Asparaginase-Phage P22 Nanoreactors: Toward a Biobetter Development for Acute Lymphoblastic Leukemia Treatment
Abstract
1. Introduction
2. Materials and Methods
2.1. ASNase-SP Expression
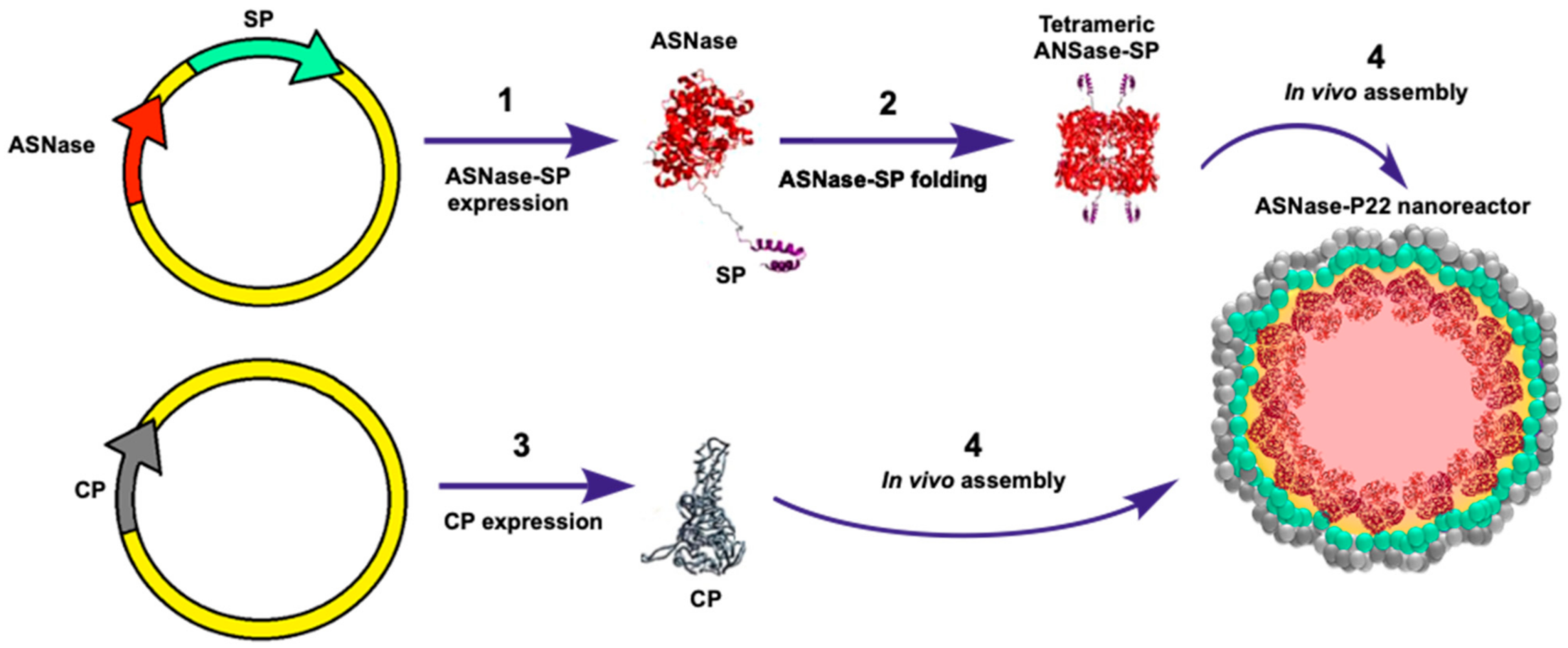
2.2. Production of ASNase-P22 Nanoreactors
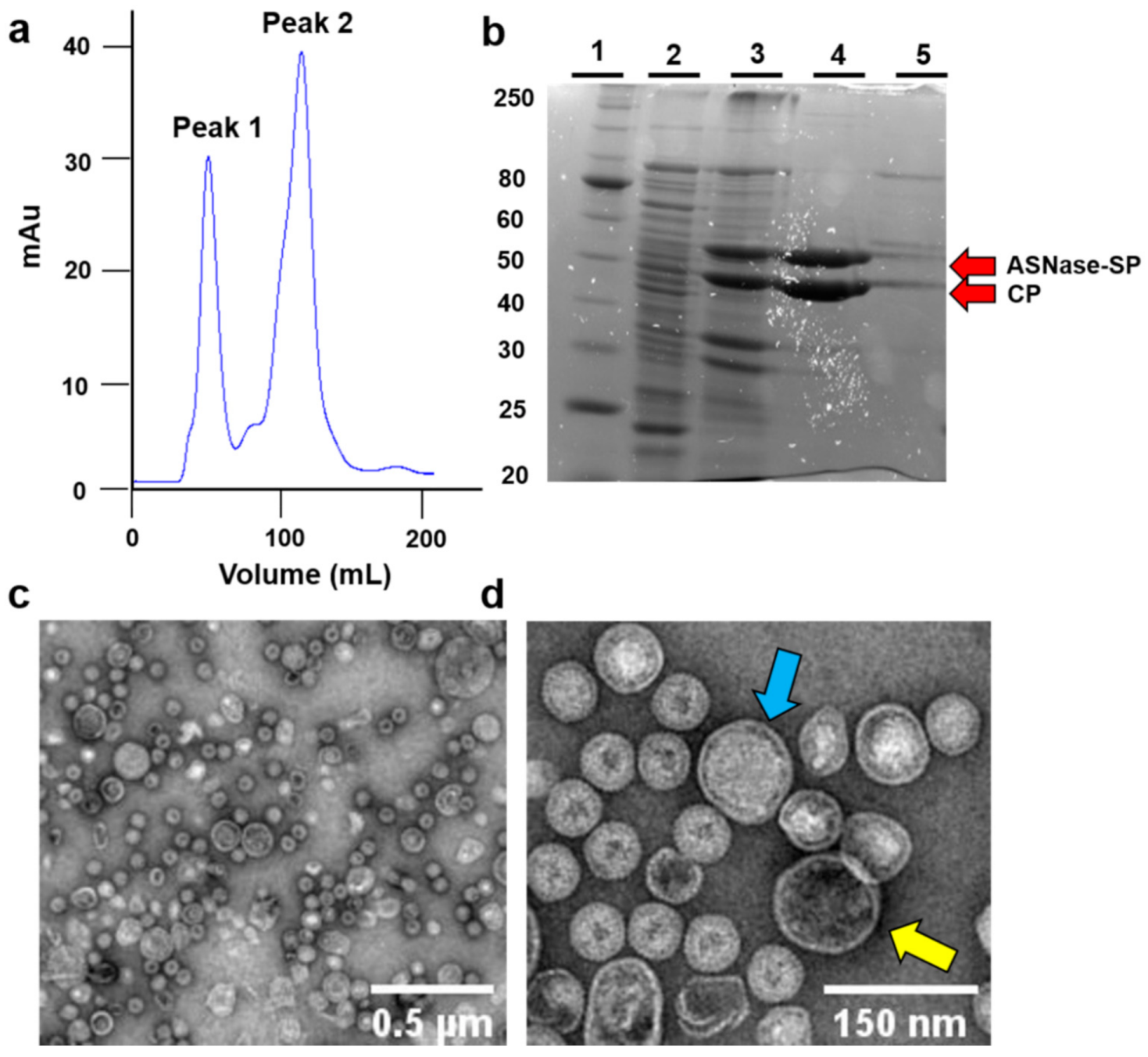
2.3. Purification of ASNase-P22 Nanoreactors
2.4. Capsid Composition
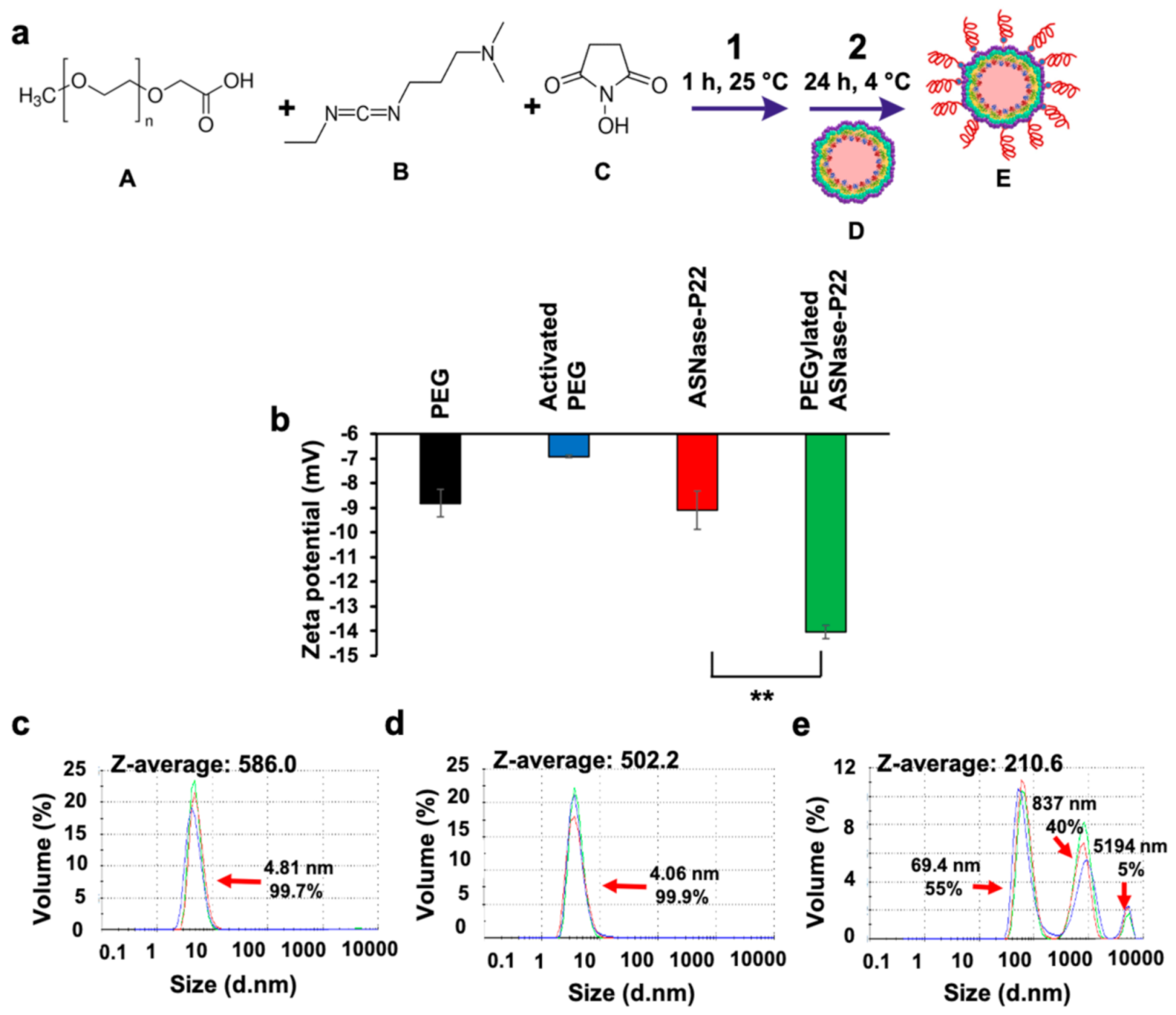
2.5. PEGylation of ASNase-P22 Nanoreactors
2.6. ASNase-P22 Nanoreactors Characterization
2.6.1. Size and Zeta Potential
2.6.2. TEM Characterization
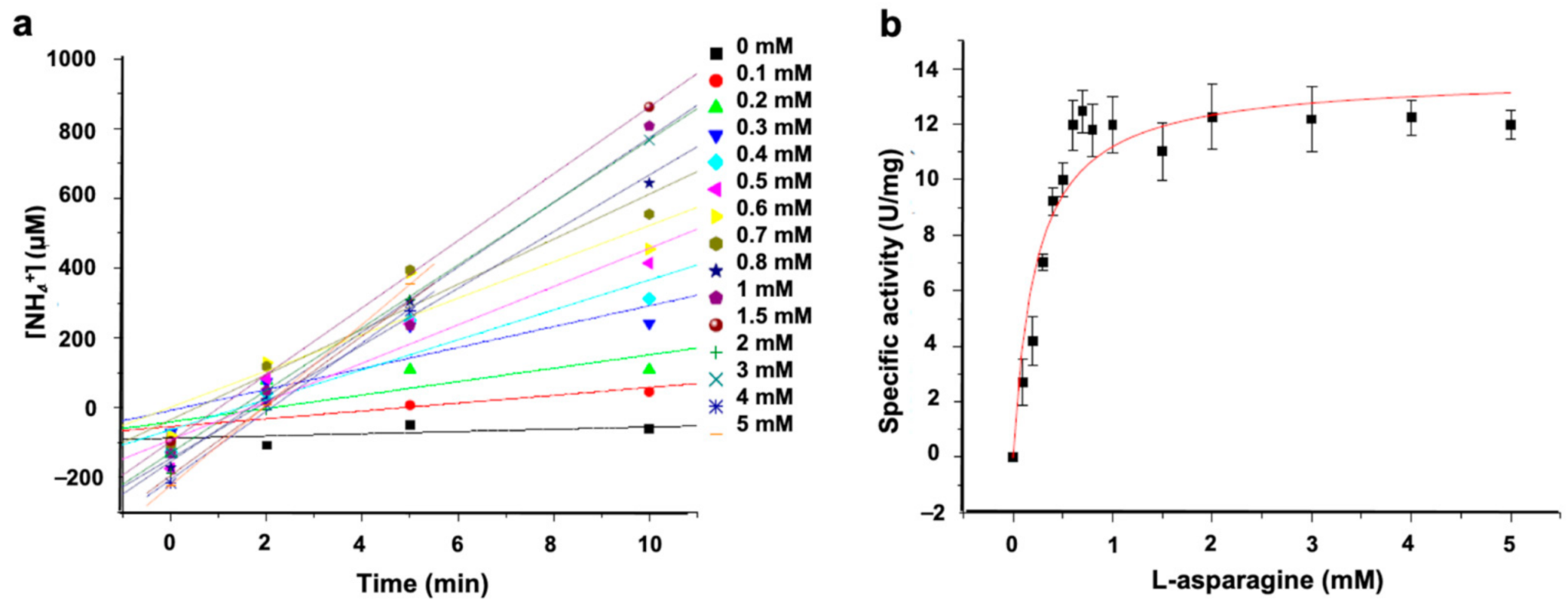
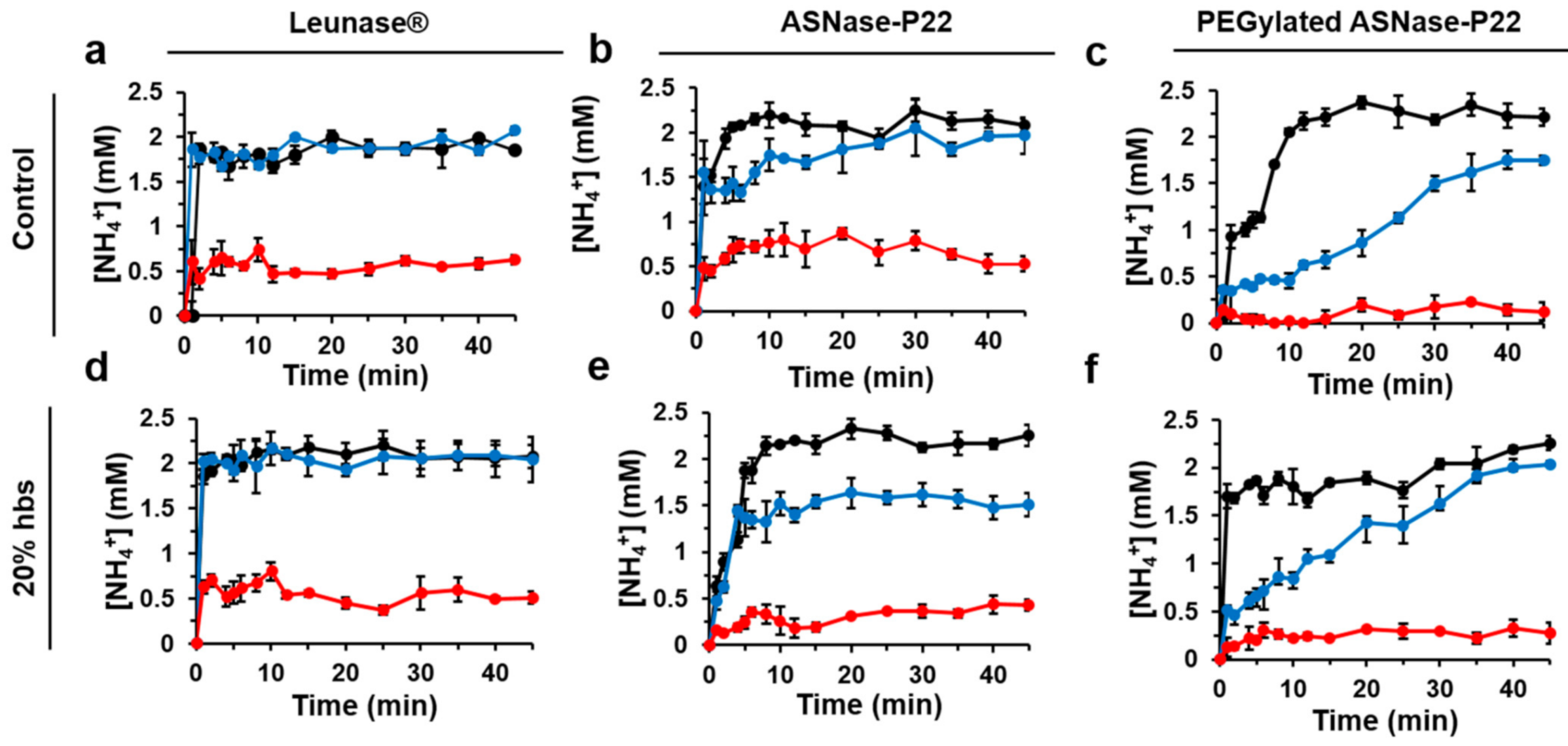
2.6.3. ASNase Activity
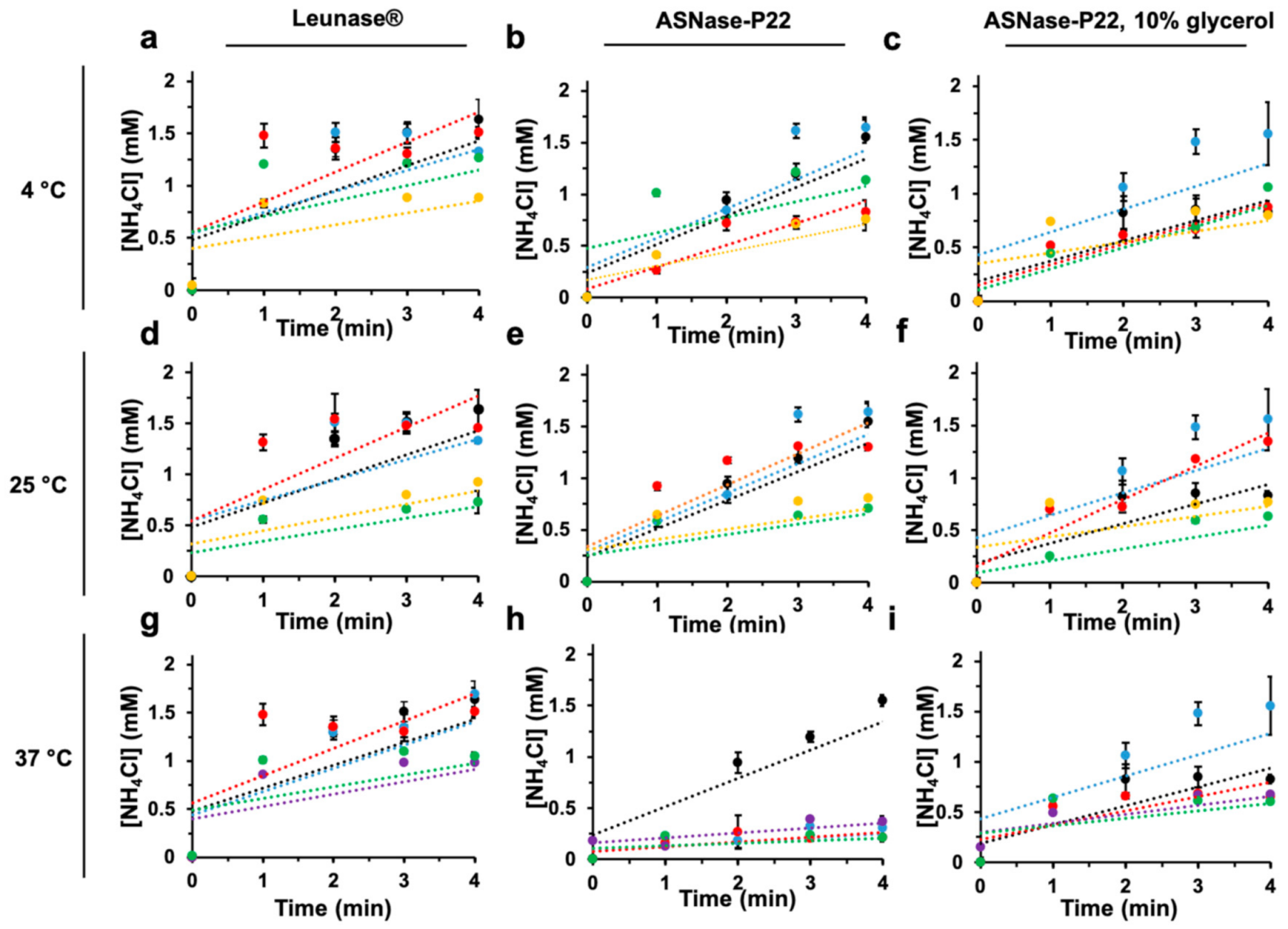
2.6.4. Thermal Stability
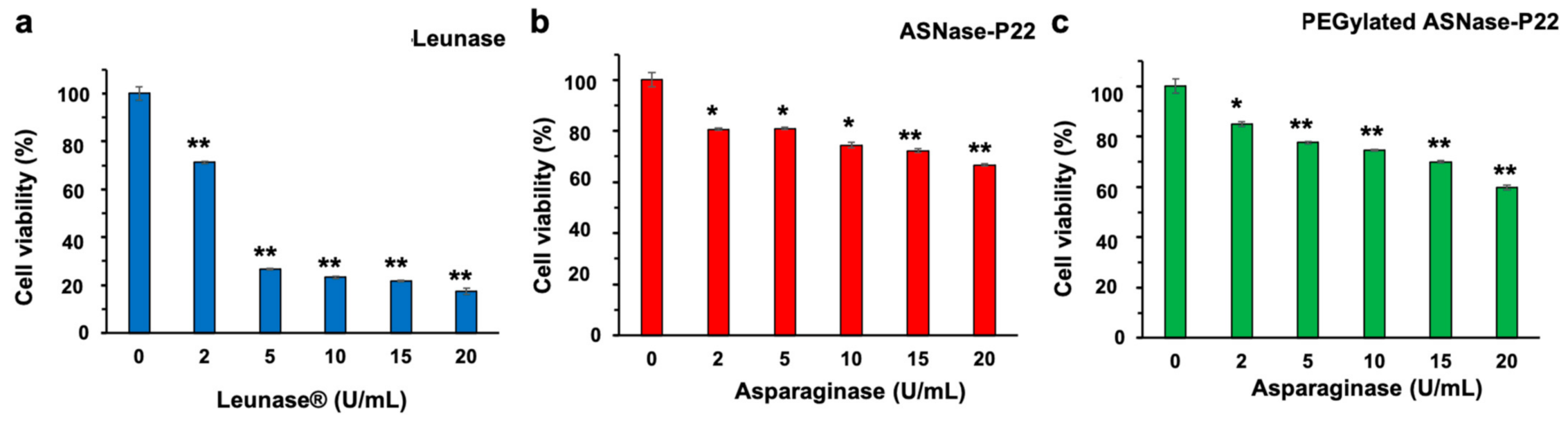
2.6.5. ASNase-P22 Cytotoxicity
2.7. Statistical Analysis
3. Results
3.1. Expression and Purification of ASNase-P22 Nanoreactors
3.2. Characterization of ASNase-P22 Nanoreactors
3.2.1. Composition
3.2.2. ASNase-P22 Nanoreactors Kinetics
3.2.3. Stability of ASNase-P22 Nanoreactors
3.2.4. Thermostability
3.2.5. Protein Aggregation
3.3. ASNase-P22 Nanoreactors PEGylation
3.4. PEGylated ASNase-P22 Nanoreactors Stability and Cytotoxicity
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Terwilliger, T.; Abdul-Hay, M. Acute lymphoblastic leukemia: A comprehensive review and 2017 update. Blood Cancer J. 2017, 7, e577. [Google Scholar] [CrossRef] [PubMed]
- Hunger, S.P.; Teachey, D.T.; Grupp, S.; Aplenc, R. Childhood Leukemia. Abeloff’s Clin. Oncol. 2020, 1748–1764.e4. [Google Scholar] [CrossRef]
- Pui, C.-H.; Robison, L.L.; Look, A.T. Acute lymphoblastic leukaemia. Lancet 2008, 371, 1030–1043. [Google Scholar] [CrossRef]
- Ortega, J.A.; Nesbit, M.E.; Donaldson, M.H.; Hittle, R.E.; Weiner, J.; Karon, M.; Hammond, D. L-Asparaginase, vincristine, and prednisone for induction of first remission in acute lymphocytic leukemia. Cancer Res. 1977, 37, 535–540. [Google Scholar]
- Esterhay, R.J.; Wiernik, P.H.; Grove, W.R.; Markus, S.D.; Wesley, M.N. Moderate dose methotrexate, vincristine, asparaginase, and dexamethasone for treatment of adult acute lymphocytic leukemia. Blood 1982, 59, 334–345. [Google Scholar] [CrossRef] [PubMed]
- Pui, C.-H.; Evans, W.E. Treatment of Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2006, 354, 166–178. [Google Scholar] [CrossRef]
- Kumar, K.; Kaur, J.; Walia, S.; Pathak, T.; Aggarwal, D. l-asparaginase: An effective agent in the treatment of acute lymphoblastic leukemia. Leuk. Lymphoma 2013, 55, 256–262. [Google Scholar] [CrossRef]
- Broome, J.D. Evidence that the L-Asparaginase Activity of Guinea Pig Serum is responsible for its Antilymphoma Effects. Nat. Cell Biol. 1961, 191, 1114–1115. [Google Scholar] [CrossRef]
- Bussolati, O.; Belletti, S.; Uggeri, J.; Gatti, R.; Orlandini, G.; Dall’Asta, V.; Gazzola, G.C. Characterization of Apoptotic Phenomena Induced by Treatment with L-Asparaginase in NIH3T3 Cells. Exp. Cell Res. 1995, 220, 283–291. [Google Scholar] [CrossRef]
- Pieters, R.; Hunger, S.; Boos, J.; Rizzari, C.; Silverman, L.; Baruchel, A.; Goekbuget, N.; Schrappe, M.; Pui, C. L-asparaginase treatment in acute lymphoblastic leukemia. Cancer 2010, 117, 238–249. [Google Scholar] [CrossRef]
- Tong, W.; Pieters, R.; Kaspers, G.; Loo, D.; Bierings, M.; Bos, C.V.D.; Kollen, W.J.W.; Hop, W.; Lanvers-Kaminsky, C.; Relling, M.; et al. A prospective study on drug monitoring of PEGasparaginase and Erwinia asparaginase and asparaginase antibodies in pediatric acute lymphoblastic leukemia. Blood 2014, 123, 2026–2033. [Google Scholar] [CrossRef]
- Brumano, L.P.; Da Silva, F.V.S.; Costa-Silva, T.A.; Apolinário, A.C.; Santos, J.H.P.M.; Kleingesinds, E.K.; Monteiro, G.; Rangel-Yagui, C.D.O.; Benyahia, B.; Junior, A.P. Development of L-Asparaginase Biobetters: Current Research Status and Review of the Desirable Quality Profiles. Front. Bioeng. Biotechnol. 2019, 6, 212. [Google Scholar] [CrossRef]
- Shrivastava, A.; Khan, A.A.; Khurshid, M.; Kalam, M.A.; Jain, S.K.; Singhal, P.K. Recent developments in l-asparaginase discovery and its potential as anticancer agent. Crit. Rev. Oncol. 2016, 100, 1–10. [Google Scholar] [CrossRef]
- Lanvers-Kaminsky, C. Asparaginase pharmacology: Challenges still to be faced. Cancer Chemother. Pharmacol. 2017, 79, 439–450. [Google Scholar] [CrossRef]
- Narta, U.K.; Kanwar, S.S.; Azmi, W. Pharmacological and clinical evaluation of l-asparaginase in the treatment of leukemia. Crit. Rev. Oncol. 2007, 61, 208–221. [Google Scholar] [CrossRef]
- Moreno-Enriquez, A.; Evangelista-Martínez, Z.; Gonzalez-Mondragon, E.G.; Calderon-Flores, A.; Arreguin, R.; Pérez-Rueda, E.; Huerta-Saquero, A. Biochemical characterization of recombinant L-asparaginase (AnsA) from Rhizobium etli, a member of an increasing rhizobial-type family of L-asparaginases. J. Microbiol. Biotechnol. 2012, 22, 292–300. [Google Scholar] [CrossRef]
- Huerta-Saquero, A.; Evangelista-Martínez, Z.; Moreno-Enriquez, A.; Perez-Rueda, E. Rhizobium etli asparaginase II: An alternative for acute lymphoblastic leukemia (ALL) treatment. Bioengineered 2013, 4, 30–36. [Google Scholar] [CrossRef]
- Lopes, A.M.; De Oliveira-Nascimento, L.; Ribeiro, A.; Tairum, C.A.; Breyer, C.A.; De Oliveira, M.A.; Monteiro, G.; De Souza-Motta, C.M.; Magalhães, P.D.O.; Avendaño, J.G.F.; et al. Therapeuticl-asparaginase: Upstream, downstream and beyond. Crit. Rev. Biotechnol. 2015, 37, 82–99. [Google Scholar] [CrossRef]
- Bosio, V.E.; Islan, G.A.; Martínez, Y.N.; Durán, N.; Castro, G. Nanodevices for the immobilization of therapeutic enzymes. Crit. Rev. Biotechnol. 2015, 36, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Ulu, A.; Ates, B. Immobilization of l-Asparaginase on Carrier Materials: A Comprehensive Review. Bioconjugate Chem. 2017, 28, 1598–1610. [Google Scholar] [CrossRef]
- Junior, C.D.R.; Caseli, L. Adsorption and enzyme activity of asparaginase at lipid Langmuir and Langmuir-Blodgett films. Mater. Sci. Eng. C 2017, 73, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, M.M.; Perez-Soler, R.; Cruz, M.E.M. Biological characterization of L -asparaginase liposomal formulations. Cancer Chemother. Pharmacol. 1996, 38, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; He, D.; Yuan, Y.; Yan, Z.; Zhang, X.; Zhang, J. Chitosan-modified lipid nanovesicles for efficient systemic delivery of l -asparaginase. Colloids Surfaces B Biointerfaces 2016, 143, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Kravtzoff, R.; Ropars, C.; Desbois, I.; Chassaigne, M.; Lamagnere, J.P.; Colombat, P.; Muh, J.P.; Valat, C. Improved pharmacodynamics of l-asparaginase-loaded in human red blood cells. Eur. J. Clin. Pharmacol. 1996, 49, 465–470. [Google Scholar] [CrossRef]
- Baran, E.T.; Özer, N.; Hasirci, V. In vivo half life of nanoencapsulated L-asparaginase. J. Mater. Sci. Mater. Med. 2002, 13, 1113–1121. [Google Scholar] [CrossRef]
- Ha, W.; Meng, X.-W.; Li, Q.; Fan, M.-M.; Peng, S.-L.; Ding, L.-S.; Tian, X.; Zhang, S.; Li, B.-J. Self-assembly hollow nanosphere for enzyme encapsulation. Soft Matter 2010, 6, 1405–1408. [Google Scholar] [CrossRef]
- Ortac, I.; Ruff, L.; Yeh, Y.; Esener, S.; Messmer, B.T. Nanoparticle Encapsulated L-Asparaginase. Blood 2013, 122, 2669. [Google Scholar] [CrossRef]
- Ma, Y.; Nolte, R.J.; Cornelissen, J.J. Virus-based nanocarriers for drug delivery. Adv. Drug Deliv. Rev. 2012, 64, 811–825. [Google Scholar] [CrossRef]
- Parodi, A.; Molinaro, R.; Sushnitha, M.; Evangelopoulos, M.; Martinez, J.O.; Arrighetti, N.; Corbo, C.; Tasciotti, E. Bio-inspired engineering of cell- and virus-like nanoparticles for drug delivery. Biomaterials 2017, 147, 155–168. [Google Scholar] [CrossRef]
- Shoeb, E.; Hefferon, K. Future of cancer immunotherapy using plant virus-based nanoparticles. Futur. Sci. OA 2019, 5, FSO401. [Google Scholar] [CrossRef]
- Chauhan, K.; Hernandez-Meza, J.M.; Rodríguez-Hernández, A.G.; Juarez-Moreno, K.; Sengar, P.; Vazquez-Duhalt, R. Multifunctionalized biocatalytic P22 nanoreactor for combinatory treatment of ER+ breast cancer. J. Nanobiotechnol. 2018, 16, 17. [Google Scholar] [CrossRef]
- Steinmetz, N.F. Viral nanoparticles as platforms for next-generation therapeutics and imaging devices. Nanomed. Nanotechnol. Biol. Med. 2010, 6, 634–641. [Google Scholar] [CrossRef]
- Ding, X.; Liu, D.; Booth, G.; Gao, W.; Lu, Y. Virus-Like Particle Engineering: From Rational Design to Versatile Applications. Biotechnol. J. 2018, 13, e1700324. [Google Scholar] [CrossRef]
- Teschke, C.M.; Parent, K.N. Let the phage do the work’: Using the phage P22 coat protein structures as a framework to understand its folding and assembly mutants. Virology 2010, 401, 119–130. [Google Scholar] [CrossRef]
- O’Neil, A.; Reichhardt, C.; Johnson, B.; Prevelige, P.E.; Douglas, T. Genetically Programmed In Vivo Packaging of Protein Cargo and Its Controlled Release from Bacteriophage P22. Angew. Chem. Int. Ed. 2011, 50, 7425–7428. [Google Scholar] [CrossRef]
- Patterson, D.P.; Schwarz, B.; Waters, R.S.; Gedeon, T.; Douglas, T. Encapsulation of an Enzyme Cascade within the Bacteriophage P22 Virus-Like Particle. ACS Chem. Biol. 2014, 9, 359–365. [Google Scholar] [CrossRef]
- Sanchez-Sanchez, L.; Tapia-Moreno, A.; Juárez-Moreno, K.; Patterson, D.P.; Cadena-Nava, R.D.; Douglas, T.; Vazquez-Duhalt, R. Design of a VLP-nanovehicle for CYP450 enzymatic activity delivery. J. Nanobiotechnol. 2015, 13, 1–10. [Google Scholar] [CrossRef]
- Pope, B.; Kent, H.M. High Efficiency 5 Min Transformation of Escherichia Coli. Nucleic Acids Res. 1996, 24, 536–537. [Google Scholar] [CrossRef][Green Version]
- Tapia-Moreno, A.; Juarez-Moreno, K.; Gonzalez-Davis, O.; Cadena-Nava, R.D.; Vazquez-Duhalt, R. Biocatalytic virus capsid as nanovehicle for enzymatic activation of Tamoxifen in tumor cells. Biotechnol. J. 2017, 12, 1600706. [Google Scholar] [CrossRef]
- Giessen, T.W.; Silver, P.A. A Catalytic Nanoreactor Based on in Vivo Encapsulation of Multiple Enzymes in an Engineered Protein Nanocompartment. ChemBioChem 2016, 17, 1931–1935. [Google Scholar] [CrossRef]
- Patterson, D.P.; Prevelige, P.E.; Douglas, T. Nanoreactors by Programmed Enzyme Encapsulation Inside the Capsid of the Bacteriophage P22. ACS Nano 2012, 6, 5000–5009. [Google Scholar] [CrossRef]
- Searle, P.L. The berthelot or indophenol reaction and its use in the analytical chemistry of nitrogen. A review. Analyst 1984, 109, 549–568. [Google Scholar] [CrossRef]
- Chaney, A.L.; Marbach, E.P. Modified Reagents for Determination of Urea and Ammonia. Clin. Chem. 1962, 8, 130–132. [Google Scholar] [CrossRef] [PubMed]
- Patterson, D.P.; Schwarz, B.; El-Boubbou, K.; Van Der Oost, J.; Prevelige, P.E.; Douglas, T. Virus-like particle nanoreactors: Programmed encapsulation of the thermostable CelB glycosidase inside the P22 capsid. Soft Matter 2012, 8, 10158–10166. [Google Scholar] [CrossRef]
- Zhou, H.-X.; Rivas, G.; Minton, A.P. Macromolecular Crowding and Confinement: Biochemical, Biophysical, and Potential Physiological Consequences. Annu. Rev. Biophys. 2008, 37, 375–397. [Google Scholar] [CrossRef] [PubMed]
- Norris, M.G.; Malys, N. What is the true enzyme kinetics in the biological system? An investigation of macromolecular crowding effect upon enzyme kinetics of glucose-6-phosphate dehydrogenase. Biochem. Biophys. Res. Commun. 2011, 405, 388–392. [Google Scholar] [CrossRef] [PubMed]
- Shulgin, I.L.; Ruckenstein, E. Preferential hydration and solubility of proteins in aqueous solutions of polyethylene glycol. Biophys. Chem. 2006, 120, 188–198. [Google Scholar] [CrossRef]
- Zhao, H.; Olubajo, O.; Song, Z.; Sims, A.L.; Person, T.E.; Lawal, R.A.; Holley, L.A. Effect of kosmotropicity of ionic liquids on the enzyme stability in aqueous solutions. Bioorg. Chem. 2006, 34, 15–25. [Google Scholar] [CrossRef]
- Miyawaki, O. Hydration state change of proteins upon unfolding in sugar solutions. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2007, 1774, 928–935. [Google Scholar] [CrossRef]
- Silva, C.; Martins, M.; Jing, S.; Fu, J.; Cavaco-Paulo, A. Practical insights on enzyme stabilization. Crit. Rev. Biotechnol. 2018, 38, 335–350. [Google Scholar] [CrossRef]
- Maggi, M.; Mittelman, S.D.; Parmentier, J.H.; Colombo, G.; Meli, M.; Whitmire, J.M.; Merrell, D.S.; Whitelegge, J.; Scott, M.D. A protease-resistant Escherichia coli asparaginase with outstanding stability and enhanced anti-leukaemic activity in vitro. Sci. Rep. 2017, 7, 14479. [Google Scholar] [CrossRef]
- Kasturi, S.; Sachaphibulkij, K.; Roy, K. Covalent conjugation of polyethyleneimine on biodegradable microparticles for delivery of plasmid DNA vaccines. Biomaterials 2005, 26, 6375–6385. [Google Scholar] [CrossRef]
- Cauda, V.A.; Argyo, C.; Bein, T. Impact of different PEGylation patterns on the long-term bio-stability of colloidal mesoporous silica nanoparticles. J. Mater. Chem. 2010, 20, 8693–8699. [Google Scholar] [CrossRef]
- Böttger, R.; Hoffmann, R.; Knappe, D. Differential stability of therapeutic peptides with different proteolytic cleavage sites in blood, plasma and serum. PLoS ONE 2017, 12, e0178943. [Google Scholar] [CrossRef]
- Robinson, H.M.; Martineau, M.; Harris, R.L.; E Barber, K.; Jalali, G.R.; Moorman, A.V.; Strefford, J.C.; Broadfield, Z.J.; Cheung, K.L.; Harrison, C.J. Derivative chromosome 9 deletions are a significant feature of childhood Philadelphia chromosome positive acute lymphoblastic leukaemia. Leukemia 2005, 19, 564–571. [Google Scholar] [CrossRef]
- Patel, N.; Krishnan, S.; Offman, M.N.; Krol, M.; Moss, C.X.; Leighton, C.; Van Delft, F.W.; Holland, M.; Liu, J.; Alexander, S.; et al. A dyad of lymphoblastic lysosomal cysteine proteases degrades the antileukemic drug l-asparaginase. J. Clin. Investig. 2009, 119, 1964–1973. [Google Scholar] [CrossRef]
- Harrison, C.J. Blood Spotlight on iAMP21 acute lymphoblastic leukemia (ALL), a high-risk pediatric disease. Blood 2015, 125, 1383–1386. [Google Scholar] [CrossRef]
- Van Der Meer, L.T.; Terry, S.Y.; Schenau, D.S.V.I.; Andree, K.C.; Franssen, G.M.; Roeleveld, D.M.; Metselaar, J.M.; Reinheckel, T.; Hoogerbrugge, P.M.; Boerman, O.C.; et al. In Vivo Imaging of Antileukemic Drug Asparaginase Reveals a Rapid Macrophage-Mediated Clearance from the Bone Marrow. J. Nucl. Med. 2016, 58, 214–220. [Google Scholar] [CrossRef]
- Derst, C.; Henseling, J.; Röhm, K.-H. Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase activity by amino acid replacements at position 248. Protein Sci. 2000, 9, 2009–2017. [Google Scholar] [CrossRef]

| Encoded Protein | Primer | Sequence | Restriction Site |
|---|---|---|---|
| E. coli asnB ASNase II | asnB Fw | GATATACCATGGCATTACCCAATATCACC | NcoI |
| asnB Rv | CCGGCTCGAGGTACTGATTGAAGATCTGCT | XhoI | |
| P22 Scaffold protein (sp) | sp Fw | ATATCTCGAGCTGGTGCCGCGCGGCAG | XhoI |
| sp Rv | TCTCGAATTCTTATCGGATTCCTTTAAG | EcoRI |
| Enzyme | Monomers per Capsid | Internal Radium of Capsid (nm) | Mconf | Occupancy (%) | Reference |
|---|---|---|---|---|---|
| CelB glycosidase | 87 | 24 | NR | NR | [44] |
| CYP450 | 110 | 22 | 3.14 | NR | [37] |
| ASNase | 448 | 30 | 6.58 | 26 | This work |
| Alcohol dehydrogenase | 249 | 24 | 7.16 | 27 | [41] |
| Sample | Kcat (s−1) | Km (mM) | Kcat/Km (M−1 s−1) | Specific Activity (U/mg) | Vmax (µM/min) | Reference |
|---|---|---|---|---|---|---|
| ASNase-P22 | 49 | 0.227 | 2.19 × 105 | 13.73 | 131.78 | This work. |
| ASNase (E. coli) | 24 | 0.015 | 1.60 × 106 | NR | NR | [59] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz-Barriga, C.; Villanueva-Flores, F.; Quester, K.; Zárate-Romero, A.; Cadena-Nava, R.D.; Huerta-Saquero, A. Asparaginase-Phage P22 Nanoreactors: Toward a Biobetter Development for Acute Lymphoblastic Leukemia Treatment. Pharmaceutics 2021, 13, 604. https://doi.org/10.3390/pharmaceutics13050604
Díaz-Barriga C, Villanueva-Flores F, Quester K, Zárate-Romero A, Cadena-Nava RD, Huerta-Saquero A. Asparaginase-Phage P22 Nanoreactors: Toward a Biobetter Development for Acute Lymphoblastic Leukemia Treatment. Pharmaceutics. 2021; 13(5):604. https://doi.org/10.3390/pharmaceutics13050604
Chicago/Turabian StyleDíaz-Barriga, Cristina, Francisca Villanueva-Flores, Katrin Quester, Andrés Zárate-Romero, Ruben Dario Cadena-Nava, and Alejandro Huerta-Saquero. 2021. "Asparaginase-Phage P22 Nanoreactors: Toward a Biobetter Development for Acute Lymphoblastic Leukemia Treatment" Pharmaceutics 13, no. 5: 604. https://doi.org/10.3390/pharmaceutics13050604
APA StyleDíaz-Barriga, C., Villanueva-Flores, F., Quester, K., Zárate-Romero, A., Cadena-Nava, R. D., & Huerta-Saquero, A. (2021). Asparaginase-Phage P22 Nanoreactors: Toward a Biobetter Development for Acute Lymphoblastic Leukemia Treatment. Pharmaceutics, 13(5), 604. https://doi.org/10.3390/pharmaceutics13050604






