The Combination of Cell Cultured Technology and In Silico Model to Inform the Drug Development
Abstract
1. Introduction
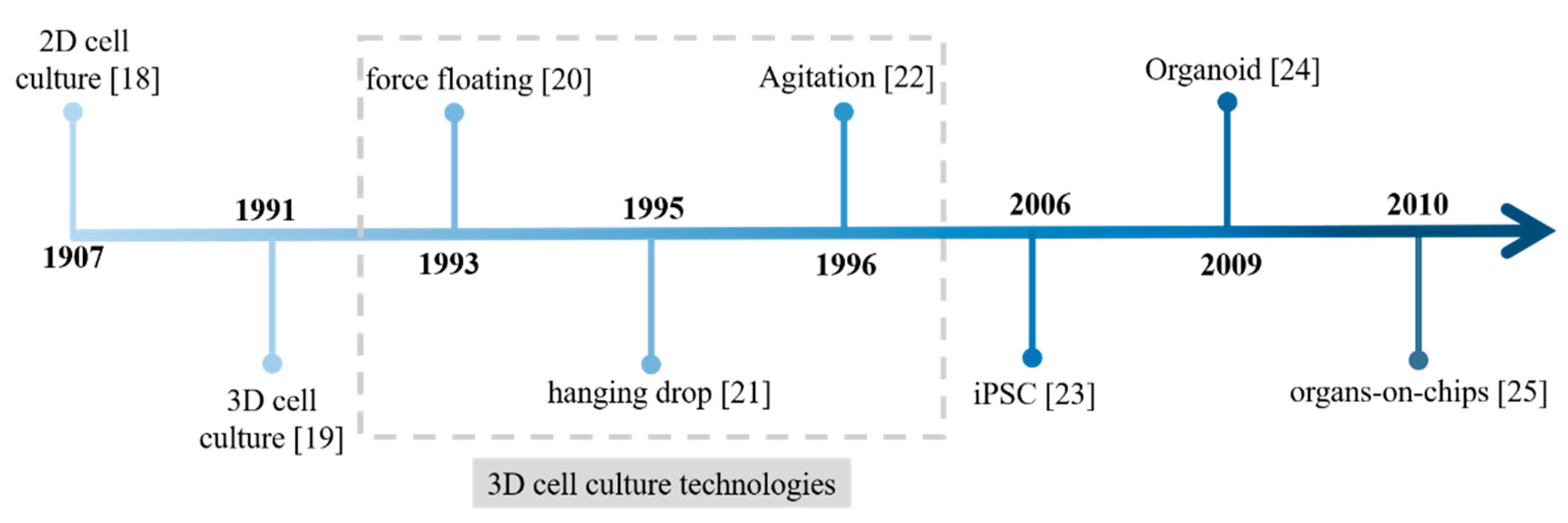
2. Cell Culture Models
2.1. Cell Origins
2.1.1. Primary Cells
2.1.2. Immortal Cells
2.1.3. Transfected Cells
2.1.4. Induced Pluripotent Stem Cells
2.2. Cell Origins
2.2.1. 2D Culture
2.2.2. 3D Culture
2.2.3. Organoids
2.2.4. Microphysiological Systems
3. Model Informed In Vitro to In Vivo Translation
3.1. Conventional PK/PD Model
| In Silico | In Vitro Assay | Examples of In Vitro to In Vivo Translation | ||||
|---|---|---|---|---|---|---|
| Technologies Used in Examples | Parameter | In Vitro to In Vivo Translation Result | Application in Drug Development | |||
| PBPK | Absorption | Caco-2 [81], MDCK [82], gut MPS [83] | MDCK-MDR1 and Caco-2 [84] | Obtaining half maximal inhibitory concentration (IC50) for P-gp and integrating it to models | Demonstrating non-interaction between Axitinib and P-gp substrate | Prediction of drug-drug interaction and exemption of related clinical trials |
| Distribution | MDCK [85], hiPSC- brain endothelial cells [86], co-culture [87] | MDCK Ⅱ [88] | Using apparent permeability coefficient (Papp) to obtain in vitro efflux transporter-mediated clearance and scaling it to the whole-brain in vivo efflux transporter-mediated clearance | Exploring the penetration of AZD1775 across BBB | Prediction of drug distribution and target concentration | |
| Metabolism | Recombinant enzymes [89], microsomes [90], primary hepatocytes [91], HepG2 [92], HepaRG [93], hESC or hiPSC-hepatocytes [94], liver MPS [95] | Primary hepatocytes [96] | Inputting the intrinsic clearance (CLint) to Simcyp software to establish PBPK model | Predicting the difference of AUC in patients with different liver damage after a single oral administration of sirolimus | Prediction of drug metabolism and inter-population extrapolations | |
| Excretion | MDCK, CHO, HEK-293, HeLa [97], primary cultured renal tubule cells [98], renal MPS [99] | renal MPS [99] | Scaling renal clearance (CLR) based on surface area | Predicting human renal excretion for cisplatin and nicotine | Prediction of excretion | |
| PBPK | Integrate ADME | MPS [99] | MPS [99] | Scaling intestinal permeability (Papp) based on absorptive surface, liver clearance (CLint, in vivo) based on the number of hepatocytes, renal clearance (CLR) based on surface area | Reproducing the clinical PK profiles for both nicotine and cisplatin at different doses and different routes of administration | Simulation of clinical PK profiles |
| PK/PD | Disease-related cell [100], 2D [80,101], 3D [102], MPS [103], organoids [104] | Six human epithelial cancer cell lines [100] | Directly combining maximal killing rate (Kmax), drug concentrations yielding 50% of Kmax (KC50) and hill index (γ) into in vivo model | Demonstrating that low doses and high dosing frequency for paclitaxel is prior to maximum tolerated doses | Dose and schedule selection | |
| L540cy cells, Karpas cells [99] | Integrating association and dissociation rate constants (Kon and Koff) to describe the interaction between ADC and target | Predicting therapy in clinical trials employing different dosing regimens | Clinical response prediction | |||
| primary liver cells, red blood cells and brain homogenates [101] | Based on the total enzyme content, scaling metabolic capacity (Vmax) and clearance (CLint); Correcting bimolecular inhibition constant (Ki) considering different states of targets in vitro and in vivo | Evaluating the biotoxicity of carbaryl and other carbamates with an anticholinesterase mode of action | Toxicity prediction | |||
| MPS [105] | Based on the number of nephrons in human kidney, scaling maximal injury rate (Emax) and drug concentrations yielding 50% of Emax (EC50) into in vivo model | Assessing renal proximal tubule injury caused by three nephrotoxic drugs | Toxicity prediction | |||
| QSP (QST) | Disease-related cell [106], 2D [106,107], 3D [107], MPS [108], organoids [109] | Primary hepatocytes [106] | Applying directly the IC50 values for the bile acid transporters to DILIsym, fitting the mitochondrial toxicity parameters (Vmax, Km) in MITOsym, and converting them to DILIsym | Explaining the liver toxicity mechanism of PF-04895162 and expound the differences of species | Characterization of target mechanism | |
| JIMT-1 cells in 2D or 3D and dynamic cell Culture [107] | Integrating drug inhibition or stimulation coefficient (S1p, S2p, Kp etc.) to describe signal pathway molecules perturbation | Optimizing the sequence and inter-dose interval of the three agents (paclitaxel, dasatinib, and everolimus) in the combination | Design of drug administration protocol and evaluation of drug combination | |||
| effector T cells (Teffs), EL4 and E.G7-OVA thymoma cells [110,111,112] | Integrating rate constants defining the half-life of engagement or dissociation between cancer cells and effector T cells (CancerTEng, CancerTInt) directly into the QSP model; scaling number of CD28 receptors expressed on each T cell during priming (CD28_receptors-per-Tcell) by the number of T cell in vivo | Predicting the checkpoint inhibitors’ therapies administered as mono-, combo- and sequential therapies | Clinical response prediction | |||
3.2. PBPK Model
3.3. QSP Model
3.4. Virtual Clinical Trials
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Waring, M.J.; Arrowsmith, J.; Leach, A.R.; Leeson, P.D.; Mandrell, S.; Owen, R.M.; Pairaudeau, G.; Pennie, W.D.; Pickett, S.D.; Wang, J.; et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat. Rev. Drug Discov. 2015, 14, 475–486. [Google Scholar] [CrossRef]
- Dowden, H.; Munro, J. Trends in clinical success rates and therapeutic focus. Nat. Rev. Drug Discov. 2019, 18, 495–496. [Google Scholar] [CrossRef]
- Arrowsmith, J.; Miller, P. Phase II and Phase III attrition rates 2011–2012. Nat. Rev. Drug Discov. 2013, 12, 569. [Google Scholar] [CrossRef]
- Hay, M.; Thomas, D.W.; Craighead, J.L.; Economides, C.; Rosenthal, J. Clinical development success rates for investigational drugs. Nat. Biotechnol. 2014, 32, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Festing, M.F. Improving Toxicity Screening and Drug Development by Using Genetically Defined Strains. Methods Mol. Biol. 2009, 602, 1–21. [Google Scholar] [CrossRef]
- Marshall, L.J.; Austin, C.P.; Casey, W.; Fitzpatrick, S.C.; Willett, C. Recommendations toward a human pathway-based approach to disease research. Drug Discov. Today 2018, 23, 1824–1832. [Google Scholar] [CrossRef]
- Hu, J.; Lin, Y.-Y.; Chen, P.-J.; Watashi, K.; Wakita, T. Cell and Animal Models for Studying Hepatitis B Virus Infection and Drug Development. Gastroenterology 2019, 156, 338–354. [Google Scholar] [CrossRef] [PubMed]
- Gamboa, J.M.; Leong, K.W. In vitro and in vivo models for the study of oral delivery of nanoparticles. Adv. Drug Deliv. Rev. 2013, 65, 800–810. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Pitt, J.M.; Daillère, L.Z.J.M.P.R.; Smyth, M.J.; Kroemer, G. Mouse models in oncoimmunology. Nat. Rev. Cancer 2016, 16, 759–773. [Google Scholar] [CrossRef]
- Marra, A. Animal Models for Drug Development for MRSA. Methods Mol. Biol. 2019, 2069, 253–266. [Google Scholar] [CrossRef]
- Couzin-Frankel, J. When Mice Mislead. Science 2013, 342, 922–925. [Google Scholar] [CrossRef] [PubMed]
- Duncombe, J.; Kitamura, A.; Hase, Y.; Ihara, M.; Kalaria, R.N.; Horsburgh, K. Chronic cerebral hypoperfusion: A key mechanism leading to vascular cognitive impairment and dementia. Closing the translational gap between rodent models and human vascular cognitive impairment and dementia. Clin. Sci. 2017, 131, 2451–2468. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Yang, B.; Wang, R.; Qin, C. Xenotransplantation: Current Status in Preclinical Research. Front. Immunol. 2020, 10, 3060. [Google Scholar] [CrossRef] [PubMed]
- Koning, M.; Berg, C.W.V.D.; Rabelink, T.J. Stem cell-derived kidney organoids: Engineering the vasculature. Cell. Mol. Life Sci. 2020, 77, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Namekawa, T.; Ikeda, K.; Horie-Inoue, K.; Inoue, S. Application of Prostate Cancer Models for Preclinical Study: Advantages and Limitations of Cell Lines, Patient-Derived Xenografts, and Three-Dimensional Culture of Patient-Derived Cells. Cells 2019, 8, 74. [Google Scholar] [CrossRef]
- Saeidnia, S.; Manayi, A.; Abdollahi, M. From in vitro Experiments to in vivo and Clinical Studies; Pros and Cons. Curr. Drug Discov. Technol. 2016, 12, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Winn, L.M. In Vitro Models in Developmental Toxicology. Breast Cancer 2019, 1965, 1–6. [Google Scholar] [CrossRef]
- Duval, C.W.; White, P.G. The histological lesions of experimental glanders. J. Exp. Med. 1907, 9, 352–380. [Google Scholar] [CrossRef][Green Version]
- Del Buono, R.; Pignatelli, M.; Bodmer, W.F.; Wright, N.A. The role of the arginine-glycine-aspartic acid-directed cellular binding to type I collagen and rat mesenchymal cells in colorectal tumour differentiation. Differentiation 1991, 46, 97–103. [Google Scholar] [CrossRef]
- Moes, A.J. Gastroretentive dosage forms. Crit. Rev. Ther. Drug Carr. Syst. 1993, 10, 143–195. [Google Scholar]
- Tsung, H.C.; Xia, S.H.; Xu, L.X.; Li, X.L.; Shi, W.K.; Yao, Z. Expression of exogenous porcine transforming growth factor beta-1 gene in ES cells and its effect on their differentiation in vitro. Shi Yan Sheng Wu Xue Bao 1995, 28, 173–189. [Google Scholar]
- Venkat, R.V.; Chalmers, J.J. Characterization of agitation environments in 250 ml spinner vessel, 3 L, and 20 L reactor vessels used for animal cell microcarrier culture. Cytotechnology 1996, 22, 95–102. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Sato, T.; Vries, R.G.; Snippert, H.J.; Van De Wetering, M.; Barker, N.; Stange, D.E.; Van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting Organ-Level Lung Functions on a Chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef]
- E Mager, D.; Jusko, W.J. Development of Translational Pharmacokinetic–Pharmacodynamic Models. Clin. Pharm. Ther. 2008, 83, 909–912. [Google Scholar] [CrossRef]
- Kuepfer, L.; Niederalt, C.; Wendl, T.; Schlender, J.; Willmann, S.; Lippert, J.; Block, M.; Eissing, T.; Teutonico, D. Applied Concepts in PBPK Modeling: How to Build a PBPK/PD Model. CPT Pharm. Syst. Pharm. 2016, 5, 516–531. [Google Scholar] [CrossRef]
- Lavé, T.; Caruso, A.; Parrott, N.; Walz, A. Translational PK/PD modeling to increase probability of success in drug discovery and early development. Drug Discov. Today Technol. 2016, 21-22, 27–34. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, H.; Madabushi, R.; Liu, Q.; Huang, S.; Zineh, I. Model-Informed Drug Development: Current US Regulatory Practice and Future Considerations. Clin. Pharm. Ther. 2019, 105, 899–911. [Google Scholar] [CrossRef]
- Bianchi, F.; Malboubi, M.; Li, Y.; George, J.H.; Jerusalem, A.; Szele, F.; Thompson, M.S.; Ye, H. Rapid and efficient differentiation of functional motor neurons from human iPSC for neural injury modelling. Stem Cell Res. 2018, 32, 126–134. [Google Scholar] [CrossRef]
- Pan, C.; Kumar, C.; Bohl, S.; Klingmueller, U.; Mann, M. Comparative Proteomic Phenotyping of Cell Lines and Primary Cells to Assess Preservation of Cell Type-specific Functions. Mol. Cell. Proteom. 2009, 8, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Sander, V.; Suñe, G.; Jopling, C.; Morera, C.; Belmonte, J.C.I. Isolation and in vitro culture of primary cardiomyocytes from adult zebrafish hearts. Nat. Protoc. 2013, 8, 800–809. [Google Scholar] [CrossRef]
- Ramboer, E.; Vanhaecke, T.; Rogiers, V.; Vinken, M. Primary hepatocyte cultures as prominentin vitrotools to study hepatic drug transporters. Drug Metab. Rev. 2013, 45, 196–217. [Google Scholar] [CrossRef]
- Lv, N.; Yuan, J.; Ji, A.; Shi, L.; Gao, M.; Cui, L.; Jiang, Q. Perfluorooctanoic acid-induced toxicities in chicken embryo primary cardiomyocytes: Roles of PPAR alpha and Wnt5a/Frizzled2. Toxicol. Appl. Pharm. 2019, 381, 114716. [Google Scholar] [CrossRef]
- Godoy, P.; Hewitt, N.J.; Albrecht, U.; Andersen, M.E.; Ansari, N.; Bhattacharya, S.; Bode, J.G.; Bolleyn, J.; Borner, C.; Böttger, J.; et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch. Toxicol. 2013, 87, 1315–1530. [Google Scholar] [CrossRef]
- Maqsood, M.I.; Matin, M.M.; Bahrami, A.R.; Ghasroldasht, M.M. Immortality of cell lines: Challenges and advantages of establishment. Cell Biol. Int. 2013, 37, 1038–1045. [Google Scholar] [CrossRef]
- Schlaermann, P.; Toelle, B.; Berger, H.; Schmidt, S.C.; Glanemann, M.; Ordemann, J.; Bartfeld, S.; Mollenkopf, H.J.; Meyer, T.F. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut 2014, 65, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.K.; Eberwine, J.H. Mammalian cell transfection: The present and the future. Anal. Bioanal. Chem. 2010, 397, 3173–3178. [Google Scholar] [CrossRef]
- Gartzke, D.; Fricker, G. Establishment of Optimized MDCK Cell Lines for Reliable Efflux Transport Studies. J. Pharm. Sci. 2014, 103, 1298–1304. [Google Scholar] [CrossRef]
- Huang, F.; Wang, M.; Liu, R.; Wang, J.-Z.; Schadt, E.; Haroutunian, V.; Katsel, P.; Zhang, B.; Wang, X. CDT2-controlled cell cycle reentry regulates the pathogenesis of Alzheimer’s disease. Alzheimer’s Dement. 2018, 15, 217–231. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Lin, Y.; Gil, C.-H.; Yoder, M.C. Differentiation, Evaluation, and Application of Human Induced Pluripotent Stem Cell–Derived Endothelial Cells. Arter. Thromb. Vasc. Biol. 2017, 37, 2014–2025. [Google Scholar] [CrossRef]
- Sharma, A.; Burridge, P.W.; McKeithan, W.L.; Serrano, R.; Shukla, P.; Sayed, N.; Churko, J.M.; Kitani, T.; Wu, H.; Holmström, A.; et al. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci. Transl. Med. 2017, 9, eaaf2584. [Google Scholar] [CrossRef]
- Langhans, S.A. Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front. Pharm. 2018, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Breslin, S.; O’Driscoll, L. Three-dimensional cell culture: The missing link in drug discovery. Drug Discov. Today 2013, 18, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Pickl, M.; Ries, C.H. Comparison of 3D and 2D tumor models reveals enhanced HER2 activation in 3D associated with an increased response to trastuzumab. Oncogene 2008, 28, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.-C.; Hsiao, A.Y.; Allen, S.G.; Torisawa, Y.-S.; Ho, M.; Takayama, S. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst 2010, 136, 473–478. [Google Scholar] [CrossRef]
- Li, Q.; Chow, A.B.; Mattingly, R.R. Three-Dimensional Overlay Culture Models of Human Breast Cancer Reveal a Critical Sensitivity to Mitogen-Activated Protein Kinase Kinase Inhibitors. J. Pharm. Exp. Ther. 2009, 332, 821–828. [Google Scholar] [CrossRef]
- David, L.; Dulong, V.; Le Cerf, D.; Cazin, L.; Lamacz, M.; Vannier, J.-P. Hyaluronan hydrogel: An appropriate three-dimensional model for evaluation of anticancer drug sensitivity. Acta Biomater. 2008, 4, 256–263. [Google Scholar] [CrossRef]
- Xu, H.; Jiao, Y.; Qin, S.; Zhao, W.; Chu, Q.; Wu, K. Organoid technology in disease modelling, drug development, personalized treatment and regeneration medicine. Exp. Hematol. Oncol. 2018, 7, 1–12. [Google Scholar] [CrossRef]
- Onozato, D.; Yamashita, M.; Nakanishi, A.; Akagawa, T.; Kida, Y.; Ogawa, I.; Hashita, T.; Iwao, T.; Matsunaga, T. Generation of Intestinal Organoids Suitable for Pharmacokinetic Studies from Human Induced Pluripotent Stem Cells. Drug Metab. Dispos. 2018, 46, 1572–1580. [Google Scholar] [CrossRef]
- Shinozawa, T.; Kimura, M.; Cai, Y.; Saiki, N.; Yoneyama, Y.; Ouchi, R.; Koike, H.; Maezawa, M.; Zhang, R.-R.; Dunn, A.; et al. High-Fidelity Drug-Induced Liver Injury Screen Using Human Pluripotent Stem Cell–Derived Organoids. Gastroenterology 2021, 160, 831–846. [Google Scholar] [CrossRef] [PubMed]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernández-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Low, L.A.; Mummery, C.; Berridge, B.R.; Austin, C.P.; Tagle, D.A. Organs-on-chips: Into the next decade. Nat. Rev. Drug Discov. 2020, 1–17. [Google Scholar] [CrossRef]
- Hardwick, R.N.; Betts, C.J.; Whritenour, J.; Sura, R.; Thamsen, M.; Kaufman, E.H.; Fabre, K. Drug-induced skin toxicity: Gaps in preclinical testing cascade as opportunities for complex in vitro models and assays. Lab. Chip 2019, 20, 199–214. [Google Scholar] [CrossRef]
- Ainslie, G.R.; Davis, M.; Ewart, L.; Lieberman, L.A.; Rowlands, D.J.; Thorley, A.J.; Yoder, G.; Ryan, A.M. Microphysiological lung models to evaluate the safety of new pharmaceutical modalities: A biopharmaceutical perspective. Lab. Chip 2019, 19, 3152–3161. [Google Scholar] [CrossRef]
- Peters, M.F.; Choy, A.L.; Pin, C.; Leishman, D.J.; Moisan, A.; Ewart, L.C.; Guzzie-Peck, P.J.; Sura, R.; Keller, D.A.; Scott, C.W.; et al. Developing in vitro assays to transform gastrointestinal safety assessment: Potential for microphysiological systems. Lab. Chip 2020, 20, 1177–1190. [Google Scholar] [CrossRef]
- Phillips, J.A.; Grandhi, T.S.P.; Davis, M.; Gautier, J.-C.; Hariparsad, N.; Keller, D.; Sura, R.; Van Vleet, T.R. A pharmaceutical industry perspective on microphysiological kidney systems for evaluation of safety for new therapies. Lab. Chip 2020, 20, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Baudy, A.R.; Otieno, M.A.; Hewitt, P.; Gan, J.; Roth, A.; Keller, D.; Sura, R.; Van Vleet, T.R.; Proctor, W.R. Liver microphysiological systems development guidelines for safety risk assessment in the pharmaceutical industry. Lab. Chip 2020, 20, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Novak, R.; Ingram, M.; Marquez, S.; Das, D.; Delahanty, A.; Herland, A.; Maoz, B.M.; Jeanty, S.S.F.; Somayaji, M.R.; Burt, M.; et al. Robotic fluidic coupling and interrogation of multiple vascularized organ chips. Nat. Biomed. Eng. 2020, 4, 407–420. [Google Scholar] [CrossRef]
- Li, X.; George, S.M.; Vernetti, L.; Gough, A.H.; Taylor, D.L. A glass-based, continuously zonated and vascularized human liver acinus microphysiological system (vLAMPS) designed for experimental modeling of diseases and ADME/TOX. Lab. Chip 2018, 18, 2614–2631. [Google Scholar] [CrossRef]
- Park, S.E.; Georgescu, A.; Huh, D. Organoids-on-a-chip. Science 2019, 364, 960–965. [Google Scholar] [CrossRef]
- Wikswo, J.P.; Curtis, E.L.; Eagleton, Z.E.; Evans, B.C.; Kole, A.; Hofmeister, L.H.; Matloff, W.J. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab. Chip 2013, 13, 3496–3511. [Google Scholar] [CrossRef] [PubMed]
- Wikswo, J.P.; Block, F.E.; Cliffel, D.E.; Goodwin, C.R.; Marasco, C.C.; Markov, D.A.; McLean, D.L.; McLean, J.A.; McKenzie, J.R.; Reiserer, R.S.; et al. Engineering Challenges for Instrumenting and Controlling Integrated Organ-on-Chip Systems. IEEE Trans. Biomed. Eng. 2013, 60, 682–690. [Google Scholar] [CrossRef]
- Chang, S.-Y.; Weber, E.J.; Sidorenko, V.S.; Chapron, A.; Yeung, C.K.; Gao, C.; Mao, Q.; Shen, D.; Wang, J.; Rosenquist, T.A.; et al. Human liver-kidney model elucidates the mechanisms of aristolochic acid nephrotoxicity. JCI Insight 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Edington, C.D.; Chen, W.L.K.; Geishecker, E.; Kassis, T.; Soenksen, L.R.; Bhushan, B.M.; Freake, D.; Kirschner, J.; Maass, C.; Tsamandouras, N.; et al. Interconnected Microphysiological Systems for Quantitative Biology and Pharmacology Studies. Sci. Rep. 2018, 8, 4530. [Google Scholar] [CrossRef]
- Komen, J.; Westerbeek, E.Y.; Kolkman, R.W.; Roesthuis, J.; Lievens, C.; Berg, A.V.D.; Van Der Meer, A.D. Controlled pharmacokinetic anti-cancer drug concentration profiles lead to growth inhibition of colorectal cancer cells in a microfluidic device. Lab. Chip 2020, 20, 3167–3178. [Google Scholar] [CrossRef]
- Satoh, T.; Sugiura, S.; Shin, K.; Onuki-Nagasaki, R.; Ishida, S.; Kikuchi, K.; Kakiki, M.; Kanamori, T. A multi-throughput multi-organ-on-a-chip system on a plate formatted pneumatic pressure-driven medium circulation platform. Lab. Chip 2017, 18, 115–125. [Google Scholar] [CrossRef]
- Wagner, J.G. Pharmacokinetics. Annu. Rev. Pharm. 1968, 8, 67–94. [Google Scholar] [CrossRef]
- Negus, S.S.; Banks, M.L. Pharmacokinetic-Pharmacodynamic (PKPD) Analysis with Drug Discrimination. Curr. Top. Behav. Neurosci. 2016, 39, 245–259. [Google Scholar] [CrossRef]
- Roberts, J.A.; Taccone, F.S.; Lipman, J. Understanding PK/PD. Intensiv. Care Med. 2015, 42, 1797–1800. [Google Scholar] [CrossRef]
- Li, G.-F.; Yu, G.; Li, Y.; Zheng, Y.; Zheng, Q.-S.; Derendorf, H. Quantitative Estimation of Plasma Free Drug Fraction in Patients With Varying Degrees of Hepatic Impairment: A Methodological Evaluation. J. Pharm. Sci. 2018, 107, 1948–1956. [Google Scholar] [CrossRef]
- Van Breemen, R.B.; Li, Y. Caco-2 cell permeability assays to measure drug absorption. Expert Opin. Drug Metab. Toxicol. 2005, 1, 175–185. [Google Scholar] [CrossRef] [PubMed]
- A Volpe, D. Drug-permeability and transporter assays in Caco-2 and MDCK cell lines. Futur. Med. Chem. 2011, 3, 2063–2077. [Google Scholar] [CrossRef]
- Maass, C.; Stokes, C.L.; Griffith, L.G.; Cirit, M. Multi-functional scaling methodology for translational pharmacokinetic and pharmacodynamic applications using integrated microphysiological systems (MPS). Integr. Biol. 2017, 9, 290–302. [Google Scholar] [CrossRef]
- Reyner, E.L.; Sevidal, S.; West, M.A.; Clouser-Roche, A.; Freiwald, S.; Fenner, K.; Ullah, M.; Lee, C.A.; Smith, B.J. In Vitro Characterization of Axitinib Interactions with Human Efflux and Hepatic Uptake Transporters: Implications for Disposition and Drug Interactions. Drug Metab. Dispos. 2013, 41, 1575–1583. [Google Scholar] [CrossRef]
- Youn, Y.-H.; Hong, J.; Burke, J.M. Endogenous N-cadherin in a subpopulation of MDCK cells: Distribution and catenin complex composition. Exp. Cell Res. 2005, 303, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Mantle, J.L.; Min, L.; Lee, K.H. Minimum Transendothelial Electrical Resistance Thresholds for the Study of Small and Large Molecule Drug Transport in a Human in Vitro Blood–Brain Barrier Model. Mol. Pharm. 2016, 13, 4191–4198. [Google Scholar] [CrossRef]
- Datta, D.; Vasudevan, A. Migration, Chemo-Attraction, and Co-Culture Assays for Human Stem Cell-Derived Endothelial Cells and GABAergic Neurons. J. Vis. Exp. 2020, e60295. [Google Scholar] [CrossRef]
- Li, J.; Wu, J.; Bao, X.; Honea, N.; Xie, Y.; Kim, S.; Sparreboom, A.; Sanai, N. Quantitative and Mechanistic Understanding of AZD1775 Penetration across Human Blood–Brain Barrier in Glioblastoma Patients Using an IVIVE–PBPK Modeling Approach. Clin. Cancer Res. 2017, 23, 7454–7466. [Google Scholar] [CrossRef] [PubMed]
- Blank, L.M.; Ebert, B.E.; Buehler, K.; Bühler, B. Redox Biocatalysis and Metabolism: Molecular Mechanisms and Metabolic Network Analysis. Antioxid. Redox Signal. 2010, 13, 349–394. [Google Scholar] [CrossRef]
- Cooman, T.; Bell, S. In vitro metabolism of the synthetic cannabinoids PX-1, PX-2, and PX-3 by high-resolution mass spectrometry and their clearance rates in human liver microsomes. Rapid Commun. Mass Spectrom. 2019, 33, 1816–1825. [Google Scholar] [CrossRef]
- Nagarajan, S.R.; Paul-Heng, M.; Krycer, J.R.; Fazakerley, D.J.; Sharland, A.F.; Hoy, A.J. Lipid and glucose metabolism in hepatocyte cell lines and primary mouse hepatocytes: A comprehensive resource for in vitro studies of hepatic metabolism. Am. J. Physiol. Metab. 2019, 316, E578–E589. [Google Scholar] [CrossRef] [PubMed]
- Boon, R.; Kumar, M.; Tricot, T.; Elia, I.; Ordovas, L.; Jacobs, F.; One, J.; De Smedt, J.; Eelen, G.; Bird, M.; et al. Amino acid levels determine metabolism and CYP450 function of hepatocytes and hepatoma cell lines. Nat. Commun. 2020, 11, 1393. [Google Scholar] [CrossRef] [PubMed]
- Guillouzo, A.; Corlu, A.; Aninat, C.; Glaise, D.; Morel, F.; Guguen-Guillouzo, C. The human hepatoma HepaRG cells: A highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem. Interact. 2007, 168, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Mun, S.J.; Ryu, J.-S.; Lee, M.-O.; Son, Y.S.; Oh, S.J.; Cho, H.-S.; Son, M.-Y.; Kim, D.-S.; Kim, S.J.; Yoo, H.J.; et al. Generation of expandable human pluripotent stem cell-derived hepatocyte-like liver organoids. J. Hepatol. 2019, 71, 970–985. [Google Scholar] [CrossRef]
- Marin, T.M.; Indolfo, N.D.C.; Rocco, S.A.; Basei, F.L.; De Carvalho, M.; Gonçalves, K.D.A.; Pagani, E. Acetaminophen absorption and metabolism in an intestine/liver microphysiological system. Chem. Interact. 2019, 299, 59–76. [Google Scholar] [CrossRef] [PubMed]
- Emoto, C.; Fukuda, T.; Cox, S.; Christians, U.; A Vinks, A. Development of a Physiologically-Based Pharmacokinetic Model for Sirolimus: Predicting Bioavailability Based on Intestinal CYP3A Content. CPT Pharm. Syst. Pharm. 2013, 2, 1–9. [Google Scholar] [CrossRef]
- Scotcher, D.; Jones, C.; Posada, M.; Rostami-Hodjegan, A.; Galetin, A. Key to Opening Kidney for In Vitro–In Vivo Extrapolation Entrance in Health and Disease: Part I: In Vitro Systems and Physiological Data. AAPS J. 2016, 18, 1067–1081. [Google Scholar] [CrossRef]
- Love, H.D.; Evans, R.C.; Humes, H.D.; Roy, S.; Zent, R.; Harris, R.; Wilson, M.H.; Fissell, W.H. Metformin and Inhibition of Transforming Growth Factor-Beta Stimulate In Vitro Transport in Primary Renal Tubule Cells. Tissue Eng. Part. A 2020, 26, 1091–1098. [Google Scholar] [CrossRef]
- Kumar, M.P.; Thyagarajan, B.; Haller, N.; Ciltea, D. A Diagnostic Conundrum of Distributive Shock: Autoimmune Polyglandular Syndrome Type II. Indian J. Crit. Care Med. 2019, 23, 582–583. [Google Scholar] [CrossRef]
- Shah, D.K.; Haddish-Berhane, N.; Betts, A. Bench to bedside translation of antibody drug conjugates using a multiscale mechanistic PK/PD model: A case study with brentuximab-vedotin. J. Pharm. Pharm. 2012, 39, 643–659. [Google Scholar] [CrossRef]
- He, H.; Cao, Y. Chemotherapeutic dosing implicated by pharmacodynamic modeling of in vitro cytotoxic data: A case study of paclitaxel. J. Pharm. Pharm. 2017, 44, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Yoon, M.; Kedderis, G.L.; Yan, G.Z.; Clewell, H.J. Use of in vitro data in developing a physiologically based pharmacokinetic model: Carbaryl as a case study. Toxicology 2015, 332, 52–66. [Google Scholar] [CrossRef] [PubMed]
- Barros, A.S.; Costa, A.; Sarmento, B. Building three-dimensional lung models for studying pharmacokinetics of inhaled drugs. Adv. Drug Deliv. Rev. 2021, 170, 386–395. [Google Scholar] [CrossRef]
- Guerrero, Y.A.; Desai, D.; Sullivan, C.; Kindt, E.; Spilker, M.E.; Maurer, T.S.; Solomon, D.E.; Bartlett, D.W. A Microfluidic Perfusion Platform for In Vitro Analysis of Drug Pharmacokinetic-Pharmacodynamic (PK-PD) Relationships. AAPS J. 2020, 22, 53. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Maass, C.; Sorensen, N.B.; Himmelfarb, J.; Kelly, E.J.; Stokes, C.L.; Cirit, M. Translational Assessment of Drug-Induced Proximal Tubule Injury Using a Kidney Microphysiological System. CPT Pharm. Syst. Pharm. 2019, 8, 316–325. [Google Scholar] [CrossRef]
- Generaux, G.; Lakhani, V.V.; Yang, Y.; Nadanaciva, S.; Qiu, L.; Riccardi, K.; Di, L.; Howell, B.A.; Siler, S.Q.; Watkins, P.B.; et al. Quantitative systems toxicology (QST) reproduces species differences in PF-04895162 liver safety due to combined mitochondrial and bile acid toxicity. Pharm. Res. Perspect. 2019, 7, e00523. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, T.R.; Ande, A.; Ait-Oudhia, S.; Anda, A. Combining Multiscale Experimental and Computational Systems Pharmacological Approaches to Overcome Resistance to HER2-targeted Therapy in Breast Cancer. J. Pharm. Exp. Ther. 2019, 369, 531–545. [Google Scholar] [CrossRef] [PubMed]
- Cirit, M.; Stokes, C.L. Maximizing the impact of microphysiological systems with in vitro–in vivo translation. Lab. Chip 2018, 18, 1831–1837. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Song, H.; Ming, G.-L. Brain organoids: Advances, applications and challenges. Development 2019, 146. [Google Scholar] [CrossRef]
- Milberg, O.; Gong, C.; Jafarnejad, M.; Bartelink, I.H.; Wang, B.; Vicini, P.; Narwal, R.; Roskos, L.; Popel, A.S. A QSP Model for Predicting Clinical Responses to Monotherapy, Combination and Sequential Therapy Following CTLA-4, PD-1, and PD-L1 Checkpoint Blockade. Sci. Rep. 2019, 9, 11286. [Google Scholar] [CrossRef] [PubMed]
- Mrass, P.; Takano, H.; Ng, L.G.; Daxini, S.; Lasaro, M.O.; Iparraguirre, A.; Cavanagh, L.L.; Von Andrian, U.H.; Ertl, H.C.; Haydon, P.G.; et al. Random migration precedes stable target cell interactions of tumor-infiltrating T cells. J. Exp. Med. 2006, 203, 2749–2761. [Google Scholar] [CrossRef] [PubMed]
- Bryl, E.; Vallejo, A.N.; Matteson, E.L.; Witkowski, J.M.; Weyand, C.M.; Goronzy, J.J. Modulation of CD28 expression with anti-tumor necrosis factor α therapy in rheumatoid arthritis. Arthritis Rheum. 2005, 52, 2996–3003. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, H. Pharmacological considerations for predicting PK/PD at the site of action for therapeutic proteins. Drug Discov. Today Technol. 2016, 21–22, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Colburn, W.A. Combined Pharmacokinetic/ Pharmacodynamic (PK/PD) Modeling. J. Clin. Pharm. 1988, 28, 769–771. [Google Scholar] [CrossRef]
- Hunter, R.P. Interspecies Allometric Scaling. Organotypic Models Drug Dev. 2010, 199, 139–157. [Google Scholar] [CrossRef]
- Huang, Q.; E Riviere, J. The application of allometric scaling principles to predict pharmacokinetic parameters across species. Expert Opin. Drug Metab. Toxicol. 2014, 10, 1241–1253. [Google Scholar] [CrossRef]
- Keller, F.; Ludwig, U.; Czock, D. Pharmacokinetic and pharmacodynamic considerations on the erythropoietin effect and adverse events of darbepoetin. Expert Opin. Drug Metab. Toxicol. 2014, 11, 139–147. [Google Scholar] [CrossRef]
- Lötsch, J. Pharmacokinetic–Pharmacodynamic Modeling of Opioids. J. Pain Symptom Manag. 2005, 29, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.K.; Loganzo, F.; Haddish-Berhane, N.; Musto, S.; Wald, H.S.; Barletta, F.; Lucas, J.; Clark, T.; Hansel, S.; Betts, A. Establishing in vitro–in vivo correlation for antibody drug conjugate efficacy: A PK/PD modeling approach. J. Pharm. Pharm. 2018, 45, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Zuo, P. Capturing the Magic Bullet: Pharmacokinetic Principles and Modeling of Antibody-Drug Conjugates. AAPS J. 2020, 22, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kahn, J.; Tofilon, P.J.; Camphausen, K. Preclinical models in radiation oncology. Radiat. Oncol. 2012, 7, 223. [Google Scholar] [CrossRef]
- Kamatar, A.; Gunay, G.; Acar, H. Natural and Synthetic Biomaterials for Engineering Multicellular Tumor Spheroids. Polymers 2020, 12, 2506. [Google Scholar] [CrossRef] [PubMed]
- Lorenzutti, A.M.; Litterio, N.J.; Himelfarb, M.A.; Zarazaga, M.D.P.; Andrés, M.I.S.; De Lucas, J.J. Pharmacokinetics, milk penetration and PK/PD analysis by Monte Carlo simulation of marbofloxacin, after intravenous and intramuscular administration to lactating goats. J. Veter. Pharm. Ther. 2017, 40, 629–640. [Google Scholar] [CrossRef]
- Ayyar, V.S.; Jusko, W.J. Transitioning from Basic toward Systems Pharmacodynamic Models: Lessons from Corticosteroids. Pharm. Rev. 2020, 72, 414–438. [Google Scholar] [CrossRef]
- Espié, P.; Tytgat, D.; Sargentini-Maier, M.-L.; Poggesi, I.; Watelet, J.-B. Physiologically based pharmacokinetics (PBPK). Drug Metab. Rev. 2009, 41, 391–407. [Google Scholar] [CrossRef]
- Sager, J.E.; Yu, J.; Ragueneau-Majlessi, I.; Isoherranen, N. Physiologically Based Pharmacokinetic (PBPK) Modeling and Simulation Approaches: A Systematic Review of Published Models, Applications, and Model Verification. Drug Metab. Dispos. 2015, 43, 1823–1837. [Google Scholar] [CrossRef]
- Sung, J.H.; Srinivasan, B.; Esch, M.B.; McLamb, W.T.; Bernabini, C.; Shuler, M.L.; Hickman, J.J. Using physiologically-based pharmacokinetic-guided “body-on-a-chip” systems to predict mammalian response to drug and chemical exposure. Exp. Biol. Med. 2014, 239, 1225–1239. [Google Scholar] [CrossRef]
- Hidalgo, I.J.; Raub, T.J.; Borchardt, R.T. Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability. Gastroenterology 1989, 96, 736–749. [Google Scholar] [CrossRef]
- Gomez-Lechon, M.; Donato, M.; Lahoz, A.; Castell, J. Cell Lines: A Tool for In Vitro Drug Metabolism Studies. Curr. Drug Metab. 2008, 9, 1–11. [Google Scholar] [CrossRef]
- Wu, J.; Sanai, N.; Bao, X.; Lorusso, P.; Li, J. An aqueous normal-phase chromatography coupled with tandem mass spectrometry method for determining unbound brain-to-plasma concentration ratio of AZD1775, a Wee1 kinase inhibitor, in patients with glioblastoma. J. Chromatogr. B 2016, 1028, 25–32. [Google Scholar] [CrossRef]
- Blanchard, J.W.; Bula, M.; Davila-Velderrain, J.; Akay, L.A.; Zhu, L.; Frank, A.; Victor, M.B.; Bonner, J.M.; Mathys, H.; Lin, Y.-T.; et al. Reconstruction of the human blood–brain barrier in vitro reveals a pathogenic mechanism of APOE4 in pericytes. Nat. Med. 2020, 26, 952–963. [Google Scholar] [CrossRef]
- Min, J.S.; Bae, S.K. Prediction of drug–drug interaction potential using physiologically based pharmacokinetic modeling. Arch. Pharmacal Res. 2017, 40, 1356–1379. [Google Scholar] [CrossRef] [PubMed]
- Hanke, N.; Frechen, S.; Moj, D.; Britz, H.; Eissing, T.; Wendl, T.; Lehr, T. PBPK Models for CYP3A4 and P-gp DDI Prediction: A Modeling Network of Rifampicin, Itraconazole, Clarithromycin, Midazolam, Alfentanil, and Digoxin. CPT Pharmacomet. Syst. Pharm. 2018, 7, 647–659. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, P.; Emond, C.; McLanahan, E.D.; Joshi-Barr, S.; Mumtaz, M. Exploring Mechanistic Toxicity of Mixtures Using PBPK Modeling and Computational Systems Biology. Toxicol. Sci. 2019, 174, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Lipscomb, J.C.; Haddad, S.; Poet, T.; Krishnan, K. Physiologically-Based Pharmacokinetic (PBPK) Models in Toxicity Testing and Risk Assessment. Chem. Biol. Pteridines Folates 2012, 745, 76–95. [Google Scholar] [CrossRef]
- Zang, X.; Kagan, L. Physiologically-based modeling and interspecies prediction of paclitaxel pharmacokinetics. J. Pharm. Pharm. 2018, 45, 577–592. [Google Scholar] [CrossRef]
- Verscheijden, L.F.; Koenderink, J.B.; Johnson, T.N.; de Wildt, S.N.; Russel, F.G. Physiologically-based pharmacokinetic models for children: Starting to reach maturation? Pharm. Ther. 2020, 211, 107541. [Google Scholar] [CrossRef]
- Prantil-Baun, R.; Novak, R.; Das, D.; Somayaji, M.R.; Przekwas, A.; Ingber, D.E. Physiologically Based Pharmacokinetic and Pharmacodynamic Analysis Enabled by Microfluidically Linked Organs-on-Chips. Annu. Rev. Pharm. Toxicol. 2018, 58, 37–64. [Google Scholar] [CrossRef]
- Liu, L.; Koo, Y.; Akwitti, C.; Russell, T.; Gay, E.; Laskowitz, D.T.; Yun, Y. Three-dimensional (3D) brain microphysiological system for organophosphates and neurochemical agent toxicity screening. PLoS ONE 2019, 14, e0224657. [Google Scholar] [CrossRef]
- Herland, A.; Maoz, B.M.; Das, D.; Somayaji, M.R.; Prantil-Baun, R.; Novak, R.; Cronce, M.; Huffstater, T.; Jeanty, S.S.F.; Ingram, M.; et al. Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nat. Biomed. Eng. 2020, 4, 421–436. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.P.; Zheng, X.; Lin-Schmidt, X.; Chen, W.; Carpenter, T.J.; Zong, A.; Wang, W.; Heald, D.L. Development of a quantitative relationship between CAR-affinity, antigen abundance, tumor cell depletion and CAR-T cell expansion using a multiscale systems PK-PD model. MAbs 2020, 12, 1688616. [Google Scholar] [CrossRef] [PubMed]
- Caruso, H.G.; Hurton, L.V.; Najjar, A.; Rushworth, D.; Ang, S.; Olivares, S.; Mi, T.; Switzer, K.; Singh, H.; Huls, H.; et al. Tuning Sensitivity of CAR to EGFR Density Limits Recognition of Normal Tissue While Maintaining Potent Antitumor Activity. Cancer Res. 2015, 75, 3505–3518. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Nueno, V.I. Using quantitative systems pharmacology for novel drug discovery. Expert Opin. Drug Discov. 2015, 10, 1315–1331. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; Pawar, R.; Pandey, R.; Madan, J.; Pawar, S.; Lakshmi, P.; Sudheesh, M. In-vitro in-vivo correlation (IVIVC) in nanomedicine: Is protein corona the missing link? Biotechnol. Adv. 2017, 35, 889–904. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Thalhauser, C.J.; Smithline, S.; Pagidala, J.; Miladinov, M.; Vezina, H.E.; Gupta, M.; Leil, T.A.; Schmidt, B.J. QSP Toolbox: Computational Implementation of Integrated Workflow Components for Deploying Multi-Scale Mechanistic Models. AAPS J. 2017, 19, 1002–1016. [Google Scholar] [CrossRef]
- Howell, B.A.; Yang, Y.; Kumar, R.; Woodhead, J.L.; Harrill, A.H.; Clewell, H.J.; Andersen, M.E.; Siler, S.Q.; Watkins, P.B. In vitro to in vivo extrapolation and species response comparisons for drug-induced liver injury (DILI) using DILIsym™: A mechanistic, mathematical model of DILI. J. Pharm. Pharm. 2012, 39, 527–541. [Google Scholar] [CrossRef]
- Longo, D.M.; Yang, Y.; Watkins, P.B.; A Howell, B.; Siler, S.Q. Elucidating Differences in the Hepatotoxic Potential of Tolcapone and Entacapone With DILIsym®, a Mechanistic Model of Drug-Induced Liver Injury. CPT Pharmacomet. Syst. Pharm. 2016, 5, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Shoda, L.K.M.; Woodhead, J.L.; Siler, S.Q.; Watkins, P.B.; Howell, B.A. Linking physiology to toxicity using DILIsym®, a mechanistic mathematical model of drug-induced liver injury. Biopharm. Drug Dispos. 2013, 35, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Nadanaciva, S.; Will, Y.; Woodhead, J.L.; Howell, B.A.; Watkins, P.B.; Siler, S.Q. MITOsym®: A Mechanistic, Mathematical Model of Hepatocellular Respiration and Bioenergetics. Pharm. Res. 2015, 32, 1975–1992. [Google Scholar] [CrossRef] [PubMed]
- Kilickap, S.; Demirci, U.; Karadurmus, N.; Dogan, M.; Akinci, B.; Sendur, M.A.N. Endpoints in oncology clinical trials. J. BUON Off. J. Balk. Union Oncol. 2018, 23, 1–6. [Google Scholar]
- Fiteni, F.; Westeel, V.; Pivot, X.; Borg, C.; Vernerey, D.; Bonnetain, F. Endpoints in cancer clinical trials. J. Visc. Surg. 2014, 151, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Di Bisceglie, A.M.; Bruix, J.; Kramer, B.S.; Lencioni, R.; Zhu, A.X.; Sherman, M.; Schwartz, M.; Lotze, M.; Talwalkar, J.; et al. Design and Endpoints of Clinical Trials in Hepatocellular Carcinoma. J. Natl. Cancer Inst. 2008, 100, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Nieuweboer, A.J.; de Morrée, E.S.; de Graan, A.-J.M.; Sparreboom, A.; de Wit, R.; Mathijssen, R.H. Inter-patient variability in docetaxel pharmacokinetics: A review. Cancer Treat. Rev. 2015, 41, 605–613. [Google Scholar] [CrossRef]
- Chelliah, V.; Lazarou, G.; Bhatnagar, S.; Gibbs, J.P.; Nijsen, M.; Ray, A.; Stoll, B.; Thompson, R.A.; Gulati, A.; Soukharev, S.; et al. Quantitative Systems Pharmacology Approaches for Immuno-Oncology: Adding Virtual Patients to the Development Paradigm. Clin. Pharm. Ther. 2021, 109, 605–618. [Google Scholar] [CrossRef]
- Andreasen, J. New drugs are tested on virtual patients. It will be possible to test new drugs on virtual patients, existing only in a computer. Ugeskr. Laeger 2003, 165, 1961–1962. [Google Scholar]
- Polak, S.; Fijorek, K.; Glinka, A.; Wiśniowska, B.; Mendyk, A. Virtual population generator for human cardiomyocytes parameters: In silicodrug cardiotoxicity assessment. Toxicol. Mech. Methods 2011, 22, 31–40. [Google Scholar] [CrossRef]
- Clemmer, J.S.; Hester, R.L.; Pruett, W.A. Simulating a virtual population’s sensitivity to salt and uninephrectomy. Interface Focus 2017, 8, 20160134. [Google Scholar] [CrossRef]
- Hartmann, S.; Biliouris, K.; Lesko, L.J.; Nowak-Göttl, U.; Trame, M.N. Quantitative Systems Pharmacology Model-Based Predictions of Clinical Endpoints to Optimize Warfarin and Rivaroxaban Anti-Thrombosis Therapy. Front. Pharm. 2020, 11, 1041. [Google Scholar] [CrossRef] [PubMed]
- T’Jollyn, H.; Vermeulen, A.; Van Bocxlaer, J. PBPK and its Virtual Populations: The Impact of Physiology on Pediatric Pharmacokinetic Predictions of Tramadol. AAPS J. 2019, 21, 8. [Google Scholar] [CrossRef] [PubMed]
- Collins, K.P.; Jackson, K.M.; Gustafson, D.L. Hydroxychloroquine: A Physiologically-Based Pharmacokinetic Model in the Context of Cancer-Related Autophagy Modulation. J. Pharm. Exp. Ther. 2018, 365, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Lindauer, A.; Valiathan, C.R.; Mehta, K.; Sriram, V.; De Greef, R.; Elassaiss-Schaap, J.; De Alwis, D.P. Translational Pharmacokinetic/Pharmacodynamic Modeling of Tumor Growth Inhibition Supports Dose-Range Selection of the Anti-PD-1 Antibody Pembrolizumab. CPT Pharmacomet. Syst. Pharm. 2016, 6, 11–20. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Z.; Zhu, J.; Jiang, M.; Sang, L.; Hao, K.; He, H. The Combination of Cell Cultured Technology and In Silico Model to Inform the Drug Development. Pharmaceutics 2021, 13, 704. https://doi.org/10.3390/pharmaceutics13050704
Zhou Z, Zhu J, Jiang M, Sang L, Hao K, He H. The Combination of Cell Cultured Technology and In Silico Model to Inform the Drug Development. Pharmaceutics. 2021; 13(5):704. https://doi.org/10.3390/pharmaceutics13050704
Chicago/Turabian StyleZhou, Zhengying, Jinwei Zhu, Muhan Jiang, Lan Sang, Kun Hao, and Hua He. 2021. "The Combination of Cell Cultured Technology and In Silico Model to Inform the Drug Development" Pharmaceutics 13, no. 5: 704. https://doi.org/10.3390/pharmaceutics13050704
APA StyleZhou, Z., Zhu, J., Jiang, M., Sang, L., Hao, K., & He, H. (2021). The Combination of Cell Cultured Technology and In Silico Model to Inform the Drug Development. Pharmaceutics, 13(5), 704. https://doi.org/10.3390/pharmaceutics13050704






