Netanely, D.; Shamir, R.; Blau, R.; Feinmesser, M.; et al. Melanoma-Secreted Lysosomes Trigger Monocyte-Derived Dendritic Cell Apoptosis and Limit Cancer Immunotherapy. Cancer Res. 2020, 80, 1942–1956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiram, G.; Segal, E.; Krivitsky, A.; Shreberk-Hassidim, R.; Ferber, S.; Ofek, P.; Udagawa, T.; Edry, L.; Shomron, N.; Roniger, M. Identification of dormancy-associated microRNAs for the design of osteosarcoma-targeted dendritic polyglycerol nanopolyplexes. ACS Nano 2016, 10, 2028–2045. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Wang, Q.; Li, N.; Lv, J.; Ge, L.; Yang, M.; Zhang, G.; An, Y.; Wang, F.; Xie, L. OSgbm: An online consensus survival analysis web server for Glioblastoma. Front. Genet. 2020, 10, 1378. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-G.; Sim, S.J.; Kim, J.-H.; Kim, S.H.; Kim, Y.H. Synthesis and polymerization of methacryloyl-PEG-sulfonic acid as a functional macromer for biocompatible polymeric surfaces. Macromol. Res. 2004, 12, 379–383. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krivitsky, A.; Pozzi, S.; Yeini, E.; Israeli Dangoor, S.; Zur, T.; Golan, S.; Krivitsky, V.; Albeck, N.; Pisarevsky, E.; Ofek, P.; et al. Sulfonated Amphiphilic Poly(α)glutamate Amine—A Potential siRNA Nanocarrier for the Treatment of Both Chemo-Sensitive and Chemo-Resistant Glioblastoma Tumors. Pharmaceutics 2021, 13, 2199. https://doi.org/10.3390/pharmaceutics13122199
Krivitsky A, Pozzi S, Yeini E, Israeli Dangoor S, Zur T, Golan S, Krivitsky V, Albeck N, Pisarevsky E, Ofek P, et al. Sulfonated Amphiphilic Poly(α)glutamate Amine—A Potential siRNA Nanocarrier for the Treatment of Both Chemo-Sensitive and Chemo-Resistant Glioblastoma Tumors. Pharmaceutics. 2021; 13(12):2199. https://doi.org/10.3390/pharmaceutics13122199
Chicago/Turabian StyleKrivitsky, Adva, Sabina Pozzi, Eilam Yeini, Sahar Israeli Dangoor, Tal Zur, Sapir Golan, Vadim Krivitsky, Nitzan Albeck, Evgeny Pisarevsky, Paula Ofek, and et al. 2021. "Sulfonated Amphiphilic Poly(α)glutamate Amine—A Potential siRNA Nanocarrier for the Treatment of Both Chemo-Sensitive and Chemo-Resistant Glioblastoma Tumors" Pharmaceutics 13, no. 12: 2199. https://doi.org/10.3390/pharmaceutics13122199
APA StyleKrivitsky, A., Pozzi, S., Yeini, E., Israeli Dangoor, S., Zur, T., Golan, S., Krivitsky, V., Albeck, N., Pisarevsky, E., Ofek, P., Madi, A., & Satchi-Fainaro, R. (2021). Sulfonated Amphiphilic Poly(α)glutamate Amine—A Potential siRNA Nanocarrier for the Treatment of Both Chemo-Sensitive and Chemo-Resistant Glioblastoma Tumors. Pharmaceutics, 13(12), 2199. https://doi.org/10.3390/pharmaceutics13122199