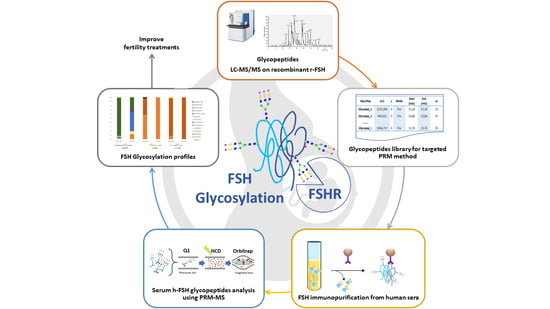
Identification and Relative Quantification of hFSH Glycoforms in Women’s Sera via MS–PRM-Based Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Immunopurification FSH from Serum
2.2. Reduction, Alkylation and Enzymatic Digestion
2.3. Mass Spectrometry Analysis
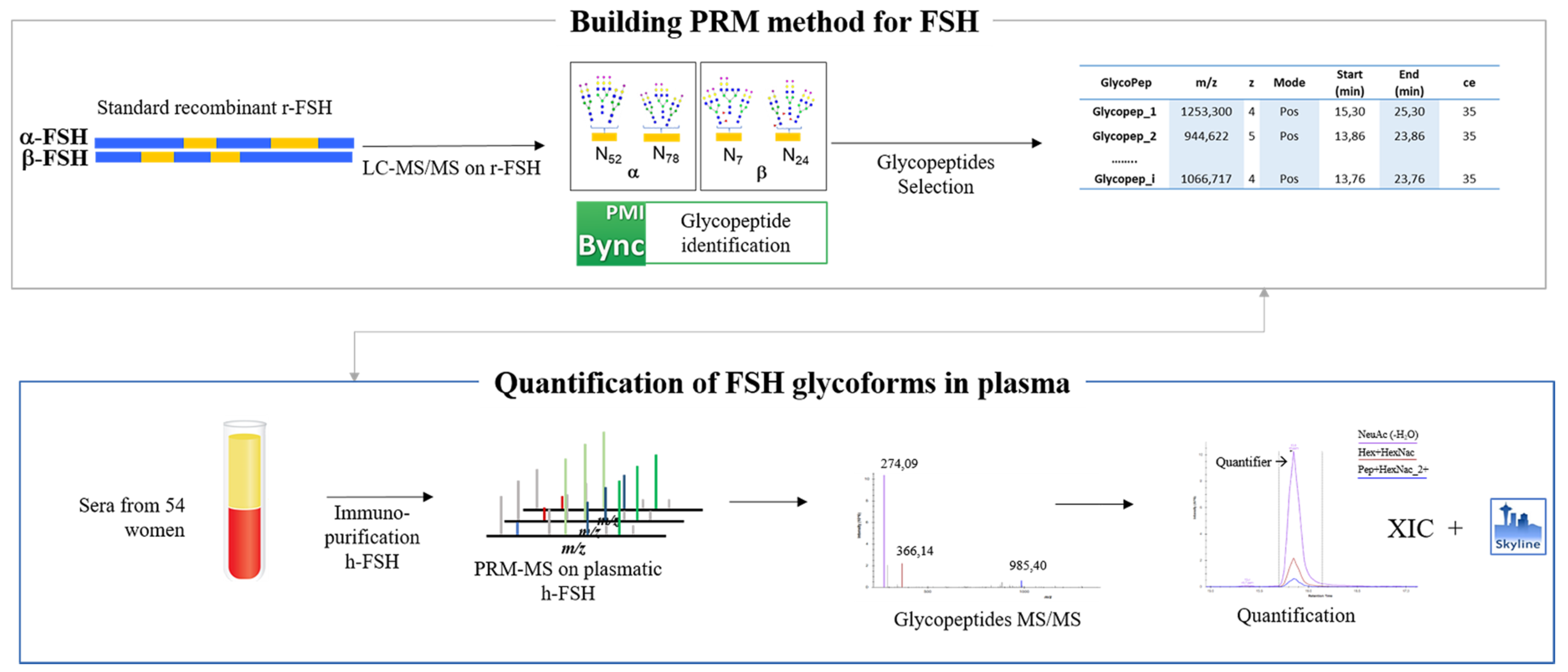
2.4. PRM Method
2.5. Glycopeptides Enrichment
3. Results and Discussion
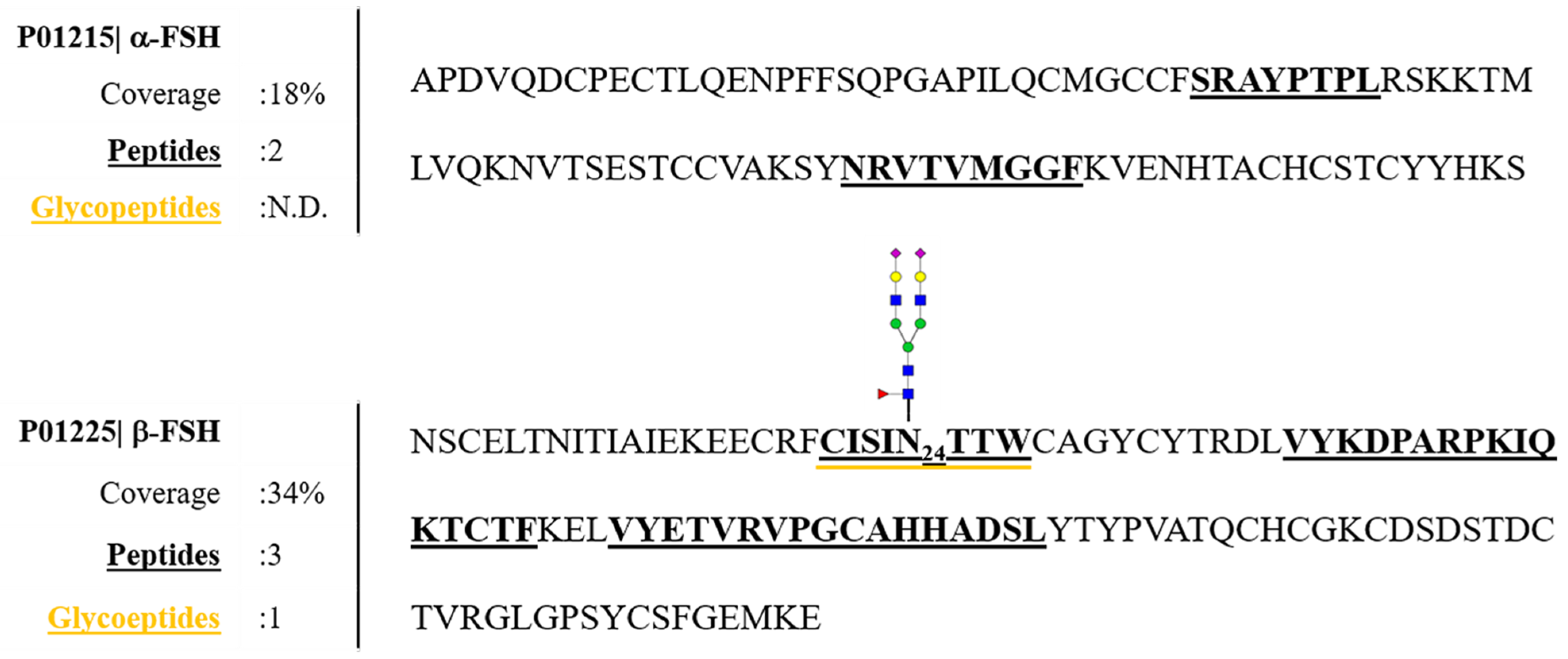
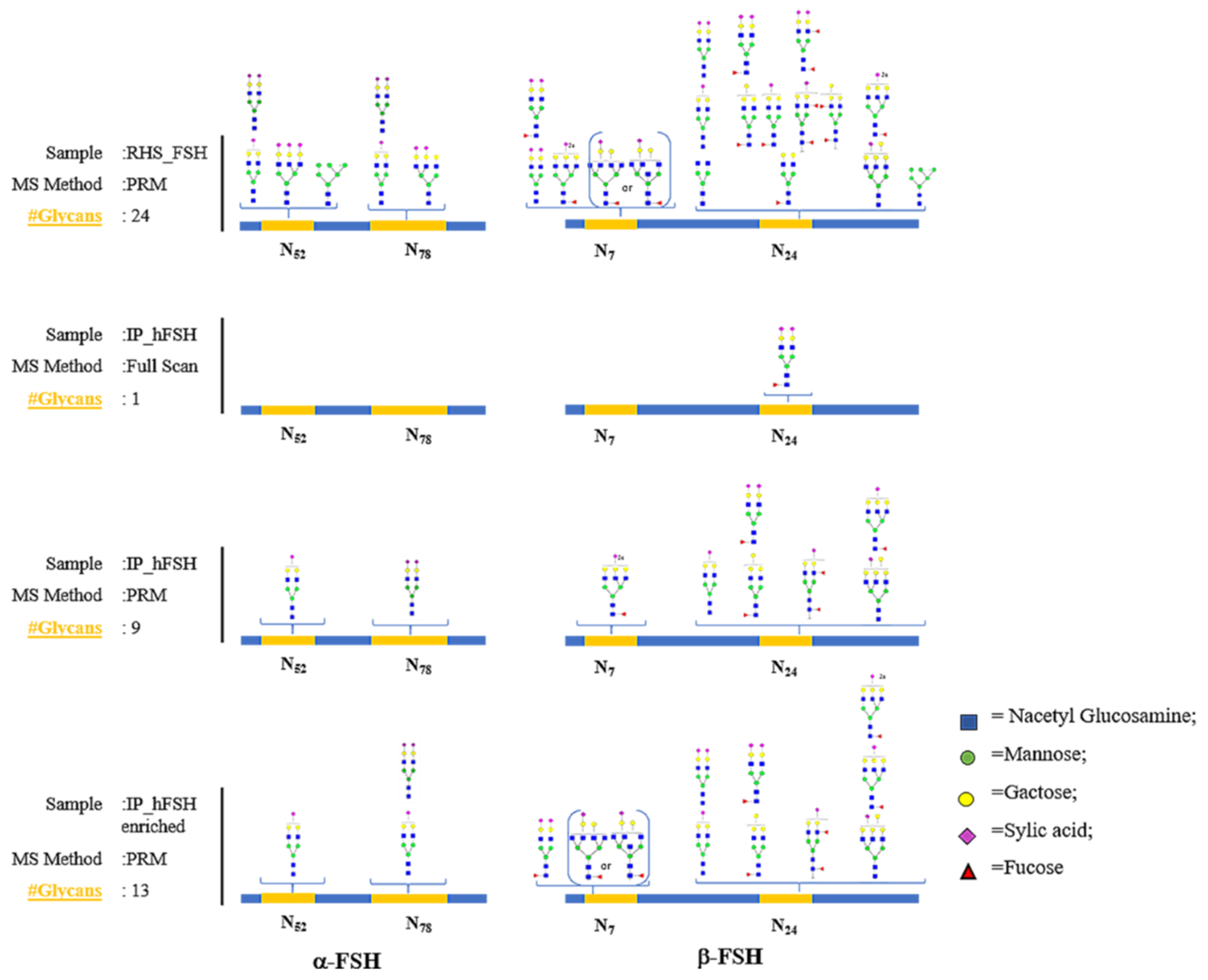
3.1. Recombinant FSH Full Scan LC–MS/MS Analysis: PRM Method Development
3.2. Characterization of hFSH Purified from Serum Using Full Scan LC–MS/MS Analysis
3.3. Detection of hFSH Glycoforms Purified from Serum by MS–PRM Analysis
3.4. Glycopeptide Enrichment Prior PRM Analysis
3.5. Comparison of the Glycoprofiles of RHS_FSH and hFSH from Serum
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Rose, M.P.; Gaines Das, R.E.; Balen, A.H. Definition and measurement of follicle stimulating hormone. Endocr. Rev. 2000, 21, 5–22. [Google Scholar] [CrossRef]
- Sam, T.; De Leeuw, R. Follicle-stimulating hormone. In Pharmaceutical Biotechnology; Springer: New York, NY, USA, 2013; pp. 277–283. [Google Scholar]
- Sairam, M.R.; Bhargavi, G.N. A role for glycosylation of the alpha subunit in transduction of biological signal in glycoprotein hormones. Science 1985, 229, 65–67. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Liu, H.; Chen, X.; Chen, P.H.; Fischer, D.; Sriraman, V.; Henry, N.Y.; Arkinstall, S.; He, X. Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor. Proc. Natl. Acad. Sci. USA 2012, 109, 12491–12496. [Google Scholar] [CrossRef] [PubMed]
- Gloaguen, P.; Crépieux, P.; Heitzler, D.; Poupon, A.; Reiter, E. Mapping the follicle-stimulating hormone-induced signaling networks. Front. Endocrinol. 2011, 2, 45. [Google Scholar] [CrossRef] [PubMed]
- Bousfield, G.R.; Butnev, V.Y.; Walton, W.J.; Nguyen, V.T.; Huneidi, J.; Singh, V.; Kolli, V.K.; Harvey, D.J.; Rance, N.E. All-or-none N-glycosylation in primate follicle-stimulating hormone β-subunits. Mol. Cell. Endocrinol. 2007, 260–262, 40–48. [Google Scholar] [CrossRef]
- Ulloa-Aguirre, A.; Dias, J.A.; Bousfield, G.R. Gonadotropins and the Importance of Glycosylation. In Post-Translational Modification of Protein Biopharmaceuticals; Walsh, G., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009; pp. 109–147. [Google Scholar]
- Walton, W.J.; Nguyen, V.T.; Butnev, V.Y.; Singh, V.; Moore, W.T.; Bousfield, G.R. Characterization of human FSH isoforms reveals a nonglycosylated beta-subunit in addition to the conventional glycosylated beta-subunit. J. Clin. Endocrinol. Metab. 2001, 8, 3675–3685. [Google Scholar] [CrossRef] [PubMed]
- Dalpathado, D.S.; Irungu, J.; Go, E.P.; Butnev, V.Y.; Norton, K.; Bousfield, G.R.; Desaire, H. Comparative Glycomics of the Glycoprotein Follicle Stimulating Hormone: Glycopeptide Analysis of Isolates from Two Mammalian Species. Biochemistry 2006, 45, 8665–8673. [Google Scholar] [CrossRef]
- Anobile, C.J.; Talbot, J.A.; McCann, S.J.; Padmanabhan, V.; Robertson, W.R. Glycoform composition of serum gonadotrophins through the normal menstrual cycle and in the post-menopausal state. Mol. Hum. Reprod. 1998, 4, 631–639. [Google Scholar] [CrossRef]
- Davis, J.S.; Kumar, T.R.; May, J.V.; Bousfield, G.R. Naturally occurring follicle-stimulating hormone glycosylation variants. J. Glycom. Lipidom. 2014, 4, e117. [Google Scholar]
- Bishop, L.A.; Robertson, D.M.; Cahir, N.; Schofield, P.R. Specific roles for the asparagine-linked carbohydrate residues of recombinant human follicle stimulating hormone in receptor binding and signal transduction. Mol. Endocrinol. 1994, 8, 722–731. [Google Scholar] [PubMed]
- Dias, J.A.; Lindau-Shepard, B.; Hauer, C.; Auger, I. Human Follicle-Stimulating Hormone Structure-Activity Relationships. Biol. Reprod. 1998, 58, 1331–1336. [Google Scholar] [CrossRef] [PubMed]
- Bousfield, G.R.; Butnev, V.Y.; Butnev, V.Y.; Hiromasa, Y.; Harvey, D.J.; May, J.V. Hypo-glycosylated human follicle-stimulating hormone (hFSH21/18) is much more active in vitro than fully-glycosylated hFSH (hFSH24). Mol. Cell. Endocrinol. 2014, 382, 989–997. [Google Scholar] [CrossRef]
- Jiang, C.; Hou, X.; Wang, C.; May, J.V.; Butnev, V.Y.; Bousfield, G.R.; Davis, J.S. Hypoglycosylated hFSH Has Greater Bioactivity Than Fully Glycosylated Recombinant hFSH in Human Granulosa Cells. J. Clin. Endocrinol. Metab. 2015, 100, E852–E860. [Google Scholar] [CrossRef] [PubMed]
- D’Antonio, M.; Borrelli, F.; Datola, A.; Bucci, R.; Mascia, M.; Polletta, P.; Piscitelli, D.; Papoian, R. Biological characterization of recombinant human follicle stimulating hormone isoforms. Hum. Reprod. 1999, 14, 1160–1167. [Google Scholar] [CrossRef]
- Emperaire, J.C. Review of Physiology, Ovulation Stimulation with Gonadotropins; Springer International Publishing: Geneva, Switzerland, 2015. [Google Scholar]
- Li, H.; D’Anjou, M. Pharmacological significance of glycosylation in therapeutic proteins. Curr. Opin. Biotechnol. 2009, 20, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, A.M.; Elliott, S. Glycoengineering: The effect of glycosylation on the properties of therapeutic proteins. J. Pharm. Sci. 2005, 94, 1626–1635. [Google Scholar] [CrossRef] [PubMed]
- Ulloa-Aguirre, A.; Timossi, C.; Damián-Matsumura, P.; Dias, J.A. Role of Glycosylation in Function of Follicle-Stimulating Hormone. Endocrine 1999, 11, 205–216. [Google Scholar] [CrossRef]
- Daya, S. Follicle-Stimulating Hormone in Clinical Practice. Treat. Endocrinol. 2004, 3, 161–171. [Google Scholar] [CrossRef]
- Mastrangeli, R.; Satwekar, A.; Cutillo, F.; Ciampolillo, C.; Palinsky, W.; Longobardi, S. In-vivo biological activity and glycosylation analysis of a biosimilar recombinant human follicle-stimulating hormone product (Bemfola) compared with its reference medicinal product (GONAL-f). PLoS ONE 2017, 12, e0184139. [Google Scholar] [CrossRef]
- Scheele, F.; Schoernaker, J. The role of follicle-stimulating hormone in the selection of follicles in human ovaries: A survey of the literature and a proposed model. Gynecol. Endocrinol. 1996, 10, 55–66. [Google Scholar] [CrossRef]
- Prakash, A.; Rezai, T.; Krastins, B.; Sarracino, D.; Athanas, M.; Russo, P.; Ross, M.M.; Zhang, H.; Tian, Y.; Kulasingam, V.; et al. Platform for Establishing Interlaboratory Reproducibility of Selected Reaction Monitoring-Based Mass Spectrometry Peptide Assays. J. Proteome Res. 2010, 9, 6678–6688. [Google Scholar] [CrossRef]
- Illiano, A.; Arpino, V.; Pinto, G.; Berti, A.; Verdoliva, V.; Peluso, G.; Pucci, P.; Amoresano, A. Multiple Reaction Monitoring Tandem Mass Spectrometry Approach for the Identification of Biological Fluids at Crime Scene Investigations. Anal. Chem. 2018, 90, 5627–5636. [Google Scholar] [CrossRef]
- Fontanarosa, C.; Pane, F.; Sepe, N.; Pinto, G.; Trifuoggi, M.; Squillace, M.; Errico, F.; Usiello, A.; Pucci, P.; Amoresano, A. Quantitative determination of free D-Asp, L-Asp and N-methyl-D-aspartate in mouse brain tissues by chiral separation and Multiple Reaction Monitoring tandem mass spectrometry. PLoS ONE 2017, 12, e0179748. [Google Scholar] [CrossRef] [PubMed]
- Rauniyar, N. Parallel Reaction Monitoring: A Targeted Experiment Performed Using High Resolution and High Mass Accuracy Mass Spectrometry. Int. J. Mol. Sci. 2015, 16, 28566–28581. [Google Scholar] [CrossRef] [PubMed]
- Harvey, D.J.; Crispin, M.; Scanlan, C.N.; Singer, B.B.; Lucka, L.; Chang, V.T.; Radcliffe, C.M.; Thobhani, S.; Yuen, C.-T.; Rudd, P.M. Differentiation between isomeric triantennaryN-linked glycans by negative ion tandem mass spectrometry and confirmation of glycans containing galactose attached to the bisecting (β1-4-GlcNAc) residue inN-glycans from IgG. Rapid Commun. Mass Spectrom. 2008, 22, 1047–1052. [Google Scholar] [CrossRef]
- Srikanth, J.; Agalyadevi, R.; Babu, P. Targeted, Site-specific quantitation of N- and O-glycopeptides using 18O–labeling and product ion based mass spectrometry. Glycoconj. J. 2016, 34, 95–105. [Google Scholar] [CrossRef]
- Kettenbach, A.N.; Rush, J.; Gerber, S.A. Absolute quantification of protein and post-translational modification abundance with stable isotope–labeled synthetic peptides. Nat. Protoc. 2011, 6, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Butnev, V.Y.; Butnev, V.Y.; May, J.V.; Shuai, B.; Tran, P.; White, W.K.; Brown, A.; Hall, A.S.; Harvey, D.J.; Bousfield, G.R. Production, purification, and characterization of recombinant hFSH glycoforms for functional studies. Mol. Cell. Endocrinol. 2015, 405, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Pekonen, F.; Williams, N.M.; Weintraub, B.D. Purification of Thyrotropin and Other Glycoprotein Hormones by Immunoaffinity Chromatography. Endocrinology 1980, 106, 1327–1332. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, S.-L.; Hancock, W.S. Approaches to the study of N-linked glycoproteins in human plasma using lectin affinity chromatography and nano-HPLC coupled to electrospray linear ion trap—Fourier transform mass spectrometry. Glycobiology 2006, 16, 514–523. [Google Scholar] [CrossRef][Green Version]
- Dufau, M.; Tsuruhara, T.; Catt, K. Interaction of glycoprotein hormones with agarose-concanavalin A. Biochim. Biophys. Acta (BBA) Protein Struct. 1972, 278, 281–292. [Google Scholar] [CrossRef]
- Idler, D.R.; Bazar, L.S.; Hwang, S.J. Fish gonadotropin (s). II. Isolation of gonadotropin (s) from chum salmon pituitary glands using affinity chromatography. Endocr. Res. Commun. 1975, 2, 215–235. [Google Scholar] [CrossRef] [PubMed]
- Ui, N.; Tamura-Takahashi, H.; Yora, T.; Condliffe, P.G. Bioaffinity chromatography of thyrotropin using immobilized concanavalin A. Biochim. Biophys. Acta (BBA) Gen. Subj. 1977, 497, 812–815. [Google Scholar] [CrossRef]
- Bloomfield, G.A.; Faith, M.R.; Pierce, J.G. Sepharose-linked concanavalin A in the purification and characterization of glycoprotein hormones of the bovine pituitary. Biochim. Biophys. Acta (BBA) Protein Struct. 1978, 533, 371–382. [Google Scholar] [CrossRef]
- Nisula, B.C.; Louvet, J.-P. Radioimmunoassay of Thyrotropin Concentrated from Serum. J. Clin. Endocrinol. Metab. 1978, 46, 729–733. [Google Scholar] [CrossRef]
- Weintraub, B.D. Concentration and purification of human chorionic somato-mammotropin (HCS) by affinity chromatography: Application to radioimmunoassay. Biochem. Biophys. Res. Commun. 1970, 39, 83–89. [Google Scholar] [CrossRef]
- Akanuma, Y.; Kuzuya, T.; Hayashi, M.; Ide, T.; Kuzuya, N. Immunological reactivity of insulin to sepharose coupled with insulin-antibody—Its use for the extraction of insulin from serum. Biochem. Biophys. Res. Commun. 1970, 38, 947–953. [Google Scholar] [CrossRef]
- Murphy, R.F.; Elmore, D.T.; Buchanan, K.D. Isolation of glucagon-like immunoreactivity of gut by affinity chromatography. Biochem. J. 1971, 125, 61P–62P. [Google Scholar] [CrossRef]
- Murphy, R.F.; Buchanan, K.D.; Elmore, D.T. Isolation of glucagon-like immunoreactivity of gut by affinity chromatography on anti-glucagon antibodies coupled to sepharose 4B. Biochim. Biophys. Acta (BBA) Protein Struct. 1973, 303, 118–127. [Google Scholar] [CrossRef]
- Gospodarowicz, D. Single Step Purification of Ovine Luteinizing Hormone by Affinity Chromatography. J. Biol. Chem. 1972, 247, 6491–6498. [Google Scholar] [CrossRef]
- Lee, C.Y.; Wong, S.; Lee, A.S.; Ma, L. Purification and properties of choriogonadotropin from human term placenta. Hoppe-Seyler´s Z. Physiol. Chem. 1977, 358, 909–914. [Google Scholar] [CrossRef]
- Yang, Z.; Hancock, W.S. Approach to the comprehensive analysis of glycoproteins isolated from human serum using a multi-lectin affinity column. J. Chromatogr. A 2004, 1053, 79–88. [Google Scholar] [CrossRef]
- Hogan, J.M.; Pitteri, S.J.; Chrisman, P.A.; McLuckey, S.A. Complementary Structural Information from a TrypticN-Linked Glycopeptide via Electron Transfer Ion/Ion Reactions and Collision-Induced Dissociation. J. Proteome Res. 2005, 4, 628–632. [Google Scholar] [CrossRef]
- Wiesner, J.; Premsler, T.; Sickmann, A. Application of electron transfer dissociation (ETD) for the analysis of posttranslational modifications. Proteom. 2008, 8, 4466–4483. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Hancock, W.S.; Chew, T.R.; Bonilla, L. A study of glycoproteins in human serum and plasma reference standards (HUPO) using multilectin affinity chromatography coupled with RPLC-MS/MS. Proteom. 2005, 5, 3353–3366. [Google Scholar] [CrossRef] [PubMed]
- Bousfield, G.R.; Butnev, V.Y.; White, W.K.; Hall, A.S.; Harvey, D.J. Comparison of Follicle-Stimulating Hormone Glycosylation Microheterogenity by Quantitative Negative Mode Nano-Electrospray Mass Spectrometry of Peptide-N-Glycanase-Released Oligosaccharides. J. Glycom. Lipidom. 2015, 5, 129. [Google Scholar] [CrossRef]
- Gallien, S.; Domon, B. Detection and quantification of proteins in clinical samples using high resolution mass spectrometry. Methods 2015, 81, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]

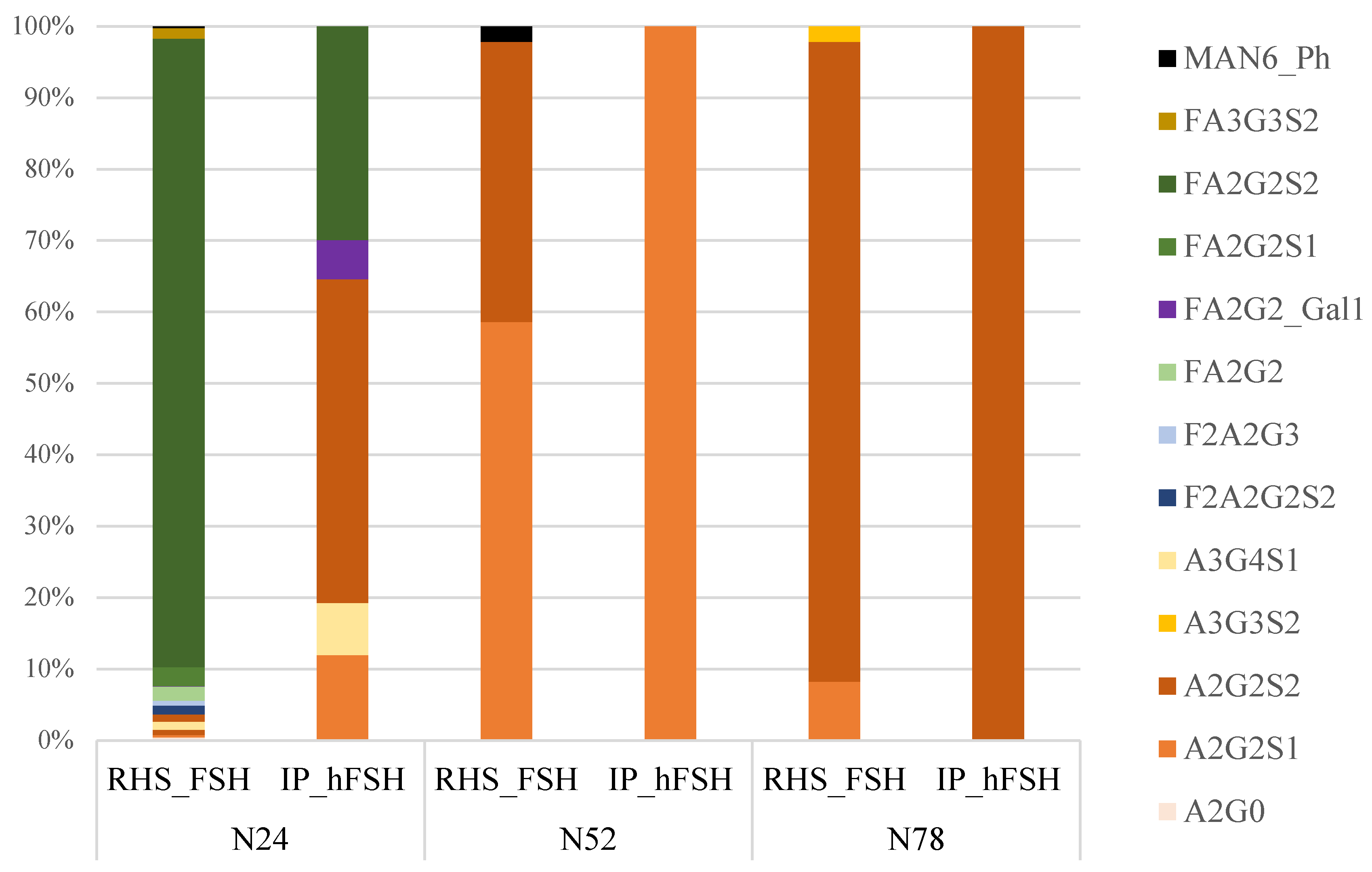
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melchiorre, C.; Chhuon, C.; Jung, V.; Lipecka, J.; Di Rella, F.; Conforti, A.; Amoresano, A.; Carpentieri, A.; Guerrera, I.C. Identification and Relative Quantification of hFSH Glycoforms in Women’s Sera via MS–PRM-Based Approach. Pharmaceutics 2021, 13, 798. https://doi.org/10.3390/pharmaceutics13060798
Melchiorre C, Chhuon C, Jung V, Lipecka J, Di Rella F, Conforti A, Amoresano A, Carpentieri A, Guerrera IC. Identification and Relative Quantification of hFSH Glycoforms in Women’s Sera via MS–PRM-Based Approach. Pharmaceutics. 2021; 13(6):798. https://doi.org/10.3390/pharmaceutics13060798
Chicago/Turabian StyleMelchiorre, Chiara, Cerina Chhuon, Vincent Jung, Joanna Lipecka, Francesca Di Rella, Alessandro Conforti, Angela Amoresano, Andrea Carpentieri, and Ida Chiara Guerrera. 2021. "Identification and Relative Quantification of hFSH Glycoforms in Women’s Sera via MS–PRM-Based Approach" Pharmaceutics 13, no. 6: 798. https://doi.org/10.3390/pharmaceutics13060798
APA StyleMelchiorre, C., Chhuon, C., Jung, V., Lipecka, J., Di Rella, F., Conforti, A., Amoresano, A., Carpentieri, A., & Guerrera, I. C. (2021). Identification and Relative Quantification of hFSH Glycoforms in Women’s Sera via MS–PRM-Based Approach. Pharmaceutics, 13(6), 798. https://doi.org/10.3390/pharmaceutics13060798