Synthesis and Characterization of Mannosylated Formulations to Deliver a Minicircle DNA Vaccine
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Amplification and Purification of mcDNA Vector
2.2.2. Agarose Gel Electrophoresis
2.2.3. Synthesis of α-d-Mannopyranosylphenyl Isothiocyanate-Octa-Arginine Conjugate
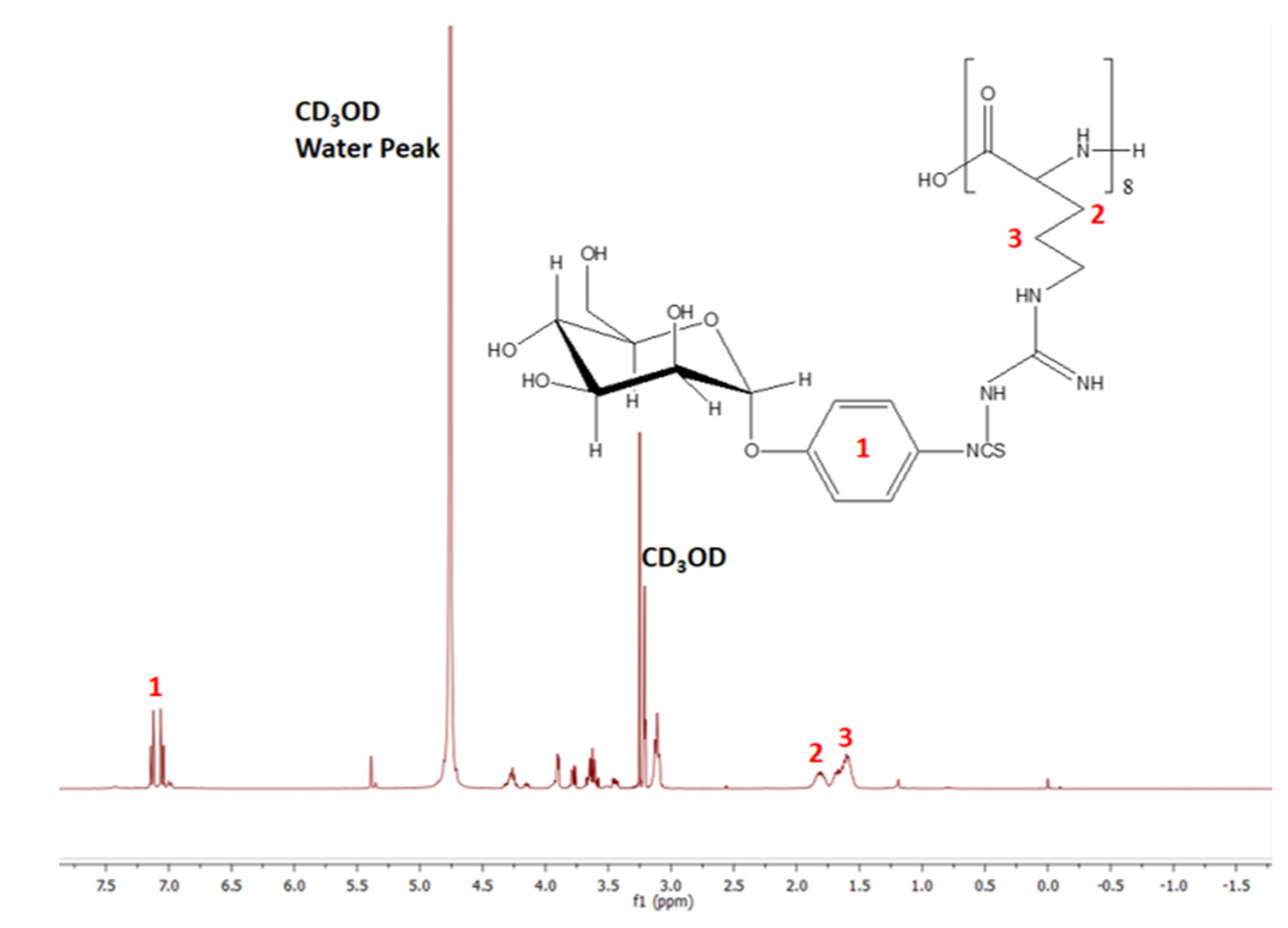
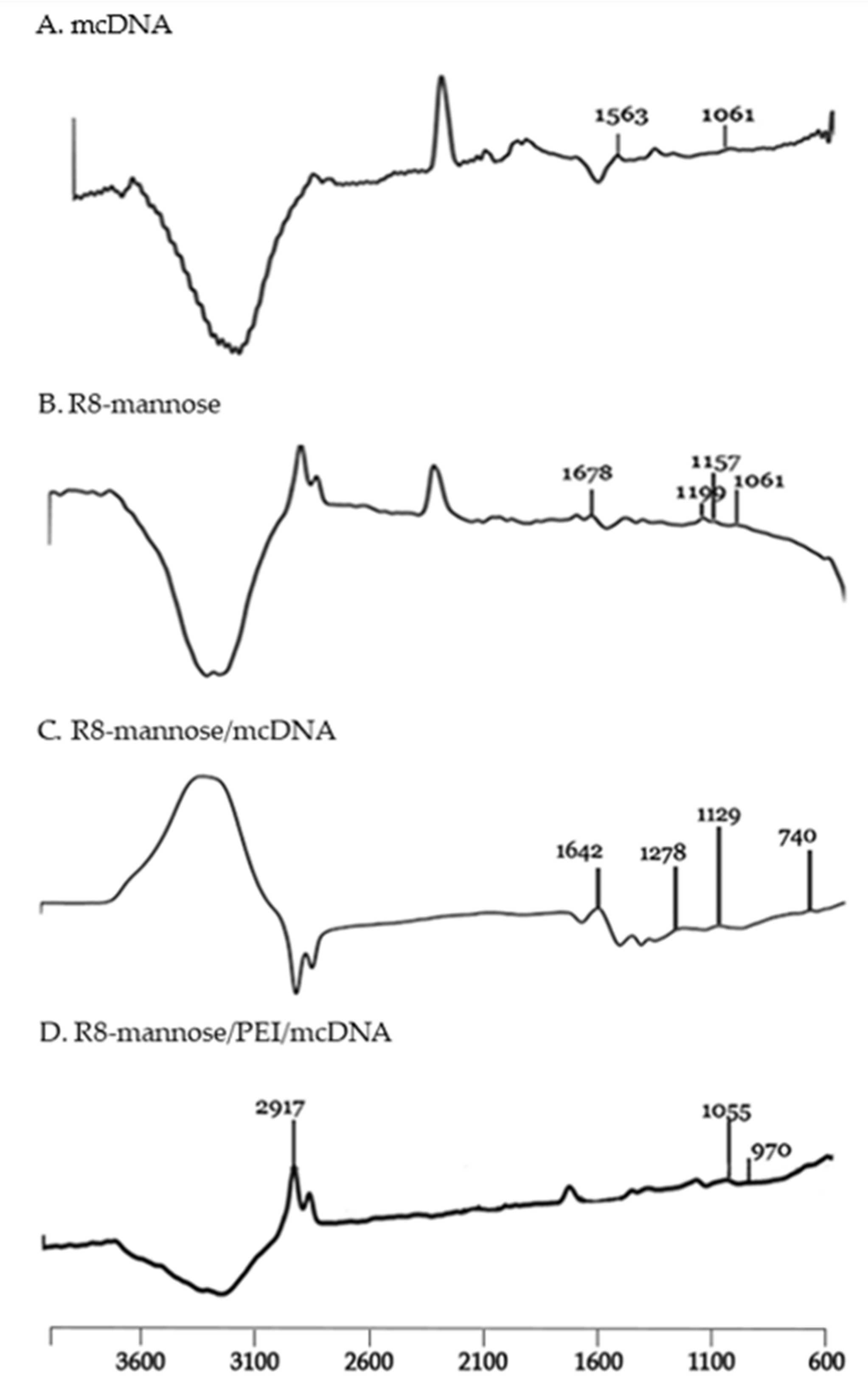
2.2.4. Characterization of Synthesized MPITC-R8 Conjugate
2.2.5. Preparation of R8-Mannose/mcDNA and R8-Mannose/PEI/mcDNA Complexes
2.2.6. Characterization of Systems
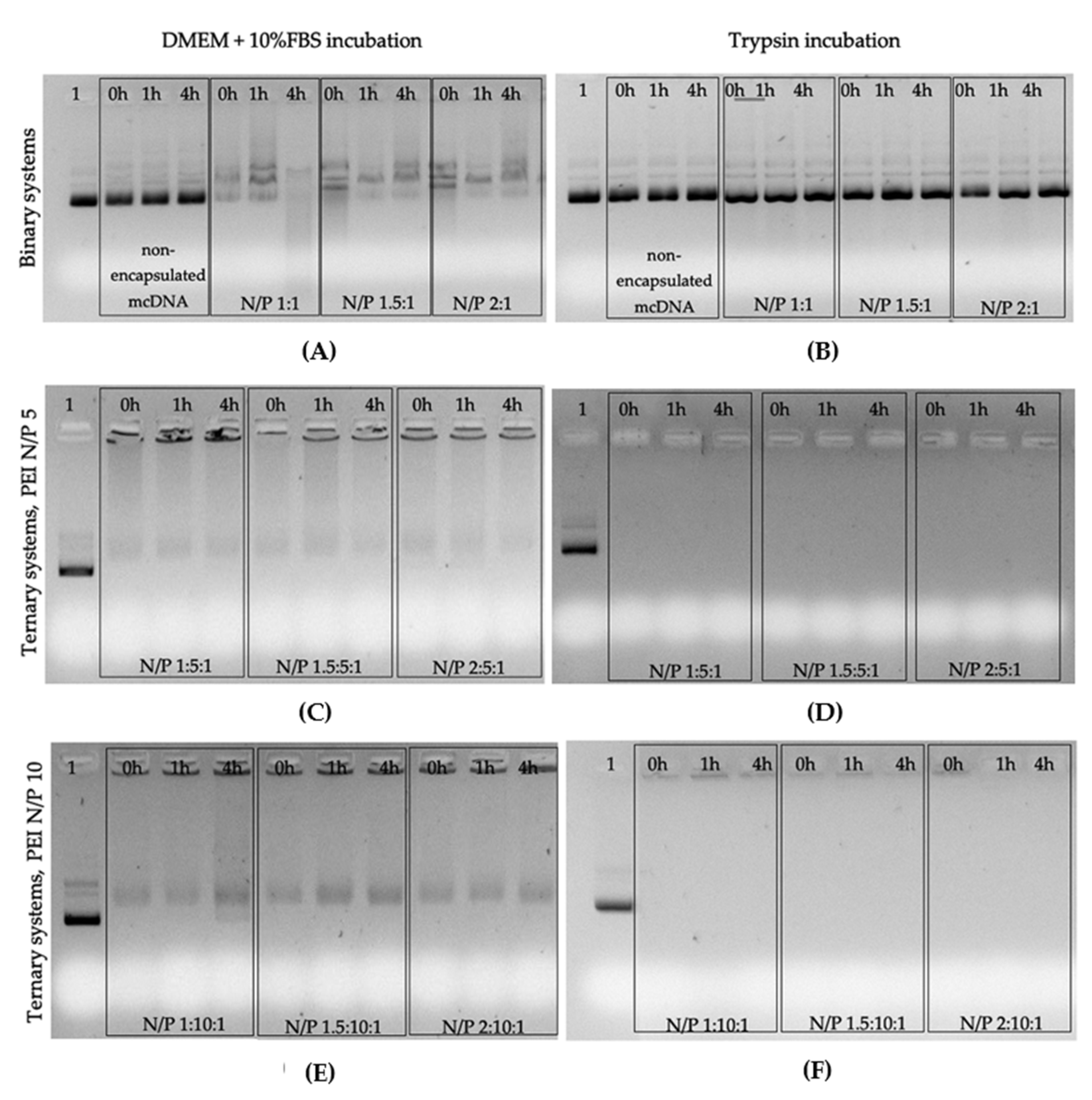
2.2.7. Stability Assays
2.2.8. In Vitro Transfection
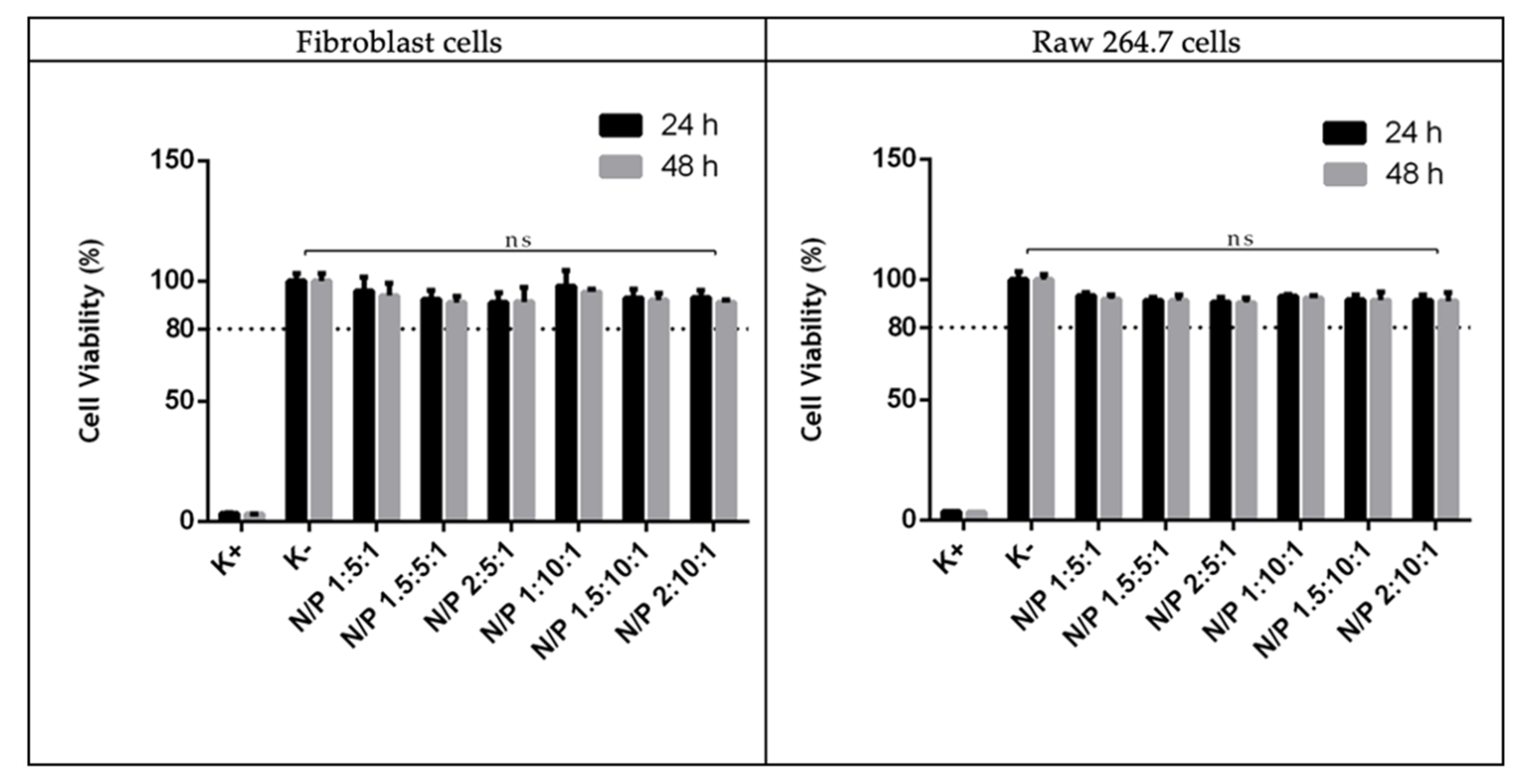
2.2.9. Biocompatibility Study
2.2.10. FITC Plasmid Labeling
2.2.11. Cellular Uptake and Internalization
2.2.12. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
2.2.13. Reverse Transcription Quantitative Real-Time PCR (RT-qPCR)
2.2.14. Statistical Analysis
3. Results and Discussion
3.1. Purification of mcDNA Vector
3.2. Synthesis and Characterization of MPITC-R8 Conjugate
3.3. The Properties of R8-Mannose/mcDNA and R8-Mannose/PEI/mcDNA Complexes
3.4. Stability Assay
3.5. Biocompatibility Evaluation
3.6. Cellular Uptake and Intracellular Location of Complexes
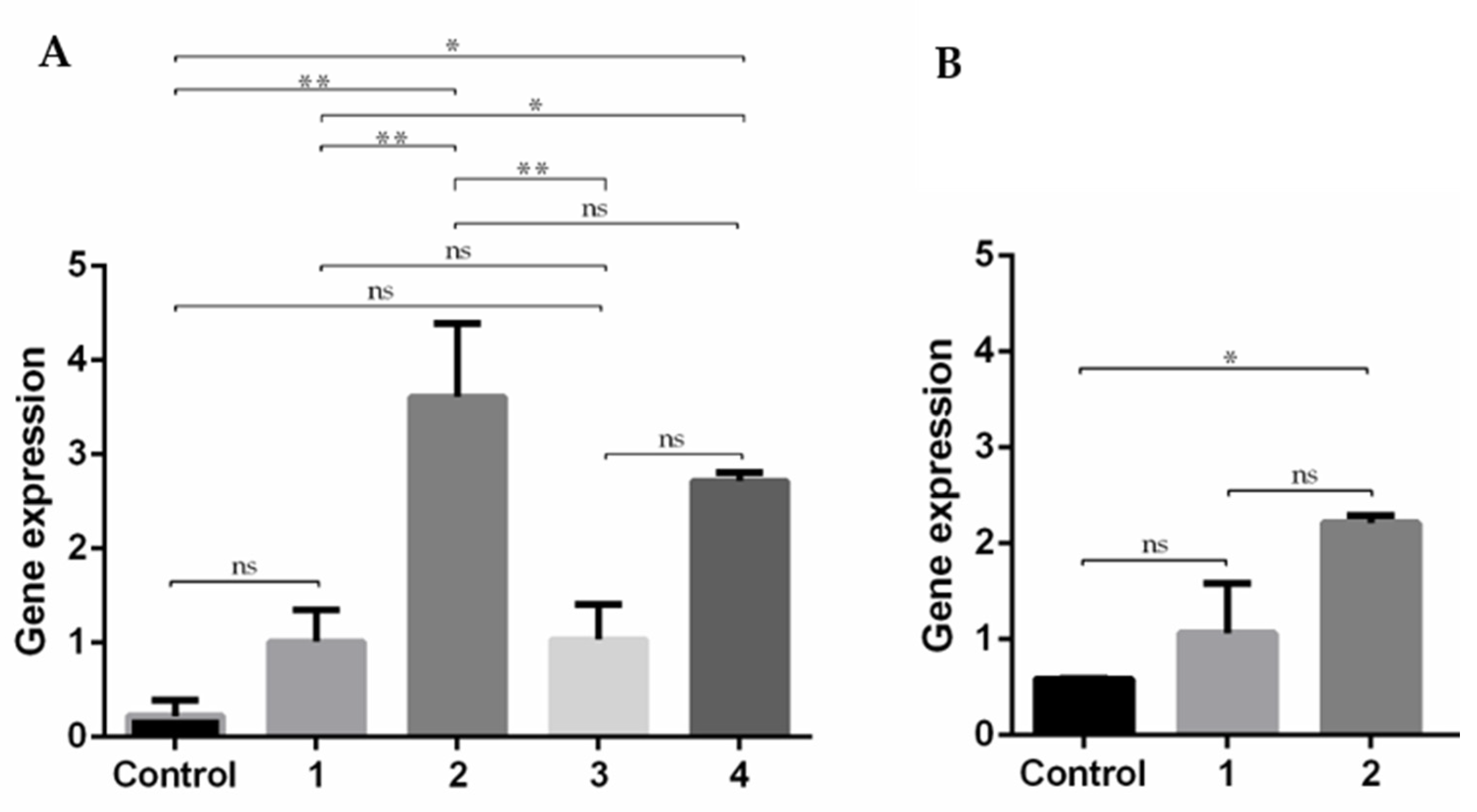
3.7. Expression of E7 Gene
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Almeida, A.M.; Queiroz, J.A.; Sousa, F.; Sousa, Â. Cervical cancer and HPV infection: Ongoing therapeutic research to counteract the action of E6 and E7 oncoproteins. Drug Discov. Today 2019, 24, 2044–2057. [Google Scholar] [CrossRef]
- Chan, C.K.; Aimagambetova, G.; Ukybassova, T.; Kongrtay, K.; Azizan, A. Human Papillomavirus Infection and Cervical Cancer: Epidemiology, Screening, and Vaccination-Review of Current Perspectives. J. Oncol. 2019, 2019, 3257939. [Google Scholar] [CrossRef] [PubMed]
- Harden, M.E.; Munger, K. Human papillomavirus molecular biology. Mutat. Res. Rev. Mutat. Res. 2017, 772, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Peralta-Zaragoza, O.; Bermúdez-Morales, V.H.; Pérez-Plasencia, C.; Salazar-León, J.; Gómez-Cerón, C.; Madrid-Marina, V. Targeted treatments for cervical cancer: A review. OncoTargets Ther. 2012, 5, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Almeida, A.M.; Queiroz, J.A.; Sousa, F.; Sousa, Â. Minicircle DNA: The Future for DNA-Based Vectors? Trends Biotechnol. 2020, 38, 1047–1051. [Google Scholar] [CrossRef] [PubMed]
- Petkar, K.C.; Patil, S.M.; Chavhan, S.S.; Kaneko, K.; Sawant, K.K.; Kunda, N.K.; Saleem, I.Y. An Overview of Nanocarrier-Based Adjuvants for Vaccine Delivery. Pharmaceutics 2021, 13, 455. [Google Scholar] [CrossRef]
- Shi, B.; Zheng, M.; Tao, W.; Chung, R.; Jin, D.; Ghaffari, D.; Farokhzad, O.C. Challenges in DNA Delivery and Recent Advances in Multifunctional Polymeric DNA Delivery Systems. Biomacromolecules 2017, 18, 2231–2246. [Google Scholar] [CrossRef]
- Thomas, T.J.; Tajmir-Riahi, H.-A.; Pillai, C.K.S. Biodegradable Polymers for Gene Delivery. Molecules 2019, 24, 3744. [Google Scholar] [CrossRef]
- Ruseska, I.; Zimmer, A. Internalization mechanisms of cell-penetrating peptides. Beilstein J. Nanotechnol. 2020, 11, 101–123. [Google Scholar] [CrossRef] [PubMed]
- Lehto, T.; Kurrikoff, K.; Langel, Ü. Cell-penetrating peptides for the delivery of nucleic acids. Expert Opin. Drug Deliv. 2012, 9, 823–836. [Google Scholar] [CrossRef]
- Böhmová, E.; Machová, D.; Pechar, M.; Pola, R.; Venclíková, K.; Janoušková, O.; Etrych, T. Cell-penetrating peptides: A useful tool for the delivery of various cargoes into cells. Physiol. Res. 2018, 67, S267–S279. [Google Scholar] [CrossRef]
- Takeuchi, T.; Futaki, S. Current Understanding of Direct Translocation of Arginine-Rich Cell-Penetrating Peptides and Its Internalization Mechanisms. Chem. Pharm. Bull. 2016, 64, 1431–1437. [Google Scholar] [CrossRef]
- Rompicharla, S.V.K.; Kumari, P.; Ghosh, B.; Biswas, S. Octa-arginine modified poly(amidoamine) dendrimers for improved delivery and cytotoxic effect of paclitaxel in cancer. Artif. Cells Nanomed. Biotechnol. 2018, 46, 847–859. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, H.; Coulter, J.A.; Yang, R. Octaarginine-modified gold nanoparticles enhance the radiosensitivity of human colorectal cancer cell line LS180 to megavoltage radiation. Int. J. Nanomed. 2018, 13, 3541–3552. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Dodwadkar, N.S.; Deshpande, P.P.; Parab, S.; Torchilin, V.P. Surface functionalization of doxorubicin-loaded liposomes with octa-arginine for enhanced anticancer activity. Eur. J. Pharm. Biopharm. 2013, 84, 517–525. [Google Scholar] [CrossRef]
- Bhatt, H.; Ghosh, B.; Biswas, S. Cell-Penetrating Peptide and α-Tocopherol-Conjugated Poly(amidoamine) Dendrimers for Improved Delivery and Anticancer Activity of Loaded Paclitaxel. ACS Appl. Bio Mater. 2020, 3, 3157–3169. [Google Scholar] [CrossRef]
- Xun, M.M.; Huang, Z.; Xiao, Y.P.; Liu, Y.H.; Zhang, J.; Zhang, J.H.; Yu, X.Q. Synthesis and Properties of Low-Molecular-Weight PEI-Based Lipopolymers for Delivery of DNA. Polymers 2018, 10, 1060. [Google Scholar] [CrossRef] [PubMed]
- Hamada, E.; Kurosaki, T.; Hashizume, J.; Harasawa, H.; Nakagawa, H.; Nakamura, T.; Kodama, Y.; Sasaki, H. Anionic Complex with Efficient Expression and Good Safety Profile for mRNA Delivery. Pharmaceutics 2021, 13, 126. [Google Scholar] [CrossRef] [PubMed]
- Costa, D.; Valente, A.J.M.; Queiroz, J.A.; Sousa, Â. Finding the ideal polyethylenimine-plasmid DNA system for co-delivery of payloads in cancer therapy. Colloids Surf. B Biointerfaces 2018, 170, 627–636. [Google Scholar] [CrossRef]
- Lim, M.; Badruddoza, A.Z.M.; Firdous, J.; Azad, M.; Mannan, A.; Al-Hilal, T.A.; Cho, C.S.; Islam, M.A. Engineered Nanodelivery Systems to Improve DNA Vaccine Technologies. Pharmaceutics 2020, 12, 30. [Google Scholar] [CrossRef] [PubMed]
- Hobernik, D.; Bros, M. DNA Vaccines-How Far From Clinical Use? Int. J. Mol. Sci. 2018, 19, 3605. [Google Scholar] [CrossRef]
- Jariwala, S.P. The role of dendritic cells in the immunopathogenesis of psoriasis. Arch. Pharmacal Res. 2007, 299, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Xu, B.-H.; Xu, J.-J.; Shou, D.; Gao, J.-Q. Synthesis of Mannosylated Polyethylenimine and Its Potential Application as Cell-Targeting Non-Viral Vector for Gene Therapy. Polymers 2014, 6, 2573–2587. [Google Scholar] [CrossRef]
- Shilakari Asthana, G.; Asthana, A.; Kohli, D.V.; Vyas, S.P. Mannosylated Chitosan Nanoparticles for Delivery of Antisense Oligonucleotides for Macrophage Targeting. BioMed Res. Int. 2014, 2014, 526391. [Google Scholar] [CrossRef]
- Hu, Z.; Shi, X.; Yu, B.; Li, N.; Huang, Y.; He, Y. Structural Insights into the pH-Dependent Conformational Change and Collagen Recognition of the Human Mannose Receptor. Structure 2018, 26, 60–71.e63. [Google Scholar] [CrossRef] [PubMed]
- Eusébio, D.; Almeida, A.M.; Alves, J.M.; Maia, C.J.; Queiroz, J.A.; Sousa, F.; Sousa, Â. The Performance of Minicircle DNA Versus Parental Plasmid in p53 Gene Delivery Into HPV-18-Infected Cervical Cancer Cells. Nucleic Acid Ther. 2021, 31, 82–91. [Google Scholar] [CrossRef]
- Diogo, M.; Queiroz, J.; Monteiro, G.; Martins, S.; Ferreira, G.; Prazeres, D. Purification of a cystic fibrosis plasmid vector for gene therapy using hydrophobic interaction chromatography. Biotechnol. Bioeng. 2000, 68, 576–583. [Google Scholar] [CrossRef]
- Almeida, A.M.; Eusébio, D.; Queiroz, J.A.; Sousa, F.; Sousa, A. The use of size-exclusion chromatography in the isolation of supercoiled minicircle DNA from Escherichia coli lysate. J. Chromatogr. A 2019, 1609, 460444. [Google Scholar] [CrossRef]
- Koshkaryev, A.; Piroyan, A.; Torchilin, V.P. Bleomycin in octaarginine-modified fusogenic liposomes results in improved tumor growth inhibition. Cancer Lett. 2013, 334, 293–301. [Google Scholar] [CrossRef]
- Futaki, S. Arginine-rich peptides: Potential for intracellular delivery of macromolecules and the mystery of the translocation mechanisms. Int. J. Pharm. 2002, 245, 1–7. [Google Scholar] [CrossRef]
- Futaki, S. Membrane-permeable arginine-rich peptides and the translocation mechanisms. Adv. Drug Deliv. Rev. 2005, 57, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Sousa, Â.; Almeida, A.M.; Faria, R.; Konate, K.; Boisguerin, P.; Queiroz, J.A.; Costa, D. Optimization of peptide-plasmid DNA vectors formulation for gene delivery in cancer therapy exploring design of experiments. Colloids Surf. B Biointerfaces 2019, 183, 110417. [Google Scholar] [CrossRef] [PubMed]
- Choosakoonkriang, S.; Lobo, B.A.; Koe, G.S.; Koe, J.G.; Middaugh, C.R. Biophysical characterization of PEI/DNA complexes. J. Pharm. Sci. 2003, 92, 1710–1722. [Google Scholar] [CrossRef] [PubMed]
- Mady, M.; Awad, W.; El-Guendy, N.; Elsayed, A. Interaction of DNA and Polyethylenimine: FTIR and DSC Studies. Phys. Sci. Int. J. 2011, 6, 7328–7334. [Google Scholar]
- Imani, R.; Hojjati Emami, S.; Faghihi, S. Synthesis and Characterization of Octaarginine Functionalized Graphene Oxide Nano-carrier for Gene Delivery Applications. Phys. Chem. Chem. Phys. 2015, 17, 6328–6339. [Google Scholar] [CrossRef]
- Costa, A.; Sarmento, B.; Seabra, V. Mannose-functionalized solid lipid nanoparticles are effective in targeting alveolar macrophages. Eur. J. Pharm. Sci. 2018, 114, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Horak, D.; Babic, M.; Jendelová, P.; Herynek, V.; Trchová, M.; Pientka, Z.; Pollert, E.; Hájek, M.; Syková, E. D-mannose-modified iron oxide nanoparticles for stem cell labeling. Bioconjug. Chem. 2007, 18, 635–644. [Google Scholar] [CrossRef]
- Bayramoglu, G.; Arica, Y. Adsorption of Cr(VI) onto PEI Immobilized Acrylate-Based Magnetic Beads: Isotherms, Kinetics and Thermodynamics Study. Chem. Eng. J. 2008, 139, 20–28. [Google Scholar] [CrossRef]
- Gaspar, V.M.; Sousa, F.; Queiroz, J.A.; Correia, I.J. Formulation of chitosan-TPP-pDNA nanocapsules for gene therapy applications. Nanotechnology 2011, 22, 015101. [Google Scholar] [CrossRef]
- Chithrani, B.D.; Chan, W.C. Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett. 2007, 7, 1542–1550. [Google Scholar] [CrossRef]
- Liu, S.; Guo, T. Polycation-Based Ternary Gene Delivery System. Curr. Drug Metab. 2015, 16, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Liang, X.; Yang, S.; Wang, N.; He, T.; Wang, Y.; Zhang, L.; Wu, Q.; Gong, C. Novel polyethyleneimine-R8-heparin nanogel for high-efficiency gene delivery in vitro and in vivo. Drug Deliv. 2018, 25, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, T.; Faria, R.; Sousa, Â.; Neves, A.R.; Queiroz, J.A.; Costa, D. Polymer-peptide ternary systems as a tool to improve the properties of plasmid DNA vectors in gene delivery. J. Mol. Liq. 2020, 309, 113157. [Google Scholar] [CrossRef]
- López-García, J.; Lehocký, M.; Humpolíček, P.; Sáha, P. HaCaT Keratinocytes Response on Antimicrobial Atelocollagen Substrates: Extent of Cytotoxicity, Cell Viability and Proliferation. J. Funct. Biomater. 2014, 5, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Conniot, J.; Scomparin, A.; Peres, C.; Yeini, E.; Pozzi, S.; Matos, A.I.; Kleiner, R.; Moura, L.I.F.; Zupančič, E.; Viana, A.S.; et al. Immunization with mannosylated nanovaccines and inhibition of the immune-suppressing microenvironment sensitizes melanoma to immune checkpoint modulators. Nat. Nanotechnol 2019, 14, 891–901. [Google Scholar] [CrossRef]
- Salatin, S.; Maleki Dizaj, S.; Yari Khosroushahi, A. Effect of the surface modification, size, and shape on cellular uptake of nanoparticles. Cell Biol. Int. 2015, 39, 881–890. [Google Scholar] [CrossRef]
- Apostolopoulos, V.; Thalhammer, T.; Tzakos, A.G.; Stojanovska, L. Targeting antigens to dendritic cell receptors for vaccine development. J. Drug Deliv. 2013, 2013, 869718. [Google Scholar] [CrossRef]
- Hossain, M.K.; Wall, K.A. Use of Dendritic Cell Receptors as Targets for Enhancing Anti-Cancer Immune Responses. Cancers 2019, 11, 418. [Google Scholar] [CrossRef]
- Tang, C.K.; Sheng, K.C.; Apostolopoulos, V.; Pietersz, G.A. Protein/peptide and DNA vaccine delivery by targeting C-type lectin receptors. Expert Rev. Vaccines 2008, 7, 1005–1018. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serra, A.S.; Eusébio, D.; Neves, A.R.; Albuquerque, T.; Bhatt, H.; Biswas, S.; Costa, D.; Sousa, Â. Synthesis and Characterization of Mannosylated Formulations to Deliver a Minicircle DNA Vaccine. Pharmaceutics 2021, 13, 673. https://doi.org/10.3390/pharmaceutics13050673
Serra AS, Eusébio D, Neves AR, Albuquerque T, Bhatt H, Biswas S, Costa D, Sousa Â. Synthesis and Characterization of Mannosylated Formulations to Deliver a Minicircle DNA Vaccine. Pharmaceutics. 2021; 13(5):673. https://doi.org/10.3390/pharmaceutics13050673
Chicago/Turabian StyleSerra, Ana Sofia, Dalinda Eusébio, Ana Raquel Neves, Tânia Albuquerque, Himanshu Bhatt, Swati Biswas, Diana Costa, and Ângela Sousa. 2021. "Synthesis and Characterization of Mannosylated Formulations to Deliver a Minicircle DNA Vaccine" Pharmaceutics 13, no. 5: 673. https://doi.org/10.3390/pharmaceutics13050673
APA StyleSerra, A. S., Eusébio, D., Neves, A. R., Albuquerque, T., Bhatt, H., Biswas, S., Costa, D., & Sousa, Â. (2021). Synthesis and Characterization of Mannosylated Formulations to Deliver a Minicircle DNA Vaccine. Pharmaceutics, 13(5), 673. https://doi.org/10.3390/pharmaceutics13050673








