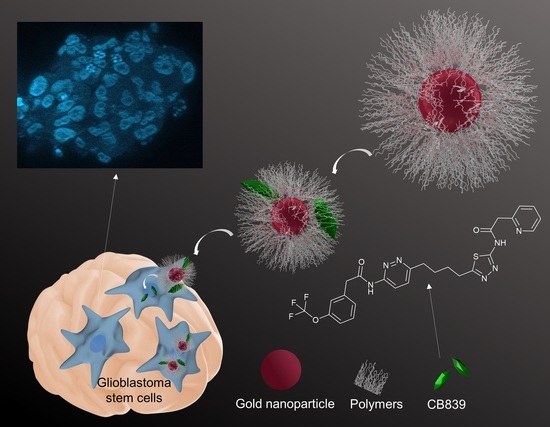
Augmented Therapeutic Potential of Glutaminase Inhibitor CB839 in Glioblastoma Stem Cells Using Gold Nanoparticle Delivery
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.3. Nanoparticle Synthesis
2.4. Loading of CB839 to Gold Nanoparticles (AuR-CB839, R: Cit, ThioPEG, PVP, PVA, PEI)
2.5. Synthesis of Fluorescent PVA Gold Nanoparticles (AuPVA-FITC)
2.6. Cell Cultures
2.7. Colony Formation Assays
2.8. Fluorescent Microscopy
3. Results and Discussion
3.1. Synthesis and Characterization of Au NPs
3.2. Quantification of CB839 Loading
3.3. Physicochemical Characterization of AuPVA-CB839 NPs
3.4. In Vitro Effect of AuPVA-CB839 NPs in GSCs
3.5. Cell Internalization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, Y.; Hsieh, I.-Y.; Huang, X.; Li, J.; Zhao, W. Glioblastoma Stem-Like Cells: Characteristics, Microenvironment and Therapy. Front. Pharmacol. 2016, 7, 477. [Google Scholar]
- Kahlert, U.D.; Mooney, S.M.; Natsumeda, M.; Steiger, H.-J.; Maciaczyk, J. Targeting cancer stem-like cells in glioblastoma and colorectal cancer through metabolic pathways. Int. J. Cancer 2017, 140, 10–22. [Google Scholar] [CrossRef]
- Harder, B.G.; Blomquist, M.R.; Wang, J.; Kim, A.J.; Woodworth, G.F.; Winkles, J.A.; Loftus, J.C.; Tran, N.L. Developments in Blood-Brain Barrier Penetrance and Drug Repurposing for Improved Treatment of Glioblastoma. Front. Oncol. 2018, 8, 462. [Google Scholar] [CrossRef]
- Garnier, D.; Renoult, O.; Alves-Guerra, M.-C.; Paris, F.; Pecqueur, C. Glioblastoma Stem-Like Cells, Metabolic Strategy to Kill a Challenging Target. Front. Oncol. 2019, 9, 118. [Google Scholar] [CrossRef]
- Cheng, L.; Wu, Q.; Guryanova, O.; Huang, Z.; Huang, Q.; Rich, J.N.; Bao, S. Elevated invasive potential of glioblastoma stem cells. Biochem. Biophys. Res. Commun. 2011, 406, 643–648. [Google Scholar] [CrossRef]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar]
- Singleton, D.C.; Dechaume, A.-L.; Murray, P.M.; Katt, W.P.; Baguley, B.C.; Leung, E.Y. Pyruvate anaplerosis is a mechanism of resistance to pharmacological glutaminase inhibition in triple-receptor negative breast cancer. BMC Cancer 2020, 20, 470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Mao, S.; Guo, Y.; Wu, Y.; Yao, X.; Huang, Y. Inhibition of GLS suppresses proliferation and promotes apoptosis in prostate cancer. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [PubMed]
- Seltzer, M.J.; Bennett, B.D.; Joshi, A.D.; Gao, P.; Thomas, A.G.; Ferraris, D.V.; Tsukamoto, T.; Rojas, C.J.; Slusher, B.S.; Rabinowitz, J.D.; et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010, 70, 8981–8987. [Google Scholar] [CrossRef]
- Choudhury, M.; Yin, X.; Schaefbauer, K.J.; Kang, J.-H.; Roy, B.; Kottom, T.J.; Limper, A.H.; Leof, E. SIRT7-mediated modulation of glutaminase 1 regulates TGF-β-induced pulmonary fibrosis. FASEB J. 2020, 34, 8920–8940. [Google Scholar] [CrossRef]
- Hoerner, C.R.; Chen, V.J.; Fan, A.C. The ‘Achilles Heel’ of Metabolism in Renal Cell Carcinoma: Glutaminase Inhibition as a Rational Treatment Strategy. Kidney Cancer 2019, 3, 5–29. [Google Scholar] [CrossRef]
- Yu, Y.; Yu, X.; Fan, C.; Wang, H.; Wang, R.; Feng, C.; Guan, H. Targeting glutaminase-mediated glutamine dependence in papillary thyroid cancer. J. Mol. Med. 2018, 96, 777–790. [Google Scholar] [CrossRef] [PubMed]
- Zacharias, N.M.; Baran, N.; Shanmugavelandy, S.S.; Lee, J.; Lujan, J.V.; Dutta, P.; Millward, S.W.; Cai, T.; Wood, C.G.; Piwnica-Worms, D.; et al. Assessing metabolic intervention with a glutaminase inhibitor in real-time by hyperpolarized magnetic resonance in acute myeloid leukemia. Mol. Cancer 2019, 18, 1937–1946. [Google Scholar] [CrossRef] [PubMed]
- Reckzeh, E.S.; Karageorgis, G.; Schwalfenberg, M.; Ceballos, J.; Nowacki, J.; Stroet, M.C.M.; Binici, A.; Knauer, L.; Brand, S.; Choidas, A.; et al. Inhibition of Glucose Transporters and Glutaminase Synergistically Impairs Tumor Cell Growth. Cell Chem. Biol. 2019, 26, 1214–1228. [Google Scholar] [CrossRef]
- De Lartigue, J. Hallmark tumor metabolism becomes a validated therapeutic target. J. Community Support. Oncol. 2018, 16, e47–e52. [Google Scholar] [CrossRef]
- Lukey, M.L.; Cerione, R.A. Starving the Devourer: Cutting Cancer Off from Its Favorite Foods. Cell Chem. Biol. 2019, 26, 1197–1199. [Google Scholar]
- Study of CB-839 in Combination w/Paclitaxel in Patients of African Ancestry and Non-African Ancestry With Advanced TNBC (Clinical Trial ID: NCT03057600). Available online: https://clinicaltrials.gov/ct2/show/NCT03057600 (accessed on 15 February 2021).
- Bennett, M.K.; Gross, M.I.; Bromley, S.D.; Li, J.; Chen, L.; Goyal, B.; Laidig, G.; Stanton, T.F.; Sjogren, E.B.; Calithera Biosciences, Inc. Treatment of Cancer with Heterocyclic Inhibitors of Glutaminase. International Publication No. WO2014/089048 Al, International Application No. PCT/US2013/072830, 12 June 2014. [Google Scholar]
- Gross, M.I.; Demo, S.D.; Dennison, J.B.; Chen, L.; Chernov-Rogan, T.; Goyal, B.; Janes, J.R.; Laidig, G.J.; Lewis, E.R.; Li, J.; et al. Antitumor Activity of the Glutaminase Inhibitor CB-839 in Triple-Negative Breast Cancer. Mol. Cancer Ther. 2014, 13, 890–901. [Google Scholar] [CrossRef]
- Restall, I.J.; Cseh, O.; Richards, L.M.; Pugh, T.J.; Luchman, H.A.; Weiss, S. Brain Tumor Stem Cell Dependence on Glutaminase Reveals a Metabolic Vulnerability through the Amino Acid Deprivation Response Pathway. Cancer Res. 2020, 80, 5478–5490. [Google Scholar] [CrossRef] [PubMed]
- Kahlert, U.D.; Cheng, M.; Koch, K.; Marchionni, L.; Fan, X.; Raabe, E.H.; Maciaczyk, J.; Glunde, K.; Eberhart, C.G. Alterations in cellular metabolome after pharmacological inhibition of Notch in glioblastoma cells. Int. J. Cancer 2016, 138, 1246–1255. [Google Scholar] [CrossRef] [PubMed]
- Koch, K.; Hartmann, R.; Tsiampali, J.; Uhlmann, C.; Nickel, A.-C.; He, X.; Kamp, M.; Sabel, M.; Barker, R.; Steiger, H.-J.; et al. A comparative pharmaco-metabolomic study of glutaminase inhibitors in glioma stem-like cells confirms biological effectiveness but reveals differences in target-specificity. Cell Death Discov. 2020, 6, 20. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, Y.; Ding, T.; Liu, J.; Zhao, H. Multifunctional Gold Nanoparticles: A Novel Nanomaterial for Various Medical Applications and Biological Activities. Front. Bioeng. Biotechnol. 2020. [Google Scholar] [CrossRef]
- Maeda, H.; Nakamura, H.; Fang, J. The EPR effect for macromolecular drug delivery to solid tumors: Improvement of tumor uptake, lowering of systemic toxicity, and distinct tumor imaging in vivo. Adv. Drug Deliv. Rev. 2013, 65, 71–79. [Google Scholar] [CrossRef]
- Giesen, B.; Nickel, A.-C.; Garzón-Manjón, A.; Vargas-Toscano, A.; Scheu, C.; Kahlert, U.D.; Janiak, C. Influence of synthesis methods on the internalization of fluorescent gold nanoparticles into glioblastoma stem-like cells. J. Inorg. Biochem. 2020, 203, 110952. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, S.C.; Wolf, E.F.; Luu, A.; Thomas, A.G.; Stathis, M.; Poore, B.; Nguyen, C.; Le, A.; Rojas, C.; Slusher, B.S.; et al. Allosteric Glutaminase Inhibitors Based on a 1,4-Di(5-amino-1,3,4-thiadiazol-2-yl)butane Scaffold. ACS Med. Chem. Lett. 2016, 7, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Mao, W.; Kim, H.S.; Son, Y.J.; Kim, S.R.; Yoo, H.S. Doxorubicin encapsulated clicked gold nanoparticle clusters exhibiting tumor-specific disassembly for enhanced tumor localization and computerized tomographic imaging. J. Control. Release 2018, 269, 52–62. [Google Scholar] [CrossRef]
- Ruan, S.; Yuan, M.; Zhang, L.; Hu, G.; Chen, J.; Cun, X.; Zhang, Q.; Yang, Y.; He, Q.; Gao, H. Tumor microenvironment sensitive doxorubicin delivery and release to glioma using angiopep-2 decorated gold nanoparticles. Biomaterials 2015, 37, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Feng, Q.; Chen, Y.; Shen, Y.; Su, Q.; Zhang, Y.; Zhou, X.; Cheng, Y. Comparison of Two Approaches for the Attachment of a Drug to Gold Nanoparticles and Their Anticancer Activities. Mol. Pharm. 2016, 13, 3308–3317. [Google Scholar] [CrossRef]
- Zhao, H.; Lin, Z.Y.; Yildirimer, L.; Dhinakar, A.; Zhao, X.; Wu, J. Polymer-based nanoparticles for protein delivery: Design, strategies and applications. J. Mater. Chem. B 2016, 4, 4060–4071. [Google Scholar] [CrossRef]
- Qin, X.; Li, Y. Strategies to Design and Synthesize Polymer-Based Stimuli-Responsive Drug-Delivery Nanosystems. ChemBioChem 2020, 21, 1236–1253. [Google Scholar] [CrossRef]
- Negron, K.; Khalasawi, N.; Lu, B.; Ho, C.-Y.; Lee, J.; Shenoy, S.; Mao, H.-Q.; Wang, T.-H.; Hanes, J.; Suk, J.S. Widespread gene transfer to malignant gliomas with In vitro-to-In vivo correlation. J. Control. Release 2019, 303, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Mhawech-Fauceglia, P.; Lee, N.; Parsanian, L.C.; Lin, Y.G.; Gayther, S.A.; Lawrenson, K. A three-dimensional microenvironment alters protein expression and chemosensitivity of epithelial ovarian cancer cells in vitro. Lab. Investig. 2013, 93, 528–542. [Google Scholar]
- Kang, M.S.; Lee, S.Y.; Kim, K.S.; Han, D.-W. State of the Art Biocompatible Gold Nanoparticles for Cancer Theragnosis. Pharmaceutics 2020, 12, 701. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.D.; Wu, D.; Shen, X.; Liu, P.X.; Yang, N.; Zhao, B.; Zhang, H.; Sun, Y.M.; Zhang, L.A.; Fan, F.Y. Size-dependent in vivo toxicity of PEG-coated gold nanoparticles. Int. J. Nanomed. 2011, 6, 2071–2081. [Google Scholar] [CrossRef]
- Paradossi, G.; Cavalieri, F.; Chiessi, E.; Spagnoli, C.; Cowman, M.K. Poly(vinyl alcohol) as versatile biomaterial for potential biomedical applications. J. Mater. Sci. Mater. Med. 2003, 14, 687–691. [Google Scholar] [CrossRef] [PubMed]
- Acharya, A.P.; Chan, S.Y.W.; Little, S.R.; University of Pittsburgh of the Commonwealth System of Higher Education. Compositions and Methods for Administering a YAP1/WWRT1 Inhibiting Composition and a GLS1 Inhibiting Composition. International Publication No. WO2019/104038 Al, International Application No. PCT/US2018/062013, 31 May 2019. [Google Scholar]
- Ruan, B.; Ruan, J. Faming Zhuanli Shenqing. Liposome containing glutamine metabolism inhibitor and pharmaceutical composition and use thereof. Patent No. CN107714650, 23 February 2018. [Google Scholar]
- Adewale, O.B.; Davids, H.; Cairncross, L.; Roux, S. Toxicological Behavior of Gold Nanoparticles on Various Models: Influence of Physicochemical Properties and Other Factors. Int. J. Toxicol. 2019, 38, 357–384. [Google Scholar] [CrossRef] [PubMed]
- Sanabria, N.M.; Gulumian, M. The presence of residual gold nanoparticles in samples interferes with the RT-qPCR assay used for gene expression profiling. J. Nanobiotechnol. 2017, 15, 1–26. [Google Scholar] [CrossRef]
- Ponti, J.; Kinsner-Ovaskainen, A.; Norlen, H.; Altmeyer, S.; Cristina, A.; Bogni, A. Interlaboratory Comparison Study of the Colony Forming Efficiency Assay for Assessing Cytotoxicity of Nanomaterials. EUR—Scientific and Technical Research Reports; Report No. 978-92-79-44677-1; Publications Office of the European Union: Luxembourg, 2014. [Google Scholar]
- Ernst Ruska-Centre for Microscopy and Spectroscopy with Electrons. FEI Tecnai G2 F20. J. Large-Scale Res. Facil. 2016, 2, 77. [Google Scholar] [CrossRef]
- Kahlert, U.D.; Suwala, A.K.; Raabe, E.H.; Siebzehnrubl, F.A.; Suarez, M.J.; Orr, B.A.; Bar, E.E.; Maciaczyk, J.; Eberhart, C.G. ZEB1 promotes invasion in human fetal neural stem cells and hypoxic glioma neurospheres. Brain Pathol. 2015, 25, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Liston, D.R.; Davis, M. Clinically Relevant Concentrations of Anticancer Drugs: A Guide for Nonclinical Studies. Clin. Cancer Res. 2017, 23, 3489–3498. [Google Scholar] [CrossRef]
- Tsiampali, J.; Neumann, S.; Giesen, B.; Koch, K.; Maciaczyk, D.; Janiak, C.; Hänggi, D.; Maciaczyk, J. Enzymatic Activity of CD73 Modulates Invasion of Gliomas via Epithelial-Mesenchymal Transition-Like Reprogramming. Pharmaceutics 2020, 13, 378. [Google Scholar] [CrossRef]
- Turkevich, J.; Stevenson, P.C.; Hillier, J. A study of the nucleation and growth processes in the synthesis of colloidal gold. Discuss. Faraday Soc. 1951, 11, 55–75. [Google Scholar] [CrossRef]
- Zhang, H.; Ren, P.; Yang, F.; Chen, J.; Wang, C.; Zhou, Y.; Fu, J. Biomimetic epidermal sensors assembled from polydopamine-modified reduced graphene oxide/polyvinyl alcohol hydrogels for the real-time monitoring of human motions. J. Mater. Chem. B 2020, 8, 10549. [Google Scholar] [CrossRef]
- McHugh, K.; Jing, L.; Severt, S.Y.; Cruz, M.; Sarmadi, M.; Jayawardena, H.S.N.; Perkinson, C.F.; Larusson, F.; Rose, S.; Tomasic, S.; et al. Biocompatible near- infrared quantum dots delivered to the skin by microneedle patches record vaccination. Sci. Transl. Med. 2019, 11, e7162. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Stalnecker, C.; Zhang, C.; McDermott, L.A.; Iyer, P.; O’Neill, J.; Reimer, S.; Cerione, R.A.; Katt, W.P. Characterization of the interactions of potent allosteric inhibitors with glutaminase C, a key enzyme in cancer cell glutamine metabolism. J. Biol. Chem. 2018, 293, 3535–3545. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Tian, X.; Wu, A.; Li, J.; Tian, J.; Chong, Y.; Chai, Z.; Zhao, Y.; Chen, C.; Ge, C. Protein Corona Influences Cellular Uptake of Gold Nanoparticles by Phagocytic; Nonphagocytic Cells in a Size-Dependent Manner. ACS Appl. Mater. Interfaces 2015, 7, 20568–20575. [Google Scholar] [CrossRef] [PubMed]
- Larsson, P.; Engqvist, H.; Biermann, J.; Rönnerman, E.W.; Forssell-Aronsson, E.; Kovács, A.; Karlsson, P.; Helou, K.; Parris, T.Z. Optimization of cell viability assays to improve replicability and reproducibility of cancer drug sensitivity screens. Sci. Rep. 2020, 10, 5798. [Google Scholar] [CrossRef] [PubMed]
- Elgogary, A.; Xu, Q.; Poore, B.; Alt, J.; Zimmermann, S.C.; Zhao, L.; Fu, J.; Chen, B.; Xia, S.; Liu, Y.; et al. Combination therapy with BPTES nanoparticles and metformin targets the metabolic heterogeneity of pancreatic cancer. Proc. Natl. Acad. Sci. USA 2016, 113, e5328–e5336. [Google Scholar] [CrossRef] [PubMed]
- Fischer, I.; Nickel, A.-C.; Qin, N.; Taban, K.; Pauck, D.; Steiger, H.-J.; Kamp, M.; Muhammad, S.; Hänggi, D.; Fritsche, E.; et al. Different Calculation Strategies Are Congruent in Determining Chemotherapy Resistance of Brain Tumors In Vitro. Cells 2020, 9, 2689. [Google Scholar] [CrossRef]
- Khadka, S.; Arthur, K.; Washington, M.; Barekatain, Y.; Ackroyd, J.; Behr, E.; Suriyamongkol, P.; Lin, Y.-H.; Crowley, K.; Pham, C.D.; et al. Impaired Anaplerosis Is a Major Contributor to Glycolysis Inhibitor Toxicity in Glioma. PREPRINT 2020. [Google Scholar] [CrossRef]
- Sokolova, V.; Mekky, G.; Van der Meer, S.B.; Seeds, M.C.; Atala, A.J.; Epple, M. Transport of ultrasmall gold nanoparticles (2 nm) across the blood-brain barrier in a six-cell brain spheroid model. Sci. Rep. 2020, 10, 18033. [Google Scholar] [CrossRef]
- Khongkow, M.; Yata, T.; Boonrungsiman, S.; Ruktanonchai, U.R.; Graham, D.; Namdee, K. Surface modification of gold nanoparticles with neuron-targeted exosome for enhanced blood-brain barrier penetration. Sci. Rep. 2019, 9, 8278. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Dai, Q.; Morshed, R.A.; Fan, X.; Wegscheid, M.L.; Wainwright, D.A.; Han, Y.; Zhang, L.; Auffinger, B.; Tobias, A.L.; et al. Blood-brain barrier permeable gold nanoparticles: An efficient delivery plat-form for enhanced malignant glioma therapy and imaging. Small 2014, 10, 5137–5150. [Google Scholar] [CrossRef]
- Sheleg, S.V.; Korotkevich, E.A.; Zhavrid, E.A.; Muravskaya, G.V.; Smeyanovich, A.F.; Shanko, Y.G.; Yurkshtovich, T.L.; Bychkovsky, P.B.; Belyaev, S.A. Local chemotherapy with cisplatin-depot for glioblastoma multiforme. J. Neurooncol. 2002, 60, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Nam, L.; Coll, C.; Erthal, L.C.S.; de la Torre, C.; Serrano, D.; Martínez-Máñez, R.; Santos-Martínez, M.J.; Ruiz-Hernández, E. Drug Delivery Nanosystems for the Localized Treatment of Glioblastoma Multiforme. Materials 2018, 11, 779. [Google Scholar] [CrossRef]
- Lenting, K.; Verhaak, R.; Ter Laan, M.; Wesseling, P.; Leenders, W. Glioma: Experimental models and reality. Acta Neuropathol. 2017, 133, 263–282. [Google Scholar] [CrossRef]
- Huszthy, P.C.; Daphu, I.; Niclou, S.P.; Stieber, D.; Nigro, J.M.; Sakariassen, P.; Miletic, H.; Thorsen, F.; Bjerkvig, R. In vivo models of primary brain tumors: Pitfalls and perspectives. Neuro. Oncol. 2012, 14, 979–993. [Google Scholar] [CrossRef] [PubMed]
- Robertson, F.L.; Marqués-Torrejón, M.A.; Morrison, G.M.; Pollard, S.M. Experimental models and tools to tackle glioblastoma. Dis. Models Mech. 2019, 12. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, U.M.; Tyler, B.; Patta, Y.; Wicks, R.; Spencer, K.; Scott, A.; Masi, B.; Hwang, L.; Grossman, R.; Cima, M.; et al. Intracranial microcapsule chemotherapy delivery for the localized treatment of rodent metastatic breast adenocarcinoma in the brain. Proc. Natl. Acad. Sci. USA 2014, 111, 16071–16076. [Google Scholar] [CrossRef]
- Semenkow, S.; Li, S.; Kahlert, U.D.; Raabe, E.H.; Xu, J.; Arnold, A.; Janowski, M.; Oh, B.C.; Brandacher, G.; Bulte, J.W.M.; et al. An immunocompetent mouse model of human glioblastoma. Oncotarget 2017, 8, 61072–61082. [Google Scholar] [CrossRef][Green Version]
- Lan, X.; Kedziorek, D.A.; Chu, C.; Jablonska, A.; Li, S.; Kai, M.; Liang, Y.; Janowski, M.; Walczak, P. Modeling human pediatric and adult gliomas in immunocompetent mice through costimulatory blockade. Oncoimmunology 2020, 9, 1776577. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.-C.; Hsu, P.-H.; Lee, Y.-F.; Lin, Y.-W.; Huang, C.-C. Selective Detection of Iodide and Cyanide Anions Using Gold-Nanoparticle-Based Fluorescent Probes. ACS Appl. Mater. Interfaces 2012, 4, 2652–2658. [Google Scholar] [CrossRef]
- Zhao, P.; Li, N.; Astruc, D. State of the art in gold nanoparticle synthesis. Coord. Chem. Rev. 2013, 257, 638–665. [Google Scholar] [CrossRef]
- Boles, M.; Ling, D.; Hyeon, T.; Talapin, D.V. The surface science of nanocrystals. Nat. Mater. 2016, 15, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Li, X.; Li, H.; Zhang, W. Quantifying thiol–gold interactions towards the efficient strength control. Nat. Commun. 2014, 5, 4348. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, X.; Zhang, Z.; Chen, L. Highly sensitive fluorescence detection of copper ion based on its catalytic oxidation to cysteine indicated by fluorescein isothiocyanate functionalized gold nanoparticles. Colloids Surf. A Physicochem. Eng. Asp. 2015, 468, 333–338. [Google Scholar] [CrossRef]
- Donahue, N.D.; Acar, H.; Wilhelm, S. Concepts of nanoparticle cellular uptake, intracellular trafficking, and kinetics in nanomedicine. Adv. Drug Deliv. Rev. 2019, 143, 68–96. [Google Scholar] [CrossRef] [PubMed]







| Au NPs | Drug Loading Efficiency (%) |
|---|---|
| AuCit | 8 |
| AuThioPEG | 4 |
| AuPVA | 12 |
| AuPVP | 0 |
| AuPEI | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giesen, B.; Nickel, A.-C.; Barthel, J.; Kahlert, U.D.; Janiak, C. Augmented Therapeutic Potential of Glutaminase Inhibitor CB839 in Glioblastoma Stem Cells Using Gold Nanoparticle Delivery. Pharmaceutics 2021, 13, 295. https://doi.org/10.3390/pharmaceutics13020295
Giesen B, Nickel A-C, Barthel J, Kahlert UD, Janiak C. Augmented Therapeutic Potential of Glutaminase Inhibitor CB839 in Glioblastoma Stem Cells Using Gold Nanoparticle Delivery. Pharmaceutics. 2021; 13(2):295. https://doi.org/10.3390/pharmaceutics13020295
Chicago/Turabian StyleGiesen, Beatriz, Ann-Christin Nickel, Juri Barthel, Ulf Dietrich Kahlert, and Christoph Janiak. 2021. "Augmented Therapeutic Potential of Glutaminase Inhibitor CB839 in Glioblastoma Stem Cells Using Gold Nanoparticle Delivery" Pharmaceutics 13, no. 2: 295. https://doi.org/10.3390/pharmaceutics13020295
APA StyleGiesen, B., Nickel, A.-C., Barthel, J., Kahlert, U. D., & Janiak, C. (2021). Augmented Therapeutic Potential of Glutaminase Inhibitor CB839 in Glioblastoma Stem Cells Using Gold Nanoparticle Delivery. Pharmaceutics, 13(2), 295. https://doi.org/10.3390/pharmaceutics13020295